Abstract
Hematopoiesis is a finely regulated process in vertebrates under both homeostatic and stress conditions. By whole exome sequencing, we studied the genomics of acute lymphoblastic leukemia (ALL) patients who needed multiple red blood cell (RBC) transfusions after intensive chemotherapy treatment. ARHGEF12, encoding a RhoA guanine nucleotide exchange factor, was found to be associated with chemotherapy-induced anemia by genome-wide association study analyses. A single nucleotide polymorphism (SNP) of ARHGEF12 located in an intron predicted to be a GATA1 binding site, rs10892563, is significantly associated with patients who need RBC transfusion (P=3.469E-03, odds ratio 5.864). A luciferase reporter assay revealed that this SNP impairs GATA1-mediated trans-regulation of ARHGEF12, and quantitative polymerase chain reaction studies confirmed that the homozygotes status is associated with an approximately 61% reduction in ARHGEF12 expression (P=0.0088). Consequently, erythropoiesis was affected at the pro-erythroblast phases. The role of ARHGEF12 and its homologs in erythroid differentiation was confirmed in human K562 cells, mouse 32D cells and primary murine bone marrow cells. We further demonstrated in zebrafish by morpholino-mediated knockdown and CRISPR/Cas9-mediated knockout of arhgef12 that its reduction resulted in erythropoiesis defects. The p38 kinase pathway was affected by the ARHGEF12-RhoA signaling in K562 cells, and consistently, the Arhgef12-RhoA-p38 pathway was also shown to be important for erythroid differentiation in zebrafish as active RhoA or p38 readily rescued the impaired erythropoiesis caused by arhgef12 knockdown. Finally, ARHGEF12-mediated p38 activity also appeared to be involved in phenotypes of patients of the rs10892563 homozygous genotype. Our findings present a novel SNP of ARHGEF12 that may involve ARHGEF12-RhoA-p38 signaling in erythroid regeneration in ALL patients after chemotherapy.
Introduction
Chemotherapy for hematologic malignancies such as acute lymphoblastic leukemia (ALL) often causes anemia. To alleviate chemotherapy-induced anemia, red blood cell (RBC) transfusion has become standard care. The need for RBC transfusion varies significantly among patients who have undergone similar treatment protocols at similar intensities. Sensitivity of erythrocytes to the cytotoxicity of chemotherapy and the recovery rate of erythropoiesis are contributing factors related to the severity and duration of the anemia. Genetic diversity in genes regulating these response processes can be a cause for the variations between patients. Uncovering the genetic basis for the variable response is important for understanding the molecular mechanisms underlying erythropoiesis and its relationship to chemotherapy-induced anemia.
In this study, we performed genome-wide association study (GWAS) analyses of samples from individuals who had undergone multiple RBC transfusions (MRT) and from those who received no RBC transfusion (NRT) when a remission was achieved. By counting the cell line, primary bone marrow (BM) cells, and considering the results of animal model and human genetic studies, we suggest a novel molecular pathway involved in erythroid regeneration in ALL patients after chemotherapy.
Methods
Patients
From January 1st 2001 to December 31st 2014, a total of 452 patients diagnosed with childhood ALL were recruited in this study. The patients included were enrolled on Shanghai Children’s Medical Center -Acute Lymphoblastic Leukemia-2005 (SCMC-ALL-2005) protocol. Standard induction and consolidation chemotherapy were used. Blood transfusion records were collected from the transfusion department of SCMC and clinical data were reviewed to exclude the events needed for additional transfusions of RBC, such as transplantation, gastrointestinal bleeding, surgery, etc. Patients who abandoned treatment or who died were not included in this study. Only total RBC transfusion units after achieving complete remission was included in the count and this number was normalized by body surface area (Unit/m2) to exclude the influence of patient’s age, mass, and the disease itself on blood transfusion units. Patients who received more than eight RBC units/m2 were defined as MRT. This study has been approved by Shanghai Children’s Medical Center Ethics Committee (n. SCMCIRB-K2018052).
Dual Luciferase reporter assays
A total of 933 bp DNA fragments surrounding rs10892563 CC and TT genotype were cloned into the firefly Luciferase vector pGl4.27 (Promega); 293T cells were transfected with 5 g PRL-TK vector and 300 ng PCDNA3-Flag expression GATA1 via lipofectemin. Luciferase activity was measured in a Varioskan Flash spectral scanning multimode reader (Thermo) using the Dual-Luciferase Reporter Assay system kit (Promega).
Targeted single nucleotide polymorphism genotyping by polymerase chain reaction
The candidate SNP rs10892563 on ARHGEF12 at position 119,729,754 bp was analyzed by polymerase chain reaction (PCR) on 381 ALL patients enrolled in the SCMC-ALL-2005 protocols for which genomic DNA samples were still available. The DNA segment containing the candidate mutation was amplified by PCR using the following primers:
5′-00ATAGGGATACCTGGCCCCTA-3′ and 5′-ndATAGGGATACCTGGCCCCTA-3′
These PCR products were subsequently Sanger sequenced.
Whole-mount in situ hybridization
The antisense probes of arhgef12a and arhgef12b were obtained by PCR with the primers
(arhgef12a forward primer, 5′-GCGGAATTCCCACCTCAAGGAGATGGAAA-3′;
reverse primer, 5′-GCGGGTACCCCAAAAGCATGCAAGAAACA-3′;
arhgef12b forward primer, 5′-GCCGAATTCTCCAGCATGAGTGGTTGGTA-3′;
reverse primer, 5′-ATTGGTACCCTCAACAGAAAGCCGA-GACC3′), and added with EcoR1/Kpn1 restriction enzyme sites for cloning into pCS2+ vector. Antisense digoxigenin (DIG)– labeled RNA probes were generated by in vitro transcription and whole-mount in situ hybridization (WISH) was performed as described previously.1 The results were imaged using a stereomicroscope Nikon SMZ1500 with a 1 x HR Plan Apo objective and ACT-1 vision software.
Micro-injection
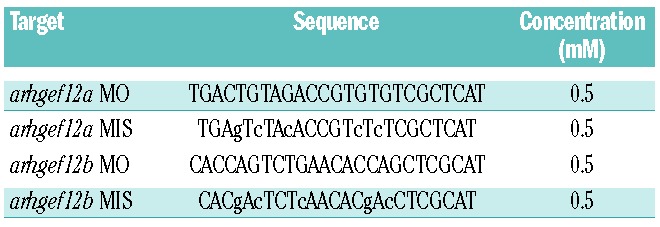
One-cell-stage embryos were injected with 2 nL of morpholino (MO) or mismatch morpholino (MIS) mixes (arhgef12a and arhgef12b) purchased from Gene-Tools. The MO sequences and concentrations are listed in Table 1.
Table 1.
Morpholino sequences and concentrations used in this study.
CRISPR/Cas9 mutagenesis
The arhgef12a gRNA (5′-GGACGTGGGTCTCGAGTCAC-3′) and arhgef12b gRNA (5'-GGAATCTGAGGCAGGCCCGG-3') were synthesized. The zebrafish optimized Cas9 mRNA was syn-thesized in vitro from the pCS2-nCas9n plasmid (addgene, #47929) as described.2 The Cas9 mRNA was synthesized in vitro by SP6 mMessage mMachine Transcription Kit (Ambion). arhgef12a gRNA (50pg), arhgef12b gRNA (50pg), and Cas9 mRNA (150pg) were co-injected into one-cell stage embryos.
Statistical analysis
Results are expressed as mean±standard deviation considering the number of experiments. Statistical comparisons between groups were performed by two-tailed t-test or one-sided t-test using Graphpad Prism version 6.0.
Other methods
Whole exome sequencing, GWAS, cell sorting, quantitative real-time (qRT)-PCR, plasmid construction, in vitro RNA synthesis, micro-injection and anisomycin treatment were performed as described in the Online Supplementary Appendix.
Results
An ARHGEF12 polymorphism in acute lymphoblastic leukemia patients is associated with susceptibility to chemotherapy-induced anemia
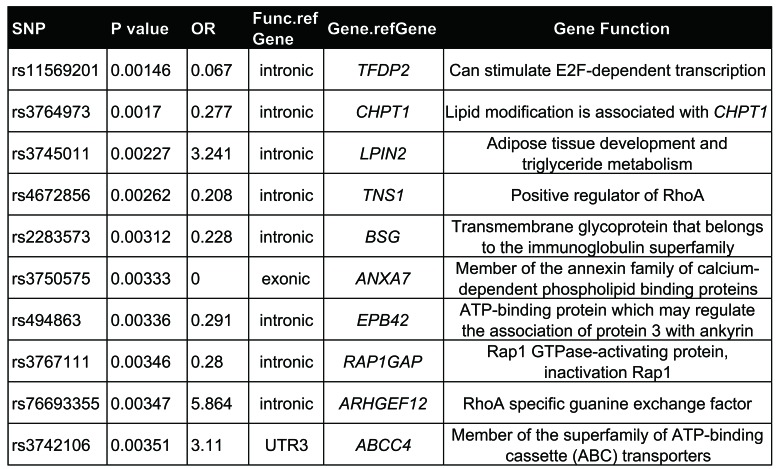
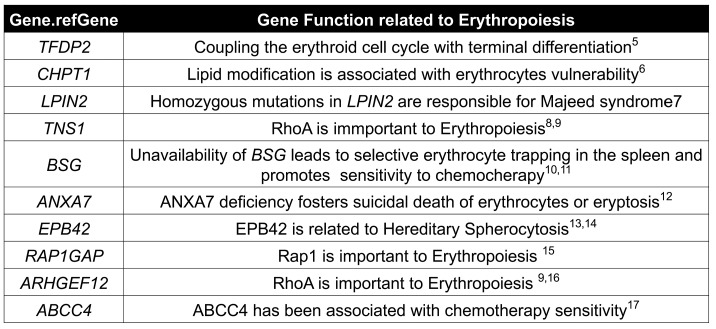
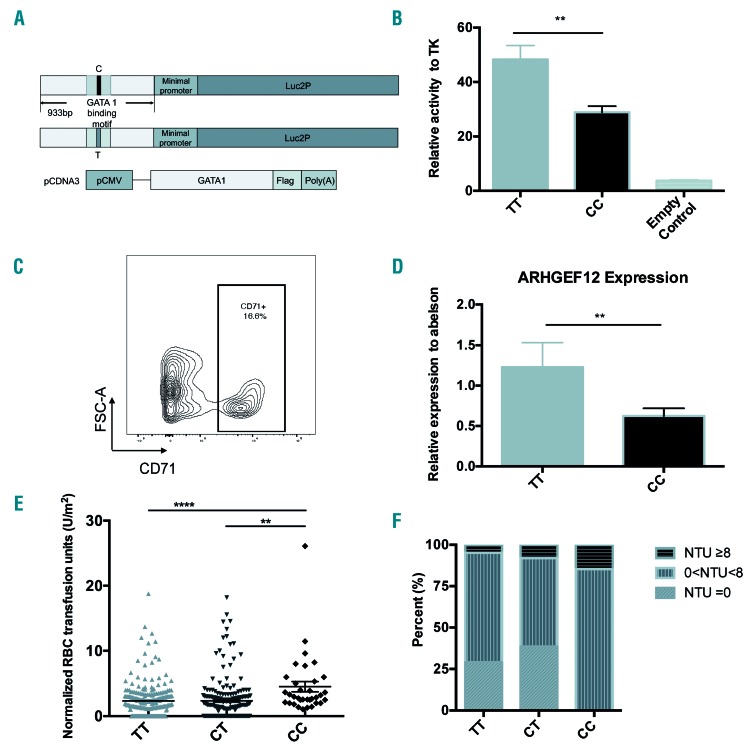
We performed whole exome sequencing in 31 individuals who had undergone at least eight RBC transfusions (MRT) and 31 patients with no RBC transfusion (NRT), all from the SCMC-ALL-2005 cohort (Figure 1A). Considering the variations in patient age and body weight, the RBC transfusion amount was normalized by patient body surface area taking into account only post-remission transfusions in order to minimize the effect of ALL itself. By a GWAS analysis, 1,708 SNPs of 281 genes passed the criterion of the primary cut: a call rate of >95% and P<0.01. Of interest, most of the SNPs were located in introns adjacent to exons, suggesting that these polymorphisms are relevant to chemotherapy-induced anemia by regulating gene expression. These genes could be highly expressed in hematopoietic cells and involved in erythroid differentiation from hematopoietic stem/progenitor cells. To address this possibility, we sorted the primary gene list with expression patterns in primitive CD34+ cells before erythroid differentiation and in erythrocytes based on the Differentiation Map Portal (DMAP) database.3 A total of 35 genes were enriched by this analysis. Among them, 12 genes were highly expressed in hematopoietic stem/progenitor cells (HSPC) and 23 genes were expressed in erythrocytes with over two times the average expressions. At the top of this enriched list were GUCY1A3,4 NUCB2, TFDP2,5 CHPT1,6 PLCB1,7 LPIN2,8 TNS1,9,10 BSG,11,12 COL5A1,13 ANXA7,14 EPB42,15,16 RAP1GAP,17 ARHGEF12,10,18 ABCC419 and FARP120 (Tables 2 and 3 and Figure 1B). Interestingly, most of these genes are well-known in association with erythropoiesis or cytotoxicity susceptibility to chemotherapy.21 Of note, four of these genes, i.e. RAP1GAP, ARHGEF12, TNS1 and FARP1, are related to small GTPase regulation (Table 2 and 3). ARHGEF12, a RhoA-specific guanine-exchange factor (GEF), can specifically activate RhoA18 which is essential for embryonic erythropoiesis.10 ARHGEF12 is thus possibly one of the associated genes involved in the regulatory mechanism of erythroid regeneration from anemia induced by chemotherapy. Among the SNP found in ARHGEF12, the most significant association Was rs76693355 (P=3.469E-03, odds ratio 5.864). All SNP were screened with linkage disequilibrium 0.2<r2<1 related to rs76693355 in the 5-kb flanking regions of ARHGEF12. We found that rs10892563 is located at a predicted binding site of the erythroid-specific transcription factor GATA1.22 To test if such a variant could disrupt this GATA1 binding site function, we employed a dual luciferase assay in 293T cells with an expression vector containing this intron motif of rs10892563 in the promoter region. The expression assay showed that the minor allele change of rs10892563 was able to down-regulate ARHGEF12 transcriptional regulation by GATA1 (Figure 2A and B). To examine if the rs10892563 SNP is actually associated with ALL patient RBC function, the CD71-positive erythroid cells from the ALL patient BM samples were isolated by flow cytometry23 for sequencing and gene expression verifications. qRT-PCR analysis found that rs10892563 homozygosity in the patients is associated with an approximately 61% reduction in ARHGEF12 expression (P=0.0088) (Figure 2C and D).
Figure 1.
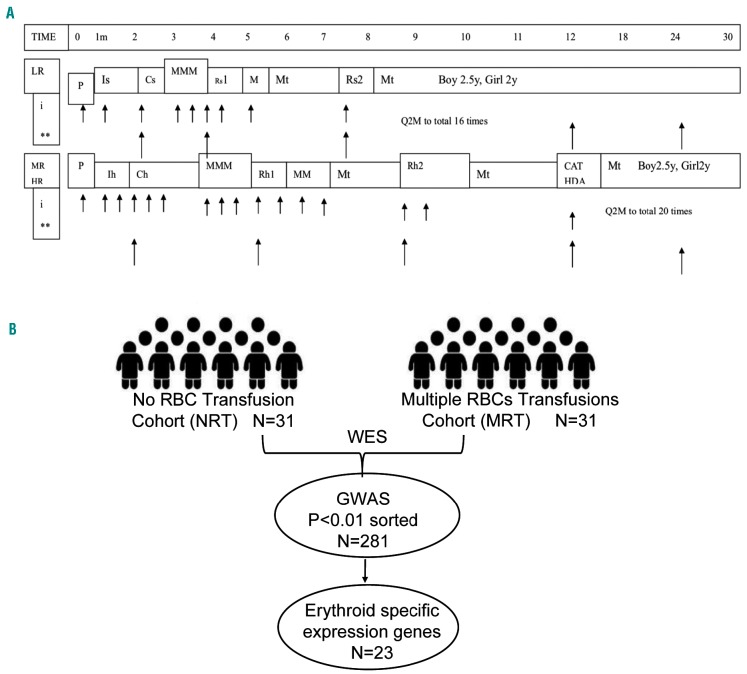
Genomic characterizations of chemotherapy-induced anemia in children with acute lymphoblastic leukemia (ALL). (A). The schematic treatment plan of SCMC-ALL 2005. LR: low-risk group; MR: medium-risk group; HR: high-risk group; P: prednisone window phase; Is: induction for standard-risk patients; Ih: induction for higher risk patients; Cs: consolidation for standard patients; Ch: consolidation for higher risk patients; M: high-dose methotrexate; Rs: re-induction for standard-risk patients; Rh: re-induction for higher risk patients; Mt: maintenance therapy; CAT HAD: chemotherapy course composed of cyclophosphamide, cytarabine, thiopurine and high-dose cytarabine. (B). An outline of the genomic characterizations. Genome-wide association study (GWAS) was performed based on whole exome sequencing data for 31 cases in each cohort of multiple red blood cell transfusion (MRT, >8 units/m2) and no red blood cell transfusion (NRT). 23 genes highly expressed in erythoid committed and 12 genes highly expressed in pre-erythroid committed were on the list of genes with significance (call rate >95% and P<0.01).
Table 2.
Gene function of top 10 genes highly expressed in hematopoietic cells along erythroid differentiation from hematopoietic stem cells.
Table 3.
Gene function related to erythropoiesis.
Figure 2.
rs10892563 may disrupt a GATA1-binding cis element and is related to chemotherapy-induced anemia. (A and B) rs10892563 is located at a binding site of erythroid-specific transcription factor GATA1. Dual luciferase assay with the vector inserted with a stretch of sequence with the motif revealed that C allele of rs10892563 down-regulated GATA1 cis-transcriptional function compared to the major allele T in 293T cells; P=0.0038. (C and D) CD71-positive nucleated erythroid cells were sorted from banked bone marrow samples of acute lymphoblastic leukemia patients in remission state by fluorescence-activated cell sorting. The relative gene expression of ARHGEF12 was reduced in CC genotype at rs10892563 versus TT genotypes by quantitative real-time polymerase chain reaction assay; P=0.0088. (E) A total of 452 children with ALL enrolled in the Shanghai Children’s Medical Center-Acute Lymphoblastic Leukemia-2005 (SCMC-ALL-2005) protocol were genotyped targeting rs10892563. The average normalized red blood cell (RBC) transfusion units was significantly higher in patients with CC genotype than the CT genotype (P=0.0011) and the TT genotype (P<0.0001); each point represents one patient’s record. (F) Distribution of RBC transfusions across different rs10892563 genotypes. All patients with CC genotype (n=34) need RBC transfusions, whereas in patients with CT and TT genotype, proportions of RBC transfusions were 61.5% and 71%, respectively. Concerning multiple red blood cell transfusions (MRT), the proportion in CC, CT and TT were 14.705%, 8.021%, and 4.762%, respectively (P<0.001).
Further verifying an involvement of rs10892563, additional targeted SNP genotyping of 452 ALL patients enrolled in the SCMC-ALL-2005 protocol showed that the genotype frequencies were CC in 7.52%, CT in 41.37%, and TT in 51.11% patients. The average normalized RBC transfusion was 4.533 units/m2 in patients with CC genotype, 2.353 and 2.335 in patients with CT and TT genotypes, respectively (Figure 2E and Online Supplementary Figure S1). All patients who were homozygous needed RBC transfusion to maintain hemoglobin >65 g/L during the course of chemotherapy, whereas among those who were heterozygous or wild-type, the frequencies were 61.497% and 70.996%, respectively. Patients who were homozygous or heterozygous had a significantly higher probability of requiring MRT than patients carrying wild-type alleles (Figure 2F). Collectively, these results suggest that the ARHGEF12 polymorphism rs10892563 is involved in the susceptibility to chemotherapy-induced anemia.
ARHGEF12 reduction blocks erythroid differentiation of K562 cells
The findings that a polymorphism of ARHGEF12 is associated with chemotherapy-induced anemia and that this gene is heavily transcribed in the human erythroid lineage (Online Supplementary Figure S2A) based on the analyses of several public databases3 suggest that ARHGEF12 is involved in erythropoiesis. As an initial test, we performed ARHGEF12 knockdown in the human erythroleukemia cell line K562 using lentiviral shRNA constructs (Online Supplementary Figure S2B). GPA expression and the benzidine cytochemical test showed that erythroid differentiation of the cells, under hemin induction, decreased significantly compared to the non-targeted (NT) cells (Online Supplementary Figure S2C).
ARHGEEF12 or its orthologs is involved in erythroid differentiation in murine progenitor cells and in a zebrafish model
To rule out potential effects by the neoplastic background of K562 cells, we knocked down Arhgef12 expression in mouse hematopoietic cell line 32D cells and in primary mouse BM cells by lentiviral shRNA transduction. Erythropoietin-induced erythroid differentiation was significantly blocked by the interference of Arhgef12 expression as by observed erythroid immunophenotyping by flow cytometry and in the burst forming units-erythroid and colony forming unit-erythroid colony forming assays (Online Supplementary Figure S3).
Zebrafish genome harbors two orthologs of ARHGEF12: arhgef12a on chromosome 15 and arhgef12b on chromosome 5. By comparing their sequences (arhgef12a, ENS-DARG00000030532; arhgef12b, ENSDARG00000067634) with human ARHGEF12, we found that the similarities were 54% and 55%, respectively. Synteny analysis also showed the relatively conserved positions for both arhgef12a and arhgef12b (Online Supplementary Figure S4A). Arhgef12a is selectively enriched in early erythroid progenitors (Online Supplementary Figure S4B) whereas arhgef12b is expressed in early erythroid progenitors (Online Supplementary Figure S4C).
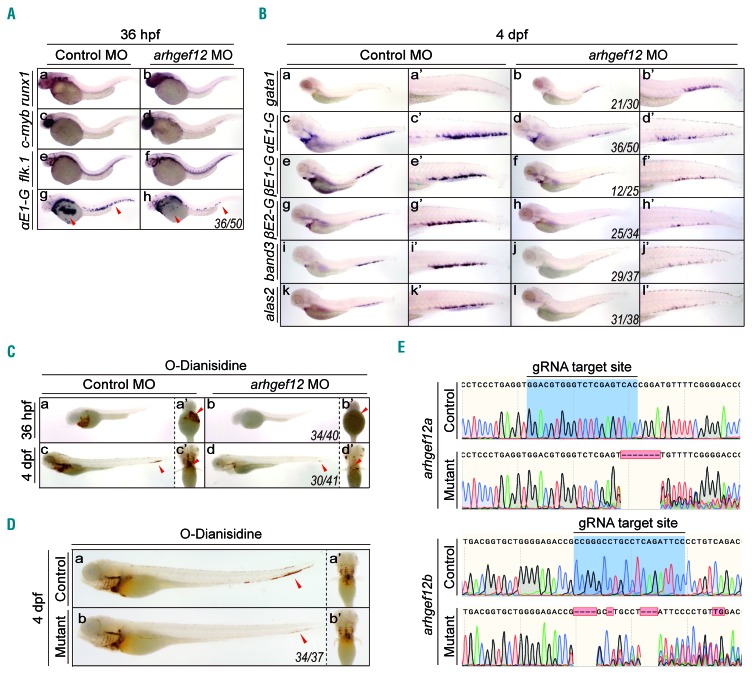
To study the role of arhgef12 in erythropoiesis, we performed microinjections of arhgef12a and arhgef12b morpholino both in combination (arhgef12 MO) and separately (arhgef12a MO and arhgef12b MO). Firstly, we performed WISH at 22 hours post fertilization (hpf) to analyze the primitive wave24 of hematopoiesis. Expressions of the erythroid progenitor marker gata1, the mature erythrocyte marker αe1-globin, the hematopoietic lineage marker scl, and the myeloid markers pu.1 and lysozyme C remained unchanged in arhgef12 MO-injected embryos (Online Supplementary Figure S5). At 36 hpf, the definitive hematopoiesis stage of zebrafish, the expression of αe1-globin was dramatically decreased in arhgef12a- and arhgef12b-deficient embryos, whereas the hematopoietic stem cells (HSC) markers runx1 and c-myb and the vascular morphology and marker flk1 were unchanged (Figure 3A). At four days post-fertilization (dpf), the markers representing mature RBC including αe1-globin, βe1-globin, βe2-globin, band3, and alas2 were severely reduced (Figure 3B). Of interest, gata1 was associated with an obvious increase in caudal hematopoietic tissue (CHT) (Figure 3B), indicating that the erythroid defect may be caused by an impaired differentiation. Of interest, gata1 was associated with an obvious CHT (Figure 3B), indicating that the erythroid defect may be caused by an impaired differentiation. Consistent with this possibility, o-Dianisidine stained hemoglobin showed that erythrocytes from arhgef12 MO-injected embryos were more immature than those from the control group at 36 hpf and 4 dpf (Figure 3C). Subsequent examinations of arhgef12a and arhgef12b double knockout mutants by using the CRISPR/Cas9 method followed by o-Dianisidine staining found that mature erythrocytes were significantly decreased at 4 dpf in the mutant CHT and heart (Figure 3D and E). These results indicate that arhgef12a and arhgef12b are required for erythroid differentiation and maturation in zebrafish.
Figure 3.
Knockdown by morpholino (MO) or knockout by CRISPR/Cas9 of arhgef12 impairs erythroid differentiation in the zebrafish model. (A) Whole-mount in situ hybridization (WISH) results of hematopoietic and vascular markers in both control and arhgef12a & arhgef12b double MO injected embryos at 36 hours post fertilization (hpf). Definitive hematopoietic markers runx1 (A-a, b) , c-myb (A-c, d) , and endothelial cell marker flk.1 (A-e, f) indicated no obvious defect of definitive hematopoiesis and vasculogenesis in arhgef12 morphants at 36 hpf. The erythrocyte marker aE1-globin (A-g, h) indicated obvious defect in erythroid differentiation in arhgef12 morphants at 36 hpf. Red arrowheads indicate the yolk sac region and posterior blood island. (B) WISH results of erythrocyte markers in both control and arhgef12a & arhgef12b double MO injected embryos at 4 dpf. The increased progenitor erythrocyte marker gata1 (a, b), and decreased mature erythrocyte markers aE1-globin (c, d), βE1-globin (e, f), βE2-globin (g, h), band3 (i, j) and alas2 (k, l) indicated obvious block in erythroid differentiation in arhgef12 morphants at 4 dpf. The corresponding magnifications are indicated as a’-l’. (C) The decreased O-Dianisidine staining of hemoglobin, which labeled mature erythrocytes, in both control and arhgef12a & arhgef12b double MO injected embryos at 36 hpf (a, a’, b, b’) and 4 dpf (c, c’, d, d’) suggested the defect in erythroid differentiation. Red arrowheads indicated the mature erythrocytes in yolk sac region (a’, b’), caudal hematopoietic tissue (CHT) (c, d) and heart (c’, d’). (D) The decreased O-Dianisidine staining of hemoglobin, in both control and arhgef12a & arhgef12b double knockout embryos at 4 dpf (a, b) suggested the defect in erythroid differentiation. Red arrowheads in a and b indicated the CHT, and the ventral view was shown in a’ and b’. (E) Genotyping of arhgef12a and arhgef12b mutants constructed by CRISPR/Cas9. The gRNA target sites are as labeled.
ARHGEF12 regulates erythroid differentiation through a RhoA-p38 pathway
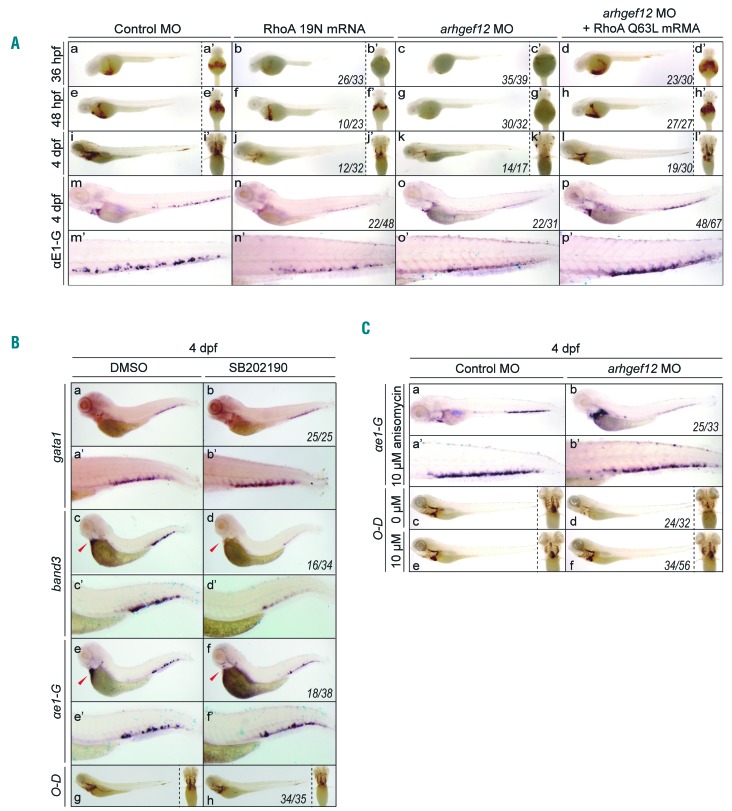
RhoA is a well-defined substrate of ARHGEF12, which activates the exchange of RhoA bound GDP in the inactivated form for GTP to yield the active RhoA-GTP.26 We hypothesized that RhoA is the key target of ARHGEF12 to mediate its function in erythrocyte maturation. Because zebrafish harbors 5 rhoa genes27 and they all have an amino acid sequence which is quite similar (identity >90%) to human RhoA, we inferred that human RhoA mutant mRNA dominant-negative (DN) mutant RhoA T19N28 and constitutively active mutant RhoA Q6329 would also function in zebrafish. In fact, injection of dominant-negative RhoA mRNA led to anemia, which mimicked the arhgef12 deficiency phenotype (Figure 4A), but the anemia seemed to be less severe than with MO injection, possibly due to mRNA instability. On the other hand, based on o-Dianisidine staining and WISH analysis of αe1-globin, a co-injection of constitutively active mutant RhoA Q63L mRNA was able to restore the erythropoiesis defect caused by arhgef12 MO (Figure 4A). These results indicate that Arhgef12 activates RhoA to control erythroid differentiation.
Figure 4.
arhgef12 regulates erythroid differentiation through RhoA and p38. (A) O-Dianisidine staining results at 36hpf, 48hpf and 4dpf of embryos injected with control morpholino (MO), RhoA 19N mRNA, arhgef12a and arhgef12b MO, RhoA Q63L mRNA co-injected with arhgef12a and arhgef12b MO (a-l). Yolk sac ventral views of the indicated microinjected embryos (a’-l’). Whole-mount in situ hybridization (WISH) results of ae1-globin in the indicated microinjected embryos at 4dpf (m-p) and the corresponding magnifications of CHT (m’-p’). (B) WISH results of gata1, band3, ae1-globin (a-f) and the corresponding magnifications of CHT (a’-f’) in control (dimethyl sulfoxide, DMSO) and SB202190 (the inhibitor of p38) treated embryos at 4 dpf. O-Dianisidine (O-D) staining results at 4dpf (g, h) with the corresponding ventral view on the right. (C) WISH results of ae1-globin in control or arhgef12a and arhgef12b MO injected embryos with anisomycin (activator of p38) treatment (a, b) and the corresponding magnifications of CHT (a’-b’). O-D staining results of control or arhgef12a and arhgef12b MO injected embryos with DMSO (c, d) or 10uM anisomycin (e, f) treatment (the corresponding ventral view on the right) showed anisomycin could rescue the blocked erythroid differentiation in the morphants at 4dpf. The indicated embryos were treated by DMSO (labeled by 0uM) and anisomycin (labeled by 10uM) for 24 hours (from 3dpf to 4dpf).
To further understand the molecular events downstream of RhoA in erythropoiesis, the K562 cell line in which ARHGEF12 is important for its erythroid differentiation was examined. An antibody microarray screen found that phosphorylation of molecules in the p38 MAPK pathway was significantly decreased in the ARHGEF12 knockdown K562 cells (Online Supplementary Figure S6A). Western blotting confirmed this effect on p38 phosphorylation (Online Supplementary Figure S6B), suggesting that the p38 MAPK signaling pathway may contribute to the ARHGEF12-regulated erythropoiesis. Further confirmation using the p38 inhibitor SB202190 in zebrafish found that p38 inhibition resulted in a similar block of erythropoiesis as in the arhgef12 morphants at 4 dpf (Figure 4B). Application of anisomycin, a p38 MAPK activator, was able to restore the erythrocyte maturation in the arhgef12 morphants (Figure 4C). Thus, an ARHGEF12-RhoA-p38 pathway is likely to be involved in erythroid differentiation.
STAT1 expression can rescue the erythroid phenotype caused by arhgef12 knockdown in zebrafish
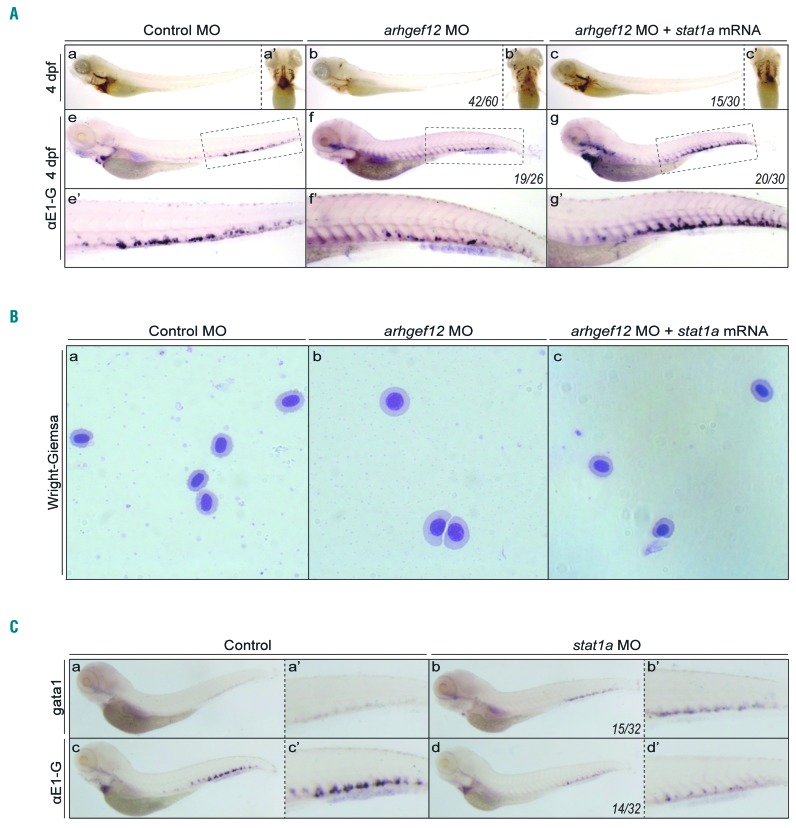
Human STAT1 produces two splicing variants that differ at their carboxy terminus. Zebrafish has two orthologous genes related to human STAT1: stat1a and stat1b. It has been shown that p38 MAPK-STAT1 pathways can regulate neutrophil development. Meanwhile, our antibody microarray screen showed that the phosphorylation of STAT1 at serine (S) 727 was decreased approximately 2-fold in ARHGEF12 knockdown K562 cells (Online Supplementary Figure S6A). We thus further examined whether STAT1 may be downstream of p38 MAPK in regulating erythropoiesis. We co-injected the HA-stat1a construct together with arhgef12 MO in zebrafish, and observed that the phenotype of erythropenia was restored (Figure 5A a-c, a′-c′) and αe1-globin expression recovered (Figure 5A e-g, e′-g′), compared with control embryos. A cytology assay by Wright-Giemsa staining showed that the co-injection of stat1a mRNA with arhgef12 MO appeared to promote the immature erythrocyte differentiation (Figure 5B). In addition, stat1 MO injection increased gata1 expression (Figure 5C a, a′, b, b′) but reduced ae1-globin expression (Figure 5C c, c′, d, d′), similar to that by arhgef12 MO injection. It is thus likely that STAT1 is involved in the ARHGEF12-p38 MAPK signaling function in erythroid differentiation.
Figure 5.
STAT1 expression rescues the erythroid differentiation phenotype by arhgef12 knockdown in zebrafish. (A) O-Dianisidine staining results of embryos injected with control morpholino (MO) (a), arhgef12a & arhgef12b MO (b), and arhgef12a & arhgef12b MO with stat1a mRNA (c) at 4dpf. Yolk sac ventral views of the indicated microinjected embryos (a’-c’). Whole-mount in situ hybridization (WISH) results of ae1-globin in the embryos injected with control MO (e), arhgef12a & arhgef12b MO (f), and arhgef12a and arhgef12b MO with stat1a mRNA (g) at 4dpf and the corresponding magnifications of CHT (e’-g’). (B) Wright-Giemsa results of erythrocytes, which were isolated from control MO (a), arhgef12a and arhgef12b MO (b), and arhgef12a & arhgef12b MO with stat1a mRNA (c) injected embryos at 4 dpf. (C) WISH results of gata1 in the embryos injected with control MO (a), stat1a MO (b) at 4dpf and the corresponding magnifications of CHT (a’,b’). WISH results of αe1-globin in the embryos injected with control MO (c), stat1a MO (d) at 4dpf and the corresponding magnifications of CHT (c’,d’).
The ARHGEF12-p38 pathway is associated with erythroid regeneration in acute lymphoblastic leukemia patients after chemotherapy
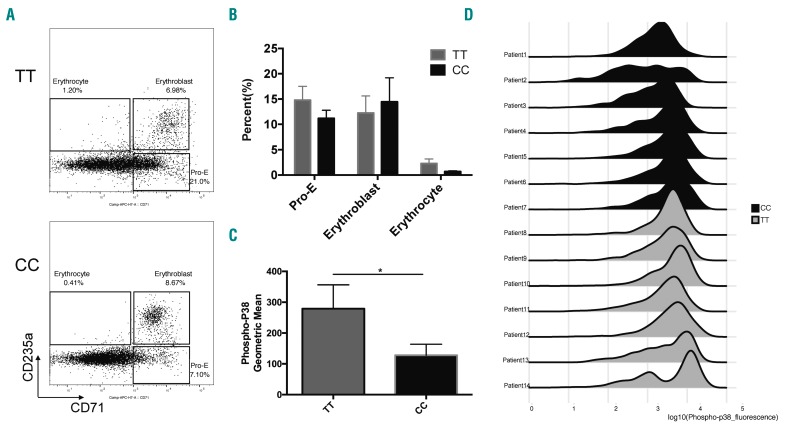
To examine whether ARHGEF12 polymorphism-associated anemia after chemotherapy in ALL patients may engage the p38 pathway, we measured p38 phosphorylation in erythroid cells in seven remission-related BM from ALL patients during maintenance therapy by phospho-flow.30 All seven patient samples with the rs10892563 CC genotype showed consistently reduced phosphorylated p38 in pro-erythroblasts (Figure 6C and D). Similar to the case of arhgef12 MO, where the p38 activity is inhibited in the zebrafish, these rs10892563 CC genotype patients showed an erythroid differentiation block at the erythroblast stage (Figure 6A and B), suggesting a strong association with the ARHGEF12-p38 pathway.
Figure 6.
The ARHGEF12-RhoA-p38 pathway is associated with erythroid regeneration from chemotherapy-induced anemia in acute lymphoblastic leukemia (ALL) patients. (A) Erythroid differentiation evaluation by flow cytometry of cryopreserved bone marrow (BM) samples from patients in remission with different rs10892563 genotypes. Three populations of different development stages, i.e. CD71+CD235a- pro-erythroblasts (Pro-E), CD71+CD235a+ erythroblasts and CD71−CD235a+ erythrocytes can be discriminated. Representative flow cytometry dot plots showing the percentage of erythroblasts in patients with CC genotype was higher than TT genotype, whereas the pro-erythroblasts (Pro-E) and erythrocytes were reduced in CC genotype. (B) The TT group and the CC group each had nine samples. Proportion statistics of each stage of erythroid differentiation are shown. (C) Statistics of mean fluorescence intensity of phospho-p38. We compared the base-2 log of phos-pho-p38 levels between the CC and TT genotypes, phospho-p38 levels are lower in the CC genotype patients (P=0.0471, one-tailed t-test). (D) Phospho-flow histograms of phospho-p38 on pro-erythroblasts with different rs10892563 genotypes. Phosphorylated p38 was stained with anti-pT180/pY182 antibody on cryopreserved BM samples from patients in remission with different rs10892563 genotypes as well as CD71 and CD235a monoclonal antibody. Histograms showing phospho-p38 peaks shifted left in CC genotype (black) comparing TT genotype (gray).
Discussion
For leukemia patients, hematologic toxicity is the most common side effect of chemotherapy as the hematopoietic cells are among the tissues most vulnerable to therapy-related damage, in part due to their active cell cycle status. Anemia is one of the most frequently recorded manifestations of the hematopoietic toxic effects during the course of chemotherapy. Chemotherapy-induced anemia can be caused by cytotoxic inhibition of normal hematopoiesis similar to chemotherapy-induced neutropenia and thrombocytopenia. Chemotherapy agent-related autoimmune hemolysis31,32 and chemotherapy-induced eryptosis can also cause anemia.33 Our current GWAS studies have found that chemotherapy-induced anemia is associated with SNP in CHPT1,6 BSG,11,12 ANAX7,14 EPB42,15,16 and ABCC417 that may be related to increased erythrocyte loss. There is evidence to suggest that: (i) ANAX7 is related to erythrocyte death;34 (ii) EPB4215,16 is related to hereditary spherocytosis in which eryptosis is an important cause of anemia; (iii) CHPT16 and BSG11,12 may be related to the vulnerability of RBC to chemotherapy; and (iv) ABCC4,17 a membrane transporter, is related to accumulation of cytotoxic agents in cells and chemo-sensitivity of hematopoietic cells. Other direct or indirect evidence based on the known functions of the gene products suggests that the remaining genes in the top ten list of GWAS in our study, TFDP2, LPIN2, TNS1, RAP1GAP, ARHGEF12, could also be related to impaired hematopoiesis in response to chemotherapy.
Erythropoiesis is a tightly regulated process in which the hemoglobin level is maintained in a narrow window between 135g/L and 155g/L under normal conditions or in response to stress such as chemotherapy. A complex lineage-specific transcription factor network underlies the homeostatic hematopoiesis and erythropoiesis mechanisms. In such a transcription network, the GATA transcription factor family plays a central role in the proper differentiation of erythroid cells together with Friend of GATA (FOG-1). The GATA family is composed of six members in mammals that are highly conserved in expression patterns in vertebrates, and GATA-1, GATA-2 and GATA-3 are classified into the hematopoietic GATA subfamily based on their expression profiles and domain structures. GATA-1 is important in adult hematopoiesis especially for erythropoiesis and regulates multiple target genes during the development and differentiation of erythroid and megakaryocytic lineages by binding to the GATA motif (A/T)GATA(A/G). In this study, we identified a novel SNP of ARHGEF12 gene, rs10892563, located in a regulatory GATA motif and found that the erythroid expression of AEHGEF12 is significantly down-regulated in rs10892563 homogeneous ALL patients who have undergone chemotherapy.
ARHGEF12 encodes a RhoA specific guanine nucleotide exchange factor which positively regulates the RhoA GDP/GTP exchange reaction. ARHGEF12 plays crucial roles in the cyclic-stretch-induced cell and stress fiber reorientation responses,35 mesenchymal stem cell fate,36 and cell migration and invasion,37 by regulating RhoA activity. ARHGEF12 is important for platelet activation and thrombosis in mice,38 but its role in erythropoiesis has not been defined. As a founding member of the Rho GTPase family, RhoA is involved in many important cellular functions, including gene transcription, survival, adhesion, and cytoskeleton reorganization. RhoA is important for hematopoiesis, regulating HSPC trafficking, interaction with the BM microenvironment, proliferation, survival, and self-renewal, and for fetal erythropoiesis in mitosis and cytokinesis.10 Interestingly, among the top ten in our GWAS list, there are three genes related to small GTPase functions: TNS1, RAP1GAP, and ARHGEF12. We focused our attention on ARHGEF12 because RhoA knockout in mice causes cytokinesis failure in erythroblasts through actomyosin and midbody dysregulation and p53 activation.10
To define the functional and mechanistic role of arhgef12 in erythropoiesis, we have used a zebrafish model to knockdown or knockout arhgef12 isoforms. We show a causal role of the ARHGEF12-RhoA signaling in this model in mediating the p38 MAPK and Stat1 pathway in erythropoiesis. In zebrafish, erythroid defects caused by arhgef12 knockdown can be rescued by p38 MAPK activator and stat1 expression. Conversely, a p38 inhibitor can induce erythropoiesis defects mimicking that of the arhgef12 knockout or knockdown. This signaling effect seems to be conserved in mammals, as the ARHGEF12-RhoA-p38 function appears to also regulate the erythroid differentiation of erythroleukemia cell line K562. A number of studies have reported that p38 MAPK is involved in erythroid differentiation,39–41 yet the role of p38 MAPK in stress erythropoiesis is still poorly understood. P38a regulates erythroblast enucleation in a cell-autonomous manner in vivo during fetal and anemic stress erythropoiesis.42 Remarkably, loss of p38a leads to downregulation of p21Cip1, and decreased activation of the p21Cip1 inactivates Rb, both of which are critical regulators of erythroblast enucleation. Hu et al. suggested P38a could act as a molecular brake to limit over-active erythropoiesis in response to stress-relief of this molecular brake by inhibiting P38-enhanced stress erythropoiesis and accelerated recovery from anemia.43 Our observed association of a down-regulated erythroid p38 phosphorylation in patients with the ARHGEF12 polymorphism who need multiple RBC transfusions to overcome chemotherapy-induced anemia also supports the involvement of such a pathway. Pharmacological activation of wild-type p53 is a logical therapeutic strategy for leukemia where the p53 pathway could be down-regulated by abnormalities in p53-regulatory proteins.44 It has been reported that p38 kinase can positively regulate p53, and activation of p38 not only promotes erythropoiesis, but also potentially maintains a higher level of p53 in cancer cells, which can be a dual benefit for cancer patients who carry wild-type p53 alleles.
Several reported GWAS studies related to hematologic traits failed to find a correlation between ARHGEF12 and erythroid phenotypes45,46 in normal populations, which may suggest there are functional redundancies to the ARHGEF12-RhoA-p38 pathway in homeostatic erythropoiesis. Suboptimal level of guanine nucleotide exchange activity may be compensated by down-regulated RhoA negative regulator, GTPase-activating proteins, or by other guanine nucleotide exchange factors, such as ARHGEF3, which was shown to be important for erythropoiesis through RhoA in a zebrafish model.47 Our GWAS results draw a clear association between ARHGEF12 at rs10892563 with erythrocyte regeneration under chemotherapy stress, suggesting that this gene/SNP status may be considered a biomarker to predict severity of chemotherapy-induced anemia among the patients.
In addition to the erythropoiesis differentiation mechanism, genes expressed in HSPC can also be associated with chemotherapy-associated anemia. To this end, we analyzed the primary gene list with expression patterns in the CD34+ cell population before erythroid differentiation with the same database as we did for the erythrocyte-specific genes. Among the genes highly expressed in HSPC, four of the top five have known functional connection with erythropoiesis (Figure 1C). While a correlation analysis between RBC transfusion and severity of neutropenia reveals that RBC transfusions had no significant correlation with neutropenia, there was a significance in correlation with thrombocytopenia (Online Supplymentary Figure S7). This suggests that it is possible that effects on HSPC such as megakaryocyte-erythroid progenitors could be a contributing factor to chemotherapy-induced anemia.
Combining our patient SNPs and phenotype observations, biochemical analyses of patient samples, and human and murine cells, together with the zebrafish genetic model characterizations, our studies unveil a novel SNP related to chemotherapy-induced anemia in ARHGEF12 and the associated signaling pathway. These findings will be useful for future consideration of strategies to overcome the chemotherapy-induced anemia in some ALL patients.
Acknowledgments
The authors would like to thank the staff of Shanghai Institute of Hematology for their assistance with zebrafish husbandry, particularly Yi Chen. We thank Yongjuan Zhang for the SNP linkage disequilibrium analysis and Professor Xiaojian Sun for helpful discussions.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/4/925
Funding
This research was supported by grants from the National Natural Science Foundation of China (n. 81270623), The National Key R&D Program of China, Stem Cell and Translation Research (n. 2016YFA0102000) and The fourth round of Three-Year Public Health Action Plan (2015-2017) (GWIV-25). This research was also supported by the National Natural Science Foundation of China (No. 81900114).
References
- 1.Sun XJ, Xu PF, Zhou T, et al. Genome-Wide Survey and Developmental Expression Mapping of Zebrafish SET Domain-Containing Genes. PLoS One. 2008;3(1): e1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110(34):13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novershtern N, Subramanian A, Lawton LN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikuta T, Sellak H, Odo N, Adekile AD, Gaensler KML. Nitric Oxide-cGMP Signaling Stimulates Erythropoiesis through Multiple Lineage- Specific Transcription Factors : Clinical Implications and a Novel Target for Erythropoiesis. Plos One. 2016;11(1):e0144561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Lodish HF. Global analysis of induced transcription factors and cofactors identifies Tfdp2 as an essential coregulator during terminal erythropoiesis. Exp Hematol. 2014;42(6):464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henneberry AL, Wistow G, Mcmaster CR. Cloning, genomic organization, and characterization of a human cholinephosphotransferase. J Biol Chem. 2000;8(4):29808–29815. [DOI] [PubMed] [Google Scholar]
- 7.Cocco L, Manzoli L, Faenza I, et al. Advances in Biological Regulation Modulation of nuclear PI-PLCbeta1 during cell differentiation. Adv Biol Regul. 2016;60:1–5. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson PJ, Chen S, Tayeh MK, et al. Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome). J Med Genet. 2005;42(7):551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih YP, Sun P, Wang A, Lo SH. Tensin1 positively regulates RhoA activity through its interaction with DLC1. Biochim Biophys Acta - Mol Cell Res. 2015;1853(12):3258–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konstantinidis DG, Giger KM, Risinger M, et al. Cytokinesis failure in RhoA-deficient mouse erythroblasts involves actomyosin and midbody dysregulation and triggers p53 activation. Blood. 2015;126(12):1473–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coste I, Gauchat JF, Wilson A, et al. Unavailability of CD147 leads to selective erythrocyte trapping in the spleen. Blood. 2001;97(12):3984–3988. [DOI] [PubMed] [Google Scholar]
- 12.Ma C, Wang J, Fan L, Guo Y. Inhibition of CD147 expression promotes chemosensitivity in HNSCC cells by deactivating MAPK/ERK signaling pathway. Exp Mol Pathol. 2017;102(1):59–64. [DOI] [PubMed] [Google Scholar]
- 13.Dorn I, Meyer G, Lindner U, Driller B, Schlenke P. The influence of extracellular matrix proteins and mesenchymal stem cells on erythropoietic cell maturation. Vox Sang. 2011;101(1):65–76. [DOI] [PubMed] [Google Scholar]
- 14.Abed M, Balasaheb S, Towhid ST, Daniel C, Amann K, Lang F. Adhesion of Annexin 7 Deficient Erythrocytes to Endothelial Cells. PLoS One. 2013;8(2):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayette S, Dhermy D, dos Santos ME, et al. A deletional frameshift mutation in protein 4.2 gene (allele 4.2 Lisboa) associated with hereditary hemolytic anemia. Blood. 1995; 85(1):250–256. [PubMed] [Google Scholar]
- 16.Yawata Y, Kanzaki A, Yawata A, Doerfler W, Ozcan R, Eber SW. Characteristic features of the genotype and phenotype of hereditary spherocytosis in the Japanese population. Int J Hematol. 2000;71(2):118–135. [PubMed] [Google Scholar]
- 17.Satyanarayana A, Gudmundsson KO, Chen X, et al. RapGEF2 is essential for embryonic hematopoiesis but dispensable for adult hematopoiesis. Blood. 2010;116(16):2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kourlas PJ, Strout MP, Becknell B, et al. Identification of a gene at 11q23 encoding a guanine nucleotide exchange factor: evidence for its fusion with MLL in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2000;97(5):2145–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drenberg C, Hu S, Li L, et al. ABCC4 Is a Determinant of Cytarabine-Induced Cytotoxicity and Myelosuppression. Clin Transl Sci. 2016;9(1):51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Wang L, Kalfa TA, et al. Cdc42 critically regulates the balance between myelopoiesis and erythropoiesis. Blood. 2007;110(12):3853–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghiaur G, Lee A, Bailey J, JA C, Zheng Y, DA W. Inhibition of RhoA GTPase activity enhances hematopoietic stem and progenitor cell proliferation and engraftment. Blood. 2006;108(6):2087–2094. [DOI] [PubMed] [Google Scholar]
- 22.Sankaran VG, Ghazvinian R, Do R, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122(7):2439–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Lochem EG, van der Velden VHJ, Wind HK, te Marvelde JG, Westerdaal NAC, van Dongen JJM. Immunophenotypic differentiation patterns of normal hematopoiesis in human bone marrow: Reference patterns for age-related changes and disease-induced shifts. Cytometry. 2004;60B(1):1–13. [DOI] [PubMed] [Google Scholar]
- 24.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paffett-Lugassy NN, Zon LI. Analysis of hematopoietic development in the zebrafish. Methods Mol Med. 2005;105: 171–198. [DOI] [PubMed] [Google Scholar]
- 26.Reuther GW, Lambert QT, Booden MA, et al. Leukemia-associated Rho Guanine Nucleotide Exchange Factor, a Dbl Family Protein Found Mutated in Leukemia, Causes Transformation by Activation of RhoA. J Biol Chem. 2001;276(29):27145–27151. [DOI] [PubMed] [Google Scholar]
- 27.Salasvidal E, Meijer AH, Cheng X, Spaink HP. Genomic annotation and expression analysis of the zebrafish Rho small GTPase family during development and bacterial infection. Genomics. 2005;86(1):25–37. [DOI] [PubMed] [Google Scholar]
- 28.Qiu RG, Chen J, Mccormick F, Symons M. A Role for Rho in Ras Transformation. Proc Natl Acad Sci U S A. 1995;92(25):11781–11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dvorsky R, Ahmadian MR. Always look on the bright site of Rho: structural implications for a conserved intermolecular interface. EMBO Rep. 2004;5(12):1130–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez OD, Mitchell D, Campos R, Gao GJ, Li L, Nolan GP. Multiparameter analysis of intracellular phosphoepitopes in immunophenotyped cell populations by flow cytometry. Curr Protoc Cytom. 2005;Chapter 6:Unit 6.20. [DOI] [PubMed] [Google Scholar]
- 31.Ahlgren J, Patel N, Simmens S, et al. O-MAX chemotherapy: high activity in metastatic esophagogastric adenocarcinoma and possible relation to subclinical hemolysis. Oncology. 2014;87(6):371–380. [DOI] [PubMed] [Google Scholar]
- 32.Betensky M, Witmer C, Fisher MJ, Nance S, Weiss MJ, Sesok-Pizzini DA. Immune hemolytic anemia with drug-induced antibodies to carboplatin and vincristine in a pediatric patient with an optic pathway glioma. Transfusion. 2015;54(11):2901–2905. [DOI] [PubMed] [Google Scholar]
- 33.Lang E, Bissinger R, Qadri SM, Lang F. Suicidal death of erythrocytes in cancer and its chemotherapy: A potential target in the treatment of tumor-associated anemia. Int J Cancer. 2017;141(8):1522–1528. [DOI] [PubMed] [Google Scholar]
- 34.Lang E, Lang PA, Shumilina E, et al. Enhanced eryptosis of erythrocytes from gene-targeted mice lacking annexin A7. Pflugers Arch. 2010;460(3):667–676. [DOI] [PubMed] [Google Scholar]
- 35.Abiko H, Fujiwara S, Ohashi K, et al. Rho guanine nucleotide exchange factors involved in cyclic-stretch-induced reorientation of vascular endothelial cells. J Cell Sci. 2015;128(9):1683–1695. [DOI] [PubMed] [Google Scholar]
- 36.Thompson WR, Yen SS, Uzer G, et al. LARG GEF and ARHGAP18 orchestrate RhoA activity to control mesenchymal stem cell lineage. Bone. 2018;107:172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi GX, Yang WS, Jin L, Matter ML, Ramos JW. RSK2 drives cell motility by serine phosphorylation of LARG and activation of Rho GTPases. Proc Natl Acad Sci U S A. 2018;115(2):E190–E199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams CM, Harper MT, Goggs R, Walsh TG, Offermanns S, Poole AW. Leukemia-associated Rho guanine-nucleotide exchange factor is not critical for RhoA regulation, yet is important for platelet activation and thrombosis in mice. J Thromb Haemost. 2015;13(11):2102–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hua WK, Chang YI, Yao CL, Hwang SM, Chang CY, Lin WJ. Protein Arginine Methyltransferase 1 Interacts with and Activates p38α to Facilitate Erythroid Differentiation. PLoS One. 2013;8(3): e56715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shahab U, Jeong AK, Jodie U, Dolores M, Amittha W. Differentiation stage-specific activation of p38 mitogen-activated protein kinase isoforms in primary human erythroid cells. Proc Natl Acad Sci U S A. 2004;101(1): 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pace BS, Qian XH, Sangerman J, et al. p38 MAP kinase activation mediates γ-globin gene induction in erythroid progenitors. Exp Hematol. 2003;31(11):1089–1096. [DOI] [PubMed] [Google Scholar]
- 42.Schultze SM, Mairhofer A, Li D, et al. p38α controls erythroblast enucleation and Rb sig-naling in stress erythropoiesis. Cell Res. 2012;22(3):539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanenberg H, Filippi M-D, Ivan M, et al. P38α/JNK signaling restrains erythropoiesis by suppressing Ezh2-mediated epigenetic silencing of Bim. Nat Commun. 2018;9(1):3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulavin DV, Saito S, Hollander MC, et al. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. Embo J. 1999;18(23):6845–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuhnel B, Aspelund T, Yang Q, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41(11):1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamatani Y, Matsuda K, Okada Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Publ Gr. 2010;42(3):210–215. [DOI] [PubMed] [Google Scholar]
- 47.Serbanovic-Canic J, Cvejic A, Soranzo N, Stemple DL, Ouwehand WH, Freson K. Silencing of RhoA nucleotide exchange factor , ARHGEF3, reveals its unexpected role in iron uptake. Blood. 2011;118(18):4967–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]