Abstract
In an effort to identify target genes in acute myeloid leukemia (AML), we compared gene expression profiles between normal and AML cells from various publicly available datasets. We identified CD99, a gene that is up-regulated in AML patients. In 186 patients from The Cancer Genome Atlas AML dataset, CD99 was over-expressed in patients with FLT3-ITD and was down-regulated in patients with TP53 mutations. CD99 is a trans-membrane protein expressed on leukocytes and plays a role in cell adhesion, trans-endothelial migration, and T-cell differentiation. The CD99 gene encodes two isoforms with distinct expression and functional profiles in both normal and malignant tissues. Here we report that, although the CD99 long isoform initially induces an increase in cell proliferation, it also induces higher levels of reactive oxygen species, DNA damage, apoptosis and a subsequent decrease in cell viability. In several leukemia murine models, the CD99 long isoform delayed disease progression and resulted in lower leukemia engraftment in the bone marrow. Furthermore, the CD99 monoclonal antibody reduced cell viability, colony formation, and cell migration, and induced cell differentiation and apoptosis in leukemia cell lines and primary blasts. Mechanistically, CD99 long isoform resulted in transient induction followed by a dramatic decrease in both ERK and SRC phosphorylation. Altogether, our study provides new insights into the role of CD99 isoforms in AML that could potentially be relevant for the preclinical development of CD99 targeted therapy.
Introduction
The outcome of patients with acute myeloid leukemia (AML) remains dismal due to the high relapse rate.1 To identify target genes that were differentially over-expressed in AML compared with normal hematopoietic cells, we leveraged genomic and transcriptomic data and discovered CD99.
CD99, a membrane glycoprotein,2 is normally expressed in cortical thymocytes, mature plasma cells, granulosa cells of the ovary, Sertoli cells of the testis, and pancreatic islet cells.3 The CD99 gene encodes two distinct proteins that are produced by alternative splicing of the CD99 transcript. The alternative spliced short isoform results from a deletion in its intracytoplasmic fragment.2 CD99 plays a role in cell migration,4 adhesion, differentiation of thymocytes and T cells,2 and regulation of diapedesis.5 In cancer cells, CD99 is highly expressed on the cell surface of Ewing’s sarcoma (EWS),6 gliomas7 and other mesenchymal,8,9 hematopoietic,10–12 and epithelial cancers.13,14 In EWS, engagement with the anti-CD99 antibody enhanced apoptosis and sensitivity to chemotherapy.15 High CD99 correlated with enhanced invasion of glioma cells.7
CD99 immunoreactivity was found in AML but rarely in myeloproliferative disorders, myelodysplastic syndromes, remission, and normal marrow samples.16 A recent study, however, showed that CD99 is a disease stem cell marker, and CD99 antibody proved beneficial in xenograft mice models of myeloid malignancies.17 With growing evidence that CD99 plays a role in cancer, and particularly in AML, and that CD99 isoforms are differentially expressed and play different roles in different hematopoietic cells,18 investigating the roles of the two isoforms is crucial for CD99 preclinical development as a therapeutic target. Here we characterize CD99 upregulation in patients with AML and its association with clinical and molecular characteristics, and determine the function of CD99 long (L) and short (S) isoforms in preclinical leukemia models.
Methods
Patients’ samples
Diagnostic or relapse blood was obtained from AML patients treated at the Norris Comprehensive Cancer Center at the University of Southern California (USC) after obtaining written informed consent. The use of human materials was approved by the Institutional Review Boards of the USC in accordance with the Declaration of Helsinki.
Patient datasets and gene expression analysis
The Cancer Genome Atlas (TCGA) AML dataset was downloaded from oncomine.19,20 Patients data from the GSE7186,21 GSE13159,22 GSE1159,23 GSE15434,24 GSE3077,25 GSE425,26 GSE12417,27 and GSE1785528 datasets were downloaded from the GEO database. Details of analysis methods are available in the Online Supplementary Appendix.
Cell lines and primary blasts
Acute myeloid leukemia cell lines were grown in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal bovine serum (FBS) and 100U/mL penicillin. Peripheral blood mononuclear cells (PBMC) from AML patients and from healthy donors were isolated by density gradient centrifugation using Ficoll-Paque. Primary cells were grown in RPMI plus 20% FBS and cytokine cocktails CC100 (Flt3L, SCF, IL-3 and IL-6).
In vivo studies
Animal protocols were approved by the Institution for Animal Care and Use Committee (IACUC) of the USC. For THP-1 and MOLM-13 xenograft experiments, 2.5×106 cells were injected via tail-vein (IV) into 4-6 week-old NOD-scid /Il2rg−/− (NSG) mice (Jackson). For the primary blasts xenograft experiment, 1×106 cells were IV engrafted into irradiated mice. Details of the methods used, including plasmids, primer sequences, antibodies and in vivo experiments are available in the Online Supplementary Appendix.
Results
CD99 is up-regulated in acute myeloid leukemia
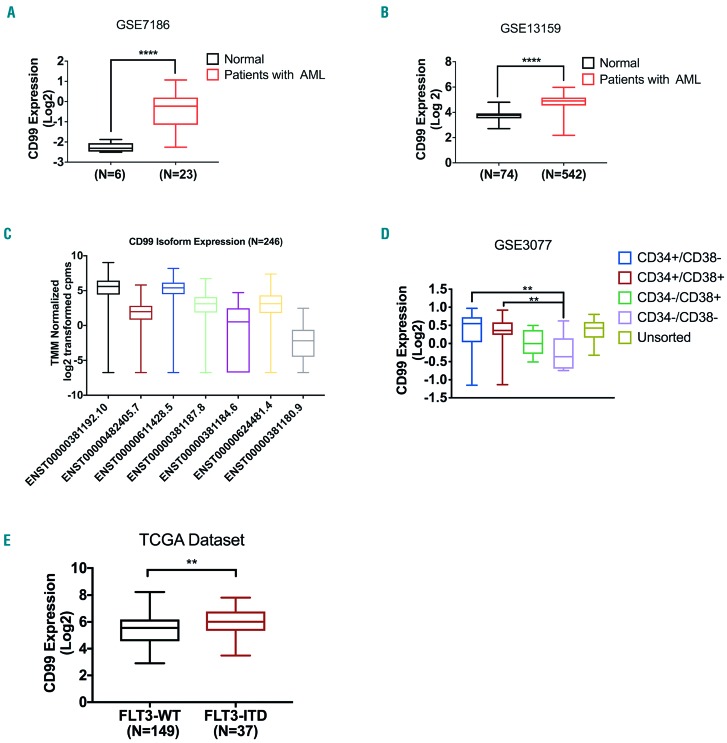
In an effort to identify target genes that were differentially over-expressed in AML, we compared gene expression profiles between normal and AML cells from various available public datasets. We found that CD99 was significantly up-regulated in AML compared with normal cells in five datasets with available measurements of CD99 RNA levels in both leukemia and normal cells (oncomine median ranking among up-regulated measured genes 155, P=0.013; TCGA data did not have CD99 levels in normal cells). CD99 was significantly higher in 23 AML samples compared with six normal bone marrow (BM) samples (GSE7186: 3.5-fold, P<0.0001) (Figure 1A). Consistently, CD99 was significantly over-expressed in blasts of 542 patients with AML compared with PBMC from 74 healthy donors (HD) (GSE13159: 2-fold, P<0.0001) (Figure 1B). Similarly, CD99 expression was 1.8-fold higher in blasts from 285 patients with AML compared with HD cells (GSE1159: P=0.003) (Online Supplementary Figure S1A). There was a 2.3-fold increase in CD99 expression (GSE13164: P<0.001) (Online Supplementary Figure S1B) in blasts of 257 AML patients compared with PBMC of 58 HD. In the GSE995 dataset, CD99 expression was 3-fold higher in blasts from nine AML patients compared with that in cells from six HD (P=0.028) (Online Supplementary Figure S1C). We also analyzed the expression of CD99 protein coding transcripts in patients with AML (GSE106291; n=246). ENST00000381192.10 (CD99-L) and ENST00000611428.5 (CD99-S) were the top expressed transcripts (Figure 1C). The combined expression of ENST00000482405.7 and ENST00000611428.5 (both code for CD99-S) was significantly higher (P=0.0073) than the level of ENST00000381192.10 (CD99-L) (Online Supplementary Figure S1D). Transcript Ensembl IDs are described in Online Supplementary Table S1. The transcript expression of the two isoforms were positively correlated (Online Supplementary Figure S1E and F).
Figure 1.
Characterization of CD99 expression in acute myeloid leukemia (AML) datasets. (A) Relative expression of CD99 in 23 AML cases compared with six healthy donors in the GSE7186 dataset. (B) Relative expression of CD99 in 542 AML cases compared with 74 healthy donors in the GSE13159 dataset. (C) Characterization of CD99 transcript expression in 246 patients with AML from the GSE106291 dataset. (D) 23 human tissue samples obtained from GSE30377 dataset were sorted into CD34+CD38−, CD34+CD38+, CD34−CD38-and CD34−CD38+ and analyzed for CD99 expression. (E) Relative expression of CD99 in 37 patients with FLT3-ITD compared with that in 149 patients without FLT3-ITD (WT) in the TCGA dataset. N: number (**** P<0.0001; *** P<0.001; ** P<0.01; *P<0.05).
Several reports suggested that CD99 is up-regulated on leukemia stem cells.29–31 To validate this, we assessed CD99 expression in the GSE3077 dataset that has gene expression data of leukemia blasts obtained from 23 patients with AML sorted according to their CD34 and CD38 expression levels. We found that CD99 expression was significantly higher (1.34-fold, P<0.001) (Figure 1D) in the CD34+CD38+ and CD34+CD38− combined subpopulation compared with CD34−CD38− and CD34−CD38+ combined.
CD99 overexpression is positively associated with FLT3-ITD and reversely with TP53 mutations
We assessed the association between CD99 upregulation and the presence of AML mutations. In the TCGA dataset, CD99 expression was significantly higher (P=0.004) in patients with FLT3-ITD (n=37) compared with patients with FLT3 point mutations (n=16) and FLT3 wild-type (n=133) (1.3 fold, P=0.002) (Figure 1E). Similarly, there was a significant association between high CD99 (above the median) and the presence of FLT3-ITD mutation (Fisher’s exact test, P=0.04) (Online Supplementary Table S2). A similar association was found in the GSE17855 and GSE15434 datasets with childhood AML and patients with cytogenetically normal karyotype, respectively. CD99 is significantly higher in patients with FLT3-ITD compared with patients with FLT3 wild-type (FLT3-WT) (1.3-fold, P=0.03, Online Supplementary Figure S1G; 1.4-fold, P<0.001, Online Supplementary Figure S1H; respectively). We next examined the association of each CD99 transcript with FLT3-ITD mutation. ENST00000381187.8 and ENST00000624481.4 were significantly higher in FLT3-ITD compared with FLT3-WT patients (P=0.001 and P=0.002) (Online Supplementary Figure S1I-N). ENST00000611428.5 (CD99-S) was not significantly associated with FLT3-ITD after adjusting for multiple hypotheses testing (unadjusted P=0.049).
CD99 expression was significantly lower (1.9-fold, P<0.001) in patients with mutated TP53 (n=15) compared with patients with TP53-WT (n=171) (Online Supplementary Figure S1O; Fisher’s Exact test, P=0.0006, Online Supplementary Table S1). CD99 expression was not associated with other mutations (Online Supplementary Table S2 and Online Supplementary Figure S2).
CD99 expression levels according to patients’ clinical characteristics
We dichotomized patients into high and low groups based on CD99 median expression. Patients with CD99-low had a significantly lower percentage of bone marrow (BM) blasts (median, 66.2 vs. 72.5, P=0.024), peripheral blood (PB) blasts (median, 23.7 vs. 50.7, P<0.0001), and white blood cell (WBC) count (median, 27.7 vs. 45.8, P=0.019). The median age was 57.8 years in CD99-low and 52.1 years in CD99-high groups (Online Supplementary Table S3).
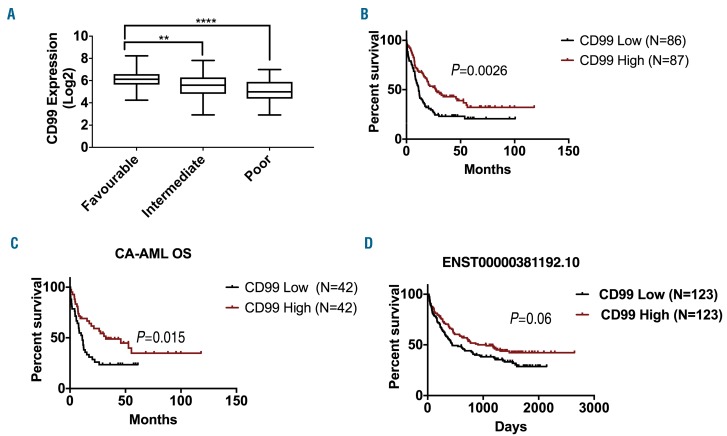
According to the National Comprehensive Cancer Network (NCCN), AML is classified into favorable, intermediate and poor-risk groups based on patients molecular and cytogenetic characteristics. CD99 expression was significantly higher in the favorable-risk group than that in the intermediate-risk (1.2-fold, P<0.01) and poor-risk (2-fold, P<0.0001) groups (Figure 2A). According to the AML French-American-British (FAB) classification data, patients with M5-AML had lower CD99 expression compared with M1, M2, M3 and M4 (Online Supplementary Figure S3A). Based on leukemia cytogenetics, no significant differences in CD99 expression was found in patients with complex karyotype, Inv (16), t (15; 17), t (8,21), del (7q) / 7q- and trisomy 8, compared with normal karyotype (Online Supplementary Figure S3B).
Figure 2.
Association of CD99 expression with patients’ clinical characteristics in acute myeloid leukemia (AML) datasets. (A) Relative expression of CD99 according to the National Comprehensive Cancer Network (NCCN) risk status classification in the The Cancer Genome Atlas (TCGA) dataset. (B) Overall survival (OS) of 173 patients grouped based on CD99 median expression into CD99 high (n=87) and CD99 low (n=86) from the TCGA dataset. (C) OS of cytogenetically abnormal (CA-AML) cases grouped based on CD99 median expression into CD99 high (n=42) and CD99 low (n=42). (D) OS of 146 patients grouped based on ENST00000381192.10 transcript isoform based on median expression into high (n=123) and low (n=123) from the GSE106291 dataset. (****P<0.0001; ** P<0.01).
CD99 overexpression is associated with better clinical outcome
The overall survival (OS) of CD99-high patients (based on median cut-off) was significantly longer than that of CD99-low patients (median: 27 vs. 11.2 months; P=0.0026) (Figure 2B). Patients with t(15;17) are treated with all-trans retinoic acid (ATRA) and have a better outcome, thus they were excluded from the survival analyses. When patients were stratified into cytogenetically normal (CN) and cytogenetically abnormal (CA), we found that in CA-AML, but not in CN-AML, CD99-high survived significantly longer than CD99-low patients (CA-AML: median OS: 32.3 vs. 11 months, P=0.02, Figure 2C; CN-AML: P=0.24, Online Supplementary Figure S4A). Yet, there was no significant difference in CD99 levels between CN-AML and CA-AML (P=0.33) (Online Supplementary Figure S4B). There was no significant difference in event-free survival (EFS) between CD99-high and CD99-low patients (median: 15.6 vs. 13.3 months; P=0.13) (Online Supplementary Figure S4C-E).
In the GSE425 dataset of 71 patients with CA-AML, CD99-high patients survived significantly longer than CD99-low patients (P=0.04) (Online Supplementary Figure S5B). No significant difference in OS was found when we included CN-AML patients or analyzed them separately (Online Supplementary Figure S5A and C). In the GSE12417 dataset, which includes only CN-AML patients, no significant difference between CD99-high and CD99-low patients (n=163) was observed (Online Supplementary Figure S5D).
In a multivariable analysis, high CD99 was not significantly associated with OS when adjusted by age, cytogenetic risk, transplant status, DNMT3A mutation status, and TP53 mutation status (P=0.364) (Online Supplementary Table S4).
We also assessed the association of CD99 transcript upregulation with OS in 246 patients with AML using median cut-off for each transcript. High ENST00000381192.10 (CD99-L) exhibited a trend of association with better OS (GSE106291: median OS: 908 vs. 445 days; P=0.06) (Figure 2D). No difference in survival between high and low expression was observed for the other transcripts (Online Supplementary Figure S6A-G).
Differential effect of CD99 isoform expression on leukemia growth
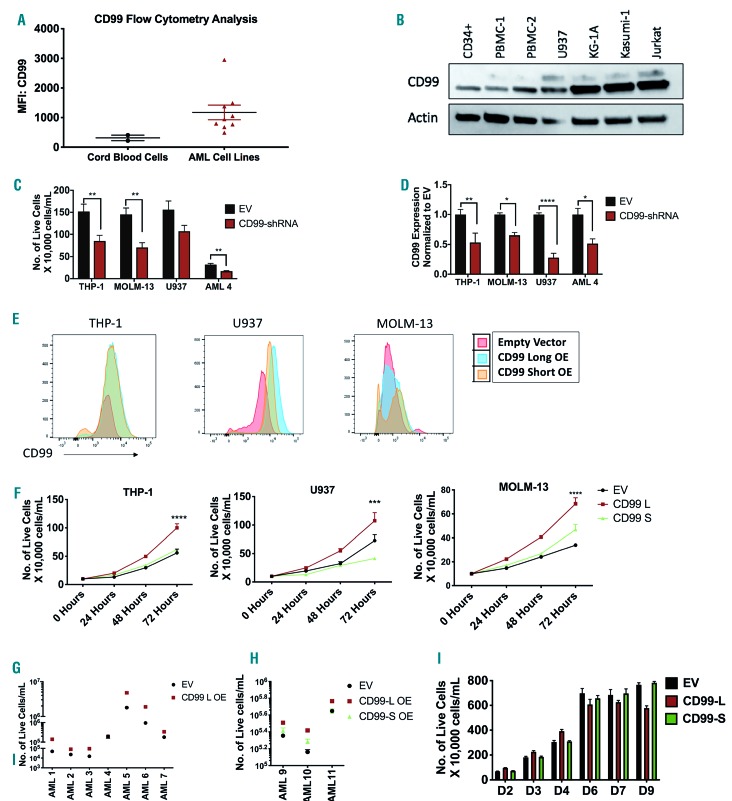
In a panel of AML cell lines (KG-1, KG-1A, MOLM13, MV4-11, Kasumi-1, THP-1, NB4, U937, UOC-M1), CD99 surface levels are higher than that of healthy cord blood cells (Figure 3A). Western blots revealed 28 and 32kDa bands corresponding to CD99-L and -S in U937, KG-1A and Kasumi-1 cells, but only the lower band was recognized in THP-1, MOLM-13, CD34+ cells and two HD PBMC lysates (Figure 3B and Online Supplementary Figure S7A). Similarly, quantitative polymerase chain reaction (qPCR) analysis showed that CD99 transcripts were up-regulated in AML blast samples (n=9) and AML cell lines (KG-1a, U937, THP-1, and MOLM-13) compared with HD PBMC (n=3) and CD34+ cells. Transcript ENST00000482405.7 is higher in CD34+ cells than in AML cell lines (Online Supplementary Figure S7B; for qPCR primers see Online Supplementary Table S5).
Figure 3.
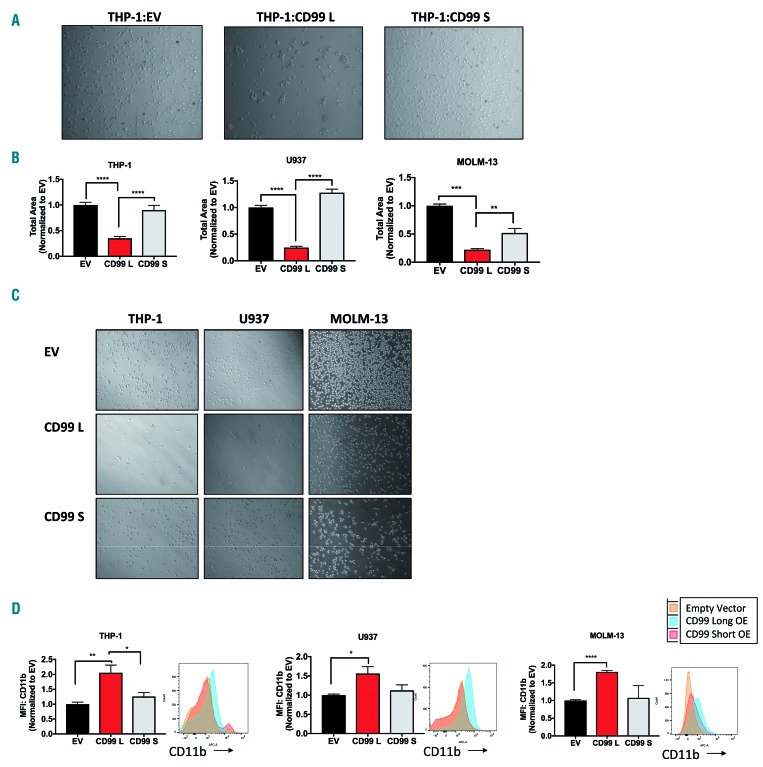
Effect of CD99 isoform expression on acute myeloid leukemia (AML) cell proliferation. (A) Relative CD99 expression in cord blood cells (n=2) and AML cell lines (n=9) measured by flow cytometry. (B) Western blot analysis of CD99 isoform expression in CD34+ cells, healthy donor peripheral blood mononuclear cells (PBMC) and AML cell lines. (C) Viability of CD99-shRNA knockdown in THP-1, MOLM-13, U937 and AML-4 cells measured 96 hours (h) after transfection using trypan-blue and alamar-blue. (D) Quantative polymerase chain reaction (qPCR) analysis using CD99 TaqMan assay to confirm CD99 knockdown in THP-1, MOLM-13, U937 and AML-4 cells transfected with CD99-shRNA plasmid (or EV) for 96 h. CD99 expression was normalized to EV cells. (E) CD99 expression in EV, CD99-L OE and CD99-S OE cells measured by flow cytometry in THP-1, U937 and MOLM-13 cells.(F) Proliferation assay of EV, CD99-L and CD99-S measured at 24,48 and 72 h by trypan-blue (n=4) in THP-1, U937, and MOLM-13 cells. Data reported as number of live cells. (G) Viability of AML blasts over-expressing CD99-L or EV measured 96 h after lentiviral transduction using trypan-blue (n=7). Data reported as total number of live cells/mL. (H) Viability of AML blasts over-expressing CD99 L, CD99-S or EV measured 96 h after lentiviral transduction using trypan-blue (n=3). Data reported as the number of live cells/mL. (I) Long-term proliferation assay of EV, CD99-L and CD99-S cells measured at days 2, 3, 4, 6, 7 and 9 by trypan-blue assay. (****P<0.0001; ***P<0.001; **P<0.01; *P<0.05).
We then examined the effect of CD99 knockdown on cell viability. Transducing cells with lentiviral-CD99-shRNA significantly reduced primary AML blast viability (n=4) (Figure 3C and Online Supplementary Figure S7C) and AML cell lines (THP-1, MOLM-13, and U937) (40-60%; P<0.05) (Figure 3C). Knockdown was confirmed by qPCR (Figure 3D) and western blot (Online Supplementary Figure S7D). THP-1 and MV4-11 cells had an approximately 35-50% decrease in cell viability when transiently transfected by elecroporation with CD99-siRNA compared with negative-control-siRNA (P=0.001 and P=0.001, respectively) (Online Supplementary Figure S7E-G).
We also established a gain-of-function approach to study CD99-L and CD99-S isoform functions. We performed lentiviral transduction to over-express CD99-L and CD99-S in THP-1, U937 and MOLM-13 cells expressing variable endogenous levels of CD99 isoforms (Figure 3E and Online Supplementary Figure S7H and I). CD99-L transduced cells had increased cell proliferation at 72 hours (h) compared with their respective empty vector (EV) controls and CD99-S transduced cells, respectively, counted by trypan-blue in THP-1 (1.78-fold, P<0.001; 1.61-fold, P<0.01), U937 (1.47-fold, P<0.001; 2.59-fold, P<0.0001), and MOLM-13 cells (2-fold, P<0.0001; 1.45-fold, P<0.0001) (Figure 3F). This was also confirmed by alamar-blue assay (Online Supplementary Figure S8A) and by BrdU staining (1.6-fold, P=0.006) (Online Supplementary Figure S8B) suggesting enhanced metabolic activity and DNA synthesis in these cells, respectively.
We also ectopically over-expressed CD99-L in AML blasts (n=7) (Online Supplementary Table S6) using lentiviral transduction. Overexpression of CD99-L was confirmed using western blot and fluorescence microscopy for green fluorescent protein (GFP) (Online Supplementary Figure S8C and D). Higher cell number was observed in lenti-CD99 blasts compared with lenti-EV transduced blasts 96 h after viral transduction (2.8-fold, P<0.0001) (Figure 3G). We also observed a modest increase in the number of colonies on day 14 in 3 of 6 patient samples over-expressing CD99-L compared with their respective controls: AML-3 (6 vs. 15, P=0.02), AML-4 (6 vs. 14, P=0.02), and AML-5 (42 vs. 128, P=0.001) (Online Supplementary Figure S8E). Furthermore, we ectopically expressed CD99-L and CD99-S in three additional AML blast samples (Online Supplementary Figure S8F). Cell viability was measured 96 h after transduction using trypan-blue and alamar-blue. A higher number of live cells was observed in CD99-L transduced blasts compared with lenti-EV (1.5-fold, P<0.0001) and CD99-S transduced blasts (1.3-fold, P<0.0001) (Figure 3H). More colonies were observed in one sample over-expressing CD99-L compared with their respective controls; AML-10 (49.5 vs. 101.5, P=0.04) (Online Supplementary Figure S8G).
However, long-term culture of CD99-L transduced cells showed a subsequent drop in cell viability, and cells could not be maintained in culture for more than 4-6 weeks. To validate this, we performed a long-term culture assay for ten days starting approximately two weeks post viral transduction. Proliferation of CD99-L cells started to decline by day 5 of initial serum stimulation compared with EV and CD99-S cells, even when cell density and nutrients were accounted for (Figure 3I). In CD34+ cells, we observed no significant change in the number of viable cells between cells transduced with CD99-L, -S isoform or EV (Online Supplementary Figure S8H) at 96 h post transduction. Transducing cells with lentiviral-CD99-shRNA resulted in a slight decrease in cell viability (Online Supplementary Figure S8I).
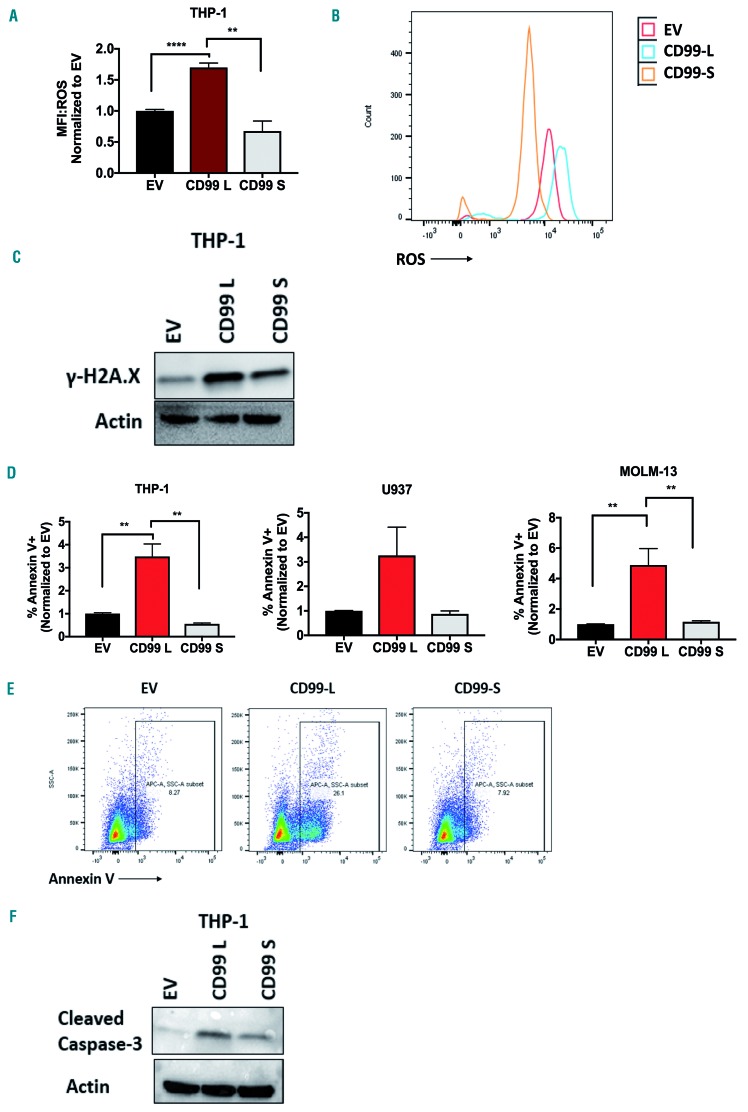
Ectopic expression of CD99 long isoform enhances reactive oxygen species levels, DNA damage, and induces cell apoptosis
Because the initial enhanced proliferation of CD99-L cells was serum induced and reversed with further expansion in vitro, we speculated that the serum-induced cell growth would stimulate higher production of reactive oxygen species (ROS) in these cells. Indeed, THP-1 cells transduced with CD99-L exhibit a 2.5- and 1.6-fold increase in ROS levels compared with CD99-S and EV cells, respectively (Figure 4A and B). Because of their higher ROS levels, we asked whether DNA damage is increased in these cells. Western blot analysis showed that CD99-L cells exhibit a higher level of the DNA damage marker H2Axγ compared with CD99-S and EV cells (Figure 4C). Furthermore, apoptosis was also enhanced in CD99-L transduced cells measured by annexin-V staining in THP-1 (CD99-L vs. EV: 3.48-fold, P=0.001; CD99-L vs. CD99-S: 6.32-fold, P=0.0027), U937(CD99-L vs. EV: 3.26-fold, P=0.07; CD99-L vs. CD99-S: 3.67-fold, P=0.10), and MOLM-13 cells (CD99-L vs. EV: 4.88-fold, P=0.0032; CD99-L vs. CD99-S: 4.2-fold, P=0.0042) (Figure 4D and E). The enhanced apoptosis was also confirmed by the increase of cleaved caspase-3 (Figure 4F).
Figure 4.
Effect of ectopic expression of CD99 long isoform on reactive oxygen species (ROS) levels, DNA damage and cell apoptosis. (A and B) ROS levels of EV, CD99-L and CD99-S cells determine by flow cytometry using the cell ROX reagent in THP-1 cells (n=4) measured by flow cytometry. (C) Western blot analysis of H2Axγ in THP-1 EV, CD99-L and CD99-S. (D and E) Apoptosis analysis of EV, CD99-L and CD99-S cells determined by annexin V+ cells through flow cytometry in THP-1, U937 and MOLM-13 cells (n=4). (F) Western blot analysis of cleaved caspase-3 in THP-1 EV, CD99-L, and CD99-S. (**** P<0.0001; ** P<0.01).
Ectopic expression of CD99-L isoform induces myeloid differentiation and reduces cell migration
Previous studies have demonstrated that CD99 homotypic interaction in CD99 expressing cells32–34 plays a role in monocyte trans-endothelial migration.4 Thus, we asked which isoform is responsible for cell homotypic interaction, and whether they affect cell migration and myeloid differentiation differently. Cells were seeded at 1×105cells/mL per well in a 6-well-plate and images were taken 6 h later; CD99-L THP-1 cells displayed higher cell aggregation compared with EV and CD99-S cells (Figure 5A). CD99-L cells exhibit decreased migration towards SDF-1α in a transwell chamber compared with EV and CD99-S cells; THP-1 (70%, P<0.0001; 66%, P<0.0001), U937 (80%, P<0.0001; 83%, P<0.0001), and MOLM-13 (80%, P<0.0001; P=0.0032) (Figure 5B and C).
Figure 5.
Functional analysis of CD99-L and CD99-S isoform overexpression in vitro. (A) Representative images for aggregation assay of THP-1 EV, CD99-L and CD99-S cells taken 6 hours (h) after seeding. (B) Migration of EV, CD99-L, and CD99-S in THP-1, U937, and MOLM 13 cells ana-lyzed by measuring cells that migrated towards SDF-1α using transwell plates (n=3). (C) Representative images for migration assay of EV, CD99-L, and CD99-S in THP-1, U937, and MOLM-13 cells. (D) CD11b analysis of EV, CD99-L, and CD99-S cells measured by flow cytometry in THP-1, U937, and MOLM-13 cells (n=4). (**** P<0.0001; *** P<0.001; ** P<0.01; * P<0.05).
CD99-L expressing cells showed an increase in CD11b surface marker measured by flow cytometry 24 h after cells were seeded in THP-1 (CD99-L vs. EV: 2-fold, P=0.0027; CD99-L vs. CD99-S: 1.63-fold, P=0.043), U937 (CD99-L vs. EV: 1.56-fold, P=0.01; CD99-L vs. CD99-S: 1.29-fold, P=0.11), and MOLM-13 (CD99-L vs. EV: 1.89-fold, P<0.0001; CD99-L vs. CD99-S: 1.68-fold, P<0.079) (Figure 5D).
Ectopic expression of CD99 long isoform delayed leukemia engraftment in acute myeloid leukemia murine models
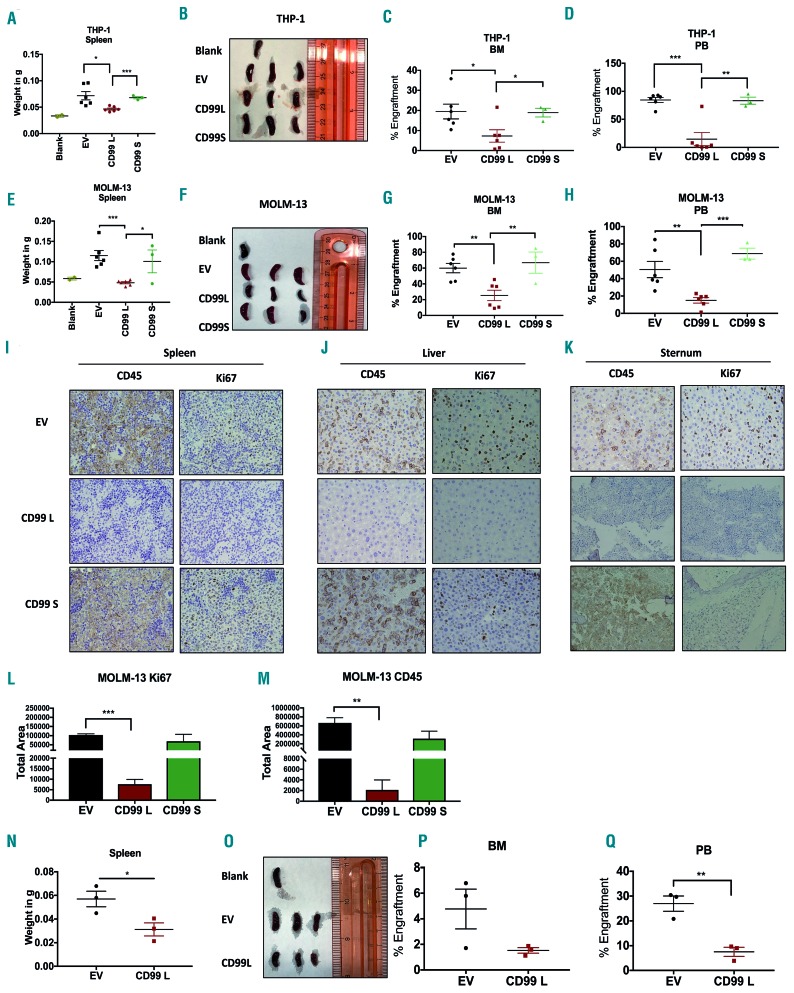
Next, we investigated the effect of ectopic expression of CD99 isoforms in murine leukemia models. THP-1 cells stably over-expressing CD99-L (n=6), CD99-S (n=3), or EV (n=6) were injected into NSG mice. Mice were sacrificed on day 30-32 post implantation. Mice engrafted with CD99-L cells had smaller spleens compared with EV mice (0.04g vs. 0.07g, P=0.010) and CD99-S mice (0.04g vs. 0.06g, P=0.0003) (Figure 6A and B). hCD45 flow analysis revealed that CD99-L mice had significantly less BM engraftment compared with EV (7.29% vs. 19.47%; P=0.02) and CD99-S mice (7.29% vs. 18.9%; P=0.04) (Figure 6C). CD99-L mice had significantly less PB engraftment compared with EV (14.73% vs. 84.52 %; P=0.0002) and CD99-S mice (14.73% vs. 83.3 %; P=0.006) (Figure 6D and Online Supplementary Figure S9). No difference in engraftment was seen in the liver (Online Supplementary Figure S10A) or spleen (Online Supplementary Figure S10B) between groups.
Figure 6.
Effect of CD99-L and CD99-S isoform overexpression in acute myeloid leukemia (AML) murine models. 2.5×106 THP-1 EV (n=6), CD99-L (n=6), or CD99-S (n=3) cells were implanted in mice and sacrificed when sick (days 30-32 post implantation). (A and B) Spleen weight and images of blank, EV, CD99-L, and CD99-S mice at time of sacrifice. (C and D) Bone marrow (BM) and peripheral blood (PB) engraftment of THP-1 cells in EV, CD99-L and CD99-S mice determined by quantitative analysis of CD45+ cells through flow cytometry. 2.5×106 MOLM-13 EV (n=6), CD99-L (n=6), or CD99-S (n=3) cells were implanted in mice and sacrificed when mice were sick. (E and F) Spleen weight and images of blank, EV, CD99-L, and CD99-S mice at time of sacrifice. (G and H) BM and PB engraftment of MOLM-13 cells in EV, CD99-L, and CD99-S mice determined by quantitative analysis of CD45+ cells through flow cytometry. (I-K) Representative images of immunostaining for CD45 and Ki67 on collected tissues of (I) spleen, (J) liver, and (K) sternum. (L and M) Quantitative analysis of Ki67 and CD45 cells in the liver tissues of mice engrafted with MOLM-13 EV, CD99-L, and CD99-S cells quantified using ImageJ. 1×106 primary AML cells over-expressing EV (n=3) or CD99-L (n=3) cells were engrafted in mice and sacrificed when mice were sick (day 19 post-cell transplantation). (N and O) Spleen weight and images of EV and CD99-L mice at the time of sacrifice (4-months post transplantation). (P and Q) BM and PB engraftment of primary AML cells in EV and CD99-L mice determined by quantitative analysis of CD45+ cells by flow cytometry. (****P<0.0001; ***P<0.001; **P<0.01; *P<0.05).
In the MOLM-13 murine model, mice were engrafted with CD99-L, CD99-S and EV mice (n=6 for EV and CD99-L, and n=3 for CD99-S) and were sacrificed on day 19 post transplantation. Mice engrafted with CD99-L cells had smaller spleens than EV mice (0.048g vs. 0.11g, P=0.004) and CD99-S mice (0.048g vs. 0.10g, P=0.026). (Figure 6E and F). CD99-L mice had significantly less engraftment than the EV mice in the BM (25.38% vs. 59.57%, P=0.003) (Figure 6G), PB (15.03% vs. 50.5%, P=0.0051) (Figure 6H), and liver (21.6% vs. 44.03%, P=0.011) (Online Supplementary Figures S10C and S11). No difference in engraftment was seen in spleen (Online Supplementary Figure S10D). Additionally, to determine whether mice engrafted with CD99-L cells developed deadly leukemia, two CD99-L engrafted mice were sacrificed only when mice appeared sick (days 29 and 44). We observed PB engraftment in both mice and in the BM of one mouse (Online Supplementary Figure S10). Hematoxylin & Eosin staining of the liver, spleen and sternum showed that EV and CD99-S tissues presented a higher percentage of infiltrated blasts compared with the CD99-L mice tissues (Online Supplementary Figure S10). We also found higher CD45 immunostaining (CD99-L vs. EV, P=0.004; CD99-L vs. CD99-S, P=0.13) and Ki67 staining (CD99-L vs. EV, P=0.0002; CD99-L vs. CD99-S, P=0.17) in EV and CD99-S tissues compared with CD99-L tissues, confirming a decrease in leukemia engraftment in mice with CD99-L cells (Figure 6I-M).
In a primary blast murine model, we engrafted primary AML samples (AML-4) transduced with CD99-L (n=3) or EV (n=3) in sub-lethally irradiated mice. Mice were sacrificed four months post engraftment (Figure 6N-O). CD99-L mice had less BM engraftment, though this was not significant compared with the EV mice (1.52 vs. 4.76%, P=0.1) (Figure 6P), and a significant decrease in hCD45+ cells in PB (7.5% vs. 26.9 %, P=0.005) (Figure 6Q and Online Supplementary Figures S10 and S12).
CD99 monoclonal antibody treatment exhibits antileukemia activity in acute myeloid leukemia cells
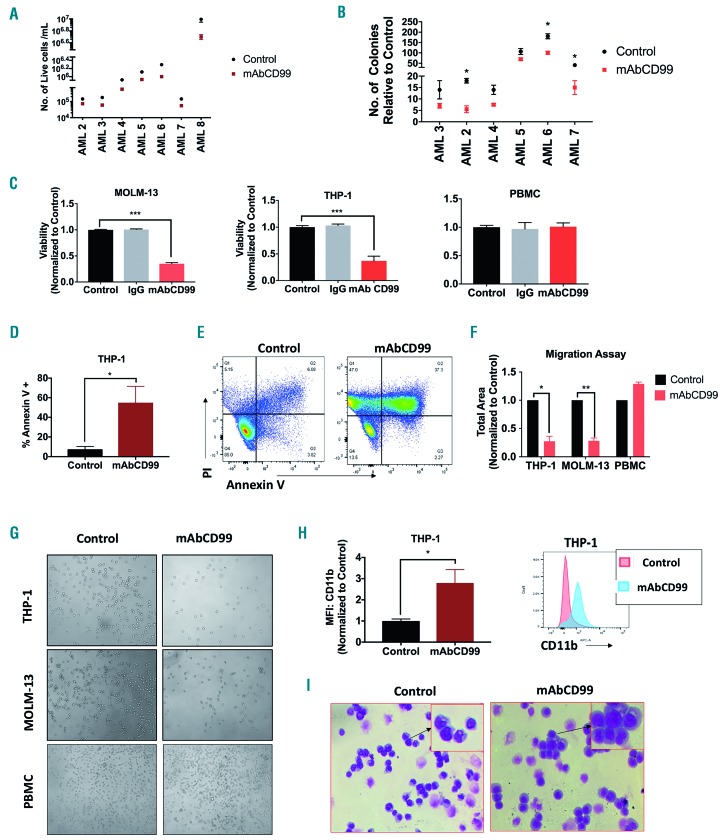
Acute myeloid leukemia patient blasts (n=7) incubated with monoclonal CD99 antibody (mAbCD99: 20 μg/mL) showed a significant decrease in cell viability (48 h: 0.5±0.02-fold, P<0.0001) (Figure 7A). Treatment with mAbCD99 caused a decrease in colony formation in 3 of 6 patient samples: AML-2 (0.5-fold, P=0.02), AML-6 (0.65-fold, P=0.03), and AML-7 (0.36-fold, P=0.01) (Figure 7B). Treatment with mAbCD99 (5 μg/mL) caused a 50% decrease in cell viability at 48 h in THP-1 and MOLM-13 cells but not in healthy donor PBMC (P<0.0001 and P<0.001, respectively) (Figure 7C). The decrease in viability was accompanied by an increase in Annexin-V apoptosis stain in THP-1 (7.35-fold, P=0.04) (Figure 7D and E). Four hours after treatment with mAbCD99 (5 μg/mL) we found an approximately 70% decrease in cell migration in both THP-1 and MOLM-13 cells (P=0.01 and P=0.004, respectively) (Figure 7F and G). Treatment of THP-1 cells with mAbCD99 triggered an increase in CD11b+ population, indicative of myeloid differentiation (2.79-fold, P=0.04) (Figure 7H). Consistently, Wright-Giemsa stain revealed that mAbCD99 (2.5 μg/mL) induced differentiation with morphology resembling more mature cell fates (Figure 7I).
Figure 7.
Effect of CD99 mAB on acute myeloid leukemia (AML) cells. (A) Cell viability of AML blasts treated with 20µg/mL of mAbCD99 for 48 hours (h) and measured using trypan-blue (n=7). (B) Total number of colonies comparison between AML blasts treated with mAb CD99 and control blasts (n=6). (C) Viability of THP-1, MOLM-13 and healthy donor peripheral blood mononuclear cells (PBMC) treated with 5µg/mL of mAbCD99 for 48 h and measured using alamar blue. (D and E) Apoptosis measured in THP-1 and MOLM-13 cells treated with mAbCD99 for 72 h and stained with Annexin V for flow cytometry analysis. (F and G) Quantitative analysis and representative images of migration of THP-1, MOLM-13 and PBMC treated with mAB CD99 towards SDF-1α performed in a transwell plate. (H) CD11b measured by flow cytometry in THP-1 cells 72 h post treatment with 2.5 µg/mL of mAB CD99. (I) Representative images for Wright-Giemsa staining of THP-1 cells treated with 2.5 µg/mL of mAb CD99 for 72 h. (***P<0.001; **P<0.01; *P<0.05).
CD99 modulates ERK and SRC signaling pathways in acute myeloid leukemia cells
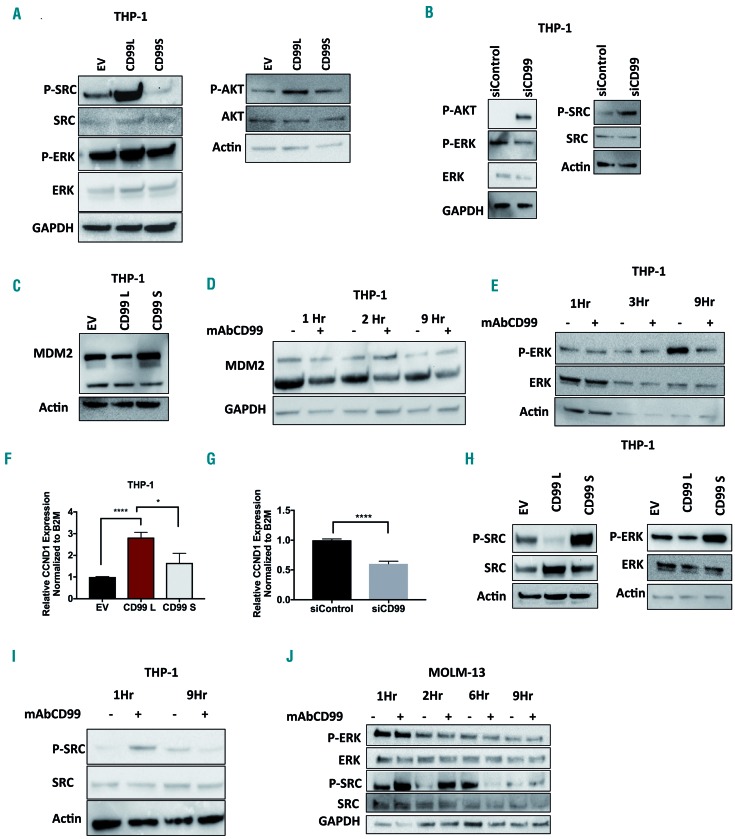
Because the initial enhanced proliferation of CD99-L expressing cells was serum stimulated and transient, we speculated that growth factor-induced signaling pathways are affected by CD99. In EWS and osteosarcoma, CD99 was found to modulate ERK pathways.35,36 Thus, we examined the effect of ectopic expression of CD99 isoforms on AKT and ERK kinase activity. We also assessed changes in SRC signaling, previously shown to be affected by CD99.17,37 CD99-L induces transient upregulation of PERK, P-AKT and P-SRC compared with CD99-S and EV expressing cells (Figure 8A and Online Supplementary Figure S13) measured 72 h post viral transduction. CD99 knockdown (against both S and L) decreased P-ERK but increased P-AKT and P-SRC (THP-1 cells express mainly CD99-S isoform) (Figure 8B). In EWS, treatment with CD99-antibody induced cell death via rapid decrease of MDM2 and activation of IGF-1R and ERK signaling.35,36 Thus, we asked whether MDM2 is affected by CD99 expression. CD99-L cells exhibit lower MDM2 protein levels compared with CD99-S and EV cells (Figure 8C). In addition, MDM2 is reduced in cells treated with CD99mAb 2-9 h post treatment (Figure 8D), yet P-ERK was not changed (Figure 8E and J). Because MDM2 is known to ubiquitinate IGF-1R, we speculated that CD99-induced reduction in MDM2 may up-regulate IGF-1R downstream target genes such as Cyclin D1.38 CCND1 mRNA levels in CD99-L cells were higher than that in CD99-S (1.75-fold, P=0.02) or EV (2.8-fold, P<0.0001) (Figure 8F). CD99 knockdown also decreased CCND1 mRNA (P<0.0001) (Figure 8G). We also examined P-ERK and P-SRC in stable cells maintained in culture for >2 weeks (Figure 8H). Contrary to the early effect, we found a dramatic decrease in P-SRC in CD99-L cells. Similarly, cells treated with anti-CD99mAb showed a rapid increase in P-SRC observed within 1-2 h followed by a decrease in P-SRC observed 3-9 h post treatment (Figure 8I and J).
Figure 8.
Effect of CD99 on ERK and SRC signaling pathways in acute myeloid leukemia (AML) cells. (A) Western blot analysis of SRC and ERK signaling pathways in THP-1 cells transduced with EV, CD99-L, and CD99-S lenti-virus for 72 hours (h). (B) Western blot analysis of SRC and ERK signaling pathways in THP-1 cells trans-fected with siCD99 for 48 h. (C) MDM2 western blot analysis in THP-1 cells transduced with EV, CD99-L, and CD99-S lentivirus. (D) MDM2 analysis performed by western blot in THP-1 cells treated with mAbCD99 for 1, 2, and 9 h. (E) ERK signaling pathway analyzed in THP-1 cells treated with mAbCD99 for 1, 3, and 9 h. (F) Cyclin D1 mRNA expression measured by taqman assay in THP-1 cells transduced with EV, CD99-L and <CD99-S lentivirus. (G) CCND1 mRNA expression measured by taqman assay in THP-1 cells transfected with siCD99. (H) Western blot analysis of SRC and ERK signaling pathways in THP-1 stable cells transduced with EV, CD99-L and CD99-S lentivirus. (I) SRC signaling pathway analyzed in THP-1 cells treated with mAbCD99 for 1 and 9 h. (J) ERK and SRC signaling pathway analyzed in MOLM-13 cells treated with mAbCD99 for 1, 2, 6, and 9 h. (****P<0.0001; *P<0.05).
Discussion
CD99 upregulation has been implicated in several malignancies and is mostly known for its role in EWS.6,39 Chung et al. have recently shown that CD99+ cells expressed an antigenic profile that enriches leukemia stem cells in the majority of human AML. They also demonstrated anti-leukemia activity of the CD99-antibody both in cellular and murine models.17 Because AML is a heterogeneous disease, it is essential to identify patients with high CD99 expression that may benefit from treatments aimed to target this gene. Our analysis revealed an association between CD99 overexpression and the presence of FLT3-ITD. A previous study has shown that CD123/CD99/CD25(+) cells in a CD34+ cell fraction predict FLT3-ITD mutations.40 Patients with FLT3-ITD have a dismal clinical outcome. Whether targeting CD99 in this patient population provides a therapeutic advantage remains to be investigated. Despite its upregulation in AML, high CD99 was associated with better outcome. Yet this association was only observed in CA-AML and was not significant in multivariate survival analysis. The inverse correlation between high CD99 and P53 mutations is likely driving its association with better clinical outcome as P53 mutations are most common in complex karyotype patients and associated with inferior outcome.41
CD99 is expressed as short and long isoforms with tissue-specific differential expression. However, the different roles of CD99 isoforms in normal and malignant tissues is only supported by limited research. Considering the increased interest in CD99 as a therapeutic target in AML, investigating the roles of its isoforms in preclinical studies is essential.18 RNA sequencing data of patients’ samples revealed that transcripts coding for CD99-S and L isoforms are both expressed in AML. We further observed varying levels of CD99 isoform protein expression in HD PBMC and AML cell lines. However, the ability of western blot to compare isoform levels is limited by the antibodies that are generated against different epitopes. Furthermore, CD99 is highly glycosylated, which also affects the size of the protein band. Due to the limited number of cells in our experiments, we were unable to distinguish CD99 isoform protein expression in primary AML blasts.
The two isoforms have distinct, and maybe opposite, functions in malignancies. Studies of the ectopic expression of CD99-L isoform supported an oncosuppressor function in osteosarcoma.42 Overexpression of CD99-S isoform, however, resulted in decreased cell aggregation and increased cell migration in osteosarcoma and prostate cancer cells.42 In EWS, CD99-S inhibited cell differentiation and contributed to the maintenance of stemness.43 Downregulation of CD99-L transformed B lymphocytes to Hodgkin and Reed-Sternberg phenotype.44 Our data suggest that CD99-L cells are more responsive to serum stimuli with increased DNA synthesis and cell growth; however, over time, these cells accumulate higher ROS and DNA damage, resulting in apoptosis. We speculate that CD99 homotypic interaction is likely driving the later reduction of cell viability in CD99-L cells. While this phenomenon resembles oncogene-induced senescence,45 only a slight increase in P16 was observed in CD99-L cells (data not shown).
CD99-L consistently delayed disease progression in AML murine models. Whether CD99 interaction with the murine microenvironment inhibits cell homing to the BM is unclear. Yet we observed no change in CXCR4 and CD49d (VLA-4a) on the surface of CD99-L cells (data not shown). Mechanistically, CD99-L induced P-SRC and PERK is likely driving the initial increase in cell growth. A CD99-derived agonist peptide that specifically interacts with conserved motifs in the CD99 extracellular domain was previously found to inhibit fibronectin-mediated β1 integrin activation through the SHP2/ERK/PTPN12/FAK signaling pathway;37 they also found that ERK is essential for CD99 homotypic aggregation.46 CD99 homotypic interactions driving a negative feedback loop or via its subsequent binding with caveolin may explain the later inhibition of ERK and SRC.42 CD99 was found to drive terminal differentiation of osteosarcoma cells via activation of membrane-bound/cytoplasmic ERK rather than affecting its nuclear localization.47 Yet, in EWS, CD99 knockdown led to prolonged nuclear phosphorylation of ERK1/2 and neural differentiation.48 In addition, CD99mAb was shown to act as an agonist, and reported similar phenotypes to those observed in our study.46,49 Both CD99-L ectopic expression and CD99mAb induced similar cellular effects, and, likewise, modulated P-SRC. However, further biochemical studies are required to establish whether CD99mAb acts as an agonist for CD99 long isoform.
In summary, our results present unique insights into the clinical, functional, and mechanistic roles of CD99 isoforms in AML. The mechanisms by which CD99 is up-regulated and those that govern its function in leukemia initiation remain to be investigated.
Acknowledgments
We would like to thank the Bioinformatics core at the Norris Medical Library, University of Southern California and TCGA. This work was supported by grant UL1TR001855 from the National Center for Advancing Translational Science (NCATS) of the US National Institutes of Health, the Southern California Clinical, Translational Science Institute KL-2 funding (KL2TR001854) and the Ming Hsieh Institute Funding. AMNB is funded by the BMBF grant 01ZZ1804B (DIFUTURE). KM is supported by the German Research Foundation (DFG SFB 1243). TH is supported by the Wilhelm Sander Foundation (2013.086.2) and by the Physician Scientists Grant (G-509200-004) from the Helmholtz Zentrum München.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/4/999
References
- 1.Lowenberg B. Acute myeloid leukemia: the challenge of capturing disease variety. Hematology Am Soc Hematol Educ Program. 2008:1–11. [DOI] [PubMed] [Google Scholar]
- 2.Gelin C, Aubrit F, Phalipon A, et al. The E2 antigen, a 32 kd glycoprotein involved in T-cell adhesion processes, is the MIC2 gene product. Embo J. 1989;8(11):3253–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dworzak MN, Fritsch G, Buchinger P, et al. Flow cytometric assessment of human MIC2 expression in bone marrow, thymus, and peripheral blood. Blood. 1994;83(2):415–425. [PubMed] [Google Scholar]
- 4.Schenkel AR, Mamdouh Z, Chen X, et al. CD99 plays a major role in the migration of monocytes through endothelial junctions. Nat Immunol. 2002;3(2):143–150. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan DP, Watson RL, Muller WA. 4D intravital microscopy uncovers critical strain differences for the roles of PECAM and CD99 in leukocyte diapedesis. Am J Physiol Heart Circ Physiol. 2016;311(3):H621–H632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambros IM, Ambros PF, Strehl S, et al. MIC2 is a specific marker for Ewing’s sarcoma and peripheral primitive neuroectodermal tumors. Evidence for a common histogenesis of Ewing’s sarcoma and peripheral primitive neuroectodermal tumors from MIC2 expression and specific chromosome aberration. Cancer. 1991;67(7):1886–1893. [DOI] [PubMed] [Google Scholar]
- 7.Seol HJ, Chang JH, Yamamoto J, et al. Overexpression of CD99 Increases the Migration and Invasiveness of Human Malignant Glioma Cells. Genes Cancer. 2012;3(9-10):535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelmus M, Guillou L, Hostein I, et al. Monophasic fibrous and poorly differentiated synovial sarcoma: immunohistochemical reassessment of 60 t(X;18)(SYT-SSX)-positive cases. Am J Surg Pathol. 2002;26(11): 1434–1440. [DOI] [PubMed] [Google Scholar]
- 9.Hartel PH, Jackson J, Ducatman BS, Zhang P. CD99 immunoreactivity in atypical fibrox-anthoma and pleomorphic malignant fibrous histiocytoma: a useful diagnostic marker. J Cutan Pathol. 2006;33 Suppl 2:24–28. [DOI] [PubMed] [Google Scholar]
- 10.Dworzak MN, Fritsch G, Fleischer C, et al. CD99 (MIC2) expression in paediatric B-lineage leukaemia/lymphoma reflects maturation-associated patterns of normal B-lymphopoiesis. Br J Haematol. 1999;105(3):690–695. [DOI] [PubMed] [Google Scholar]
- 11.Sung CO, Ko YH, Park S, Kim K, Kim W. Immunoreactivity of CD99 in non-Hodgkin’s lymphoma: unexpected frequent expression in ALK-positive anaplastic large cell lymphoma. J Korean Med Sci. 2005;20(6):952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buxton D, Bacchi CE, Gualco G, et al. Frequent expression of CD99 in anaplastic large cell lymphoma: a clinicopathologic and immunohistochemical study of 160 cases. Am J Clin Pathol. 2009;131(4):574–579. [DOI] [PubMed] [Google Scholar]
- 13.Jung KC, Park WS, Bae YM, et al. Immunoreactivity of CD99 in stomach cancer. J Korean Med Sci, 2002:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo SH, Han J, Kim TJ, Chung DH. Expression of CD99 in Pleomorphic Carcinomas of the Lung. J Korean Med Sci. 2005;20(1):50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ventura S, Aryee DNT, Felicetti F, et al. CD99 regulates neural differentiation of Ewing sarcoma cells through miR-34a-Notch-mediated control of NF-|[kappa]|B signaling. Oncogene. 2015;35(30):3944–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang PJ, Barcos M, Stewart CC, et al. Immunoreactivity of MIC2 (CD99) in acute myelogenous leukemia and related diseases. Mod Pathol. 2000(4):452–458. [DOI] [PubMed] [Google Scholar]
- 17.Chung SS, Eng WS, Hu W, et al. CD99 is a therapeutic target on disease stem cells in myeloid malignancies. Sci Transl Med. 2017;9(374). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasello M, Manara MC, Scotlandi K. CD99 at the crossroads of physiology and pathology. J Cell Commun Signal. 2018;12(1):55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):p11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersson A, Ritz C, Lindgren D, et al. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21(6):1198–1203. [DOI] [PubMed] [Google Scholar]
- 22.Haferlach T, Kohlmann A, Wieczorek L, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28(15):2529–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350(16):1617–1628. [DOI] [PubMed] [Google Scholar]
- 24.Klein HU, Ruckert C, Kohlmann A, et al. Quantitative comparison of microarray experiments with published leukemia related gene expression signatures. BMC Bioinformatics. 2009;10:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005;33(18):5914–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350(16):1605–1616. [DOI] [PubMed] [Google Scholar]
- 27.Metzeler KH, Hummel M, Bloomfield CD, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112(10):4193–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balgobind BV, Van den Heuvel-Eibrink MM, De Menezes RX, et al. Evaluation of gene expression signatures predictive of cytogenetic and molecular subtypes of pediatric acute myeloid leukemia. Haematologica. 2011;96(2):221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han L, Qiu P, Zeng Z, et al. Single-cell mass cytometry reveals intracellular survival/pro-liferative signaling in FLT3-ITD-mutated AML stem/progenitor cells. Cytometry A. 2015;87(4):346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonardi F, Fusetti F, Deelen P, van Gosliga D, Vellenga E, Schuringa JJ. A proteomics and transcriptomics approach to identify leukemic stem cell (LSC) markers. Mol Cell Proteomics. 2013;12(3):626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kikushige Y, Shima T, Takayanagi S, et al. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell. 2010;7(6):708–717. [DOI] [PubMed] [Google Scholar]
- 32.Bernard G, Zoccola D, Deckert M, et al. The E2 molecule (CD99) specifically triggers homotypic aggregation of CD4+ CD8+ thymocytes. J Immunol. 1995;154(1):26–32. [PubMed] [Google Scholar]
- 33.Cerisano V, Aalto Y, Perdichizzi S, et al. Molecular mechanisms of CD99-induced caspase-independent cell death and cell-cell adhesion in Ewing’s sarcoma cells: actin and zyxin as key intracellular mediators. Oncogene. 2004;23(33):5664–5674. [DOI] [PubMed] [Google Scholar]
- 34.Byun HJ, Hong IK, Kim E, et al. A splice variant of CD99 increases motility and MMP-9 expression of human breast cancer cells through the AKT-, ERK-, and JNK-dependent AP-1 activation signaling pathways. J Biol Chem. 2006;281(46):34833–34847. [DOI] [PubMed] [Google Scholar]
- 35.Manara MC, Terracciano M, Mancarella C, et al. CD99 triggering induces methuosis of Ewing sarcoma cells through IGF-1R/RAS/Rac1 signaling. Oncotarget. 2016;7(48):79925–79942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerzoni C, Fiori V, Terracciano M, et al. CD99 triggering in Ewing sarcoma delivers a lethal signal through p53 pathway reactivation and cooperates with doxorubicin. Clin Cancer Res. 2015;21(1):146–156. [DOI] [PubMed] [Google Scholar]
- 37.Lee KJ, Kim Y, Yoo YH, et al. CD99-Derived Agonist Ligands Inhibit Fibronectin-Induced Activation of beta1 Integrin through the Protein Kinase A/SHP2/Extracellular Signal-Regulated Kinase/PTPN12/Focal Adhesion Kinase Signaling Pathway. Mol Cell Biol. 2017;37(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warsito D, Sjostrom S, Andersson S, Larsson O, Sehat B. Nuclear IGF1R is a transcriptional co-activator of LEF1/TCF. EMBO Rep. 2012;13(3):244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kavalar R, Pohar Marinsek Z, Jereb B, Cagran B, Golouh R. Prognostic value of immunohistochemistry in the Ewing’s sarcoma family of tumors. Med Sci Monit. 2009;15(8):CR442–452. [PubMed] [Google Scholar]
- 40.Angelini DF, Ottone T, Guerrera G, et al. A Leukemia-Associated CD34/CD123/CD25/ CD99+ Immunophenotype Identifies FLT3-Mutated Clones in Acute Myeloid Leukemia. Clin Cancer Res. 2015;21(17): 3977–3985. [DOI] [PubMed] [Google Scholar]
- 41.Kadia TM, Jain P, Ravandi F, et al. TP53 mutations in newly diagnosed Acute Myeloid Leukemia –Clinico-molecular characteristics, response to therapy, and outcomes. Cancer. 2016;122(22):3484–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scotlandi K, Zuntini M, Manara MC, et al. CD99 isoforms dictate opposite functions in tumour malignancy and metastases by activating or repressing c-Src kinase activity. Oncogene. 2007;26(46):6604–6618. [DOI] [PubMed] [Google Scholar]
- 43.Lee EJ, Lee HG, Park SH, Choi EY. CD99 type II is a determining factor for the differentiation of primitive neuroectodermal cells. Exp Mol Med. 2003;35(5):438–447. [DOI] [PubMed] [Google Scholar]
- 44.Kim SH, Shin YK, Lee IS, et al. Viral latent membrane protein 1 (LMP-1)-induced CD99 down-regulation in B cells leads to the generation of cells with Hodgkin’s and Reed-Sternberg phenotype. Blood. 2000;95(1): 294–300. [PubMed] [Google Scholar]
- 45.Michaloglou C, Vredeveld LC, Soengas MS, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. [DOI] [PubMed] [Google Scholar]
- 46.Hahn MJ, Yoon SS, Sohn HW, et al. Differential activation of MAP kinase family members triggered by CD99 engagement. FEBS Lett. 2000;470(3):350–354 [DOI] [PubMed] [Google Scholar]
- 47.Sciandra M, Marino MT, Manara MC, et al. CD99 drives terminal differentiation of osteosarcoma cells by acting as a spatial regulator of ERK 1/2. J Bone Miner Res. 2014;29(5):1295–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rocchi A, Manara MC, Sciandra M, et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J Clin Invest. 2010;120(3):668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HJ, Kim E, Jee B, et al. Functional involvement of src and focal adhesion kinase in a CD99 splice variant-induced motility of human breast cancer cells. Exp Mol Med. 2002;34(3):177–183. [DOI] [PubMed] [Google Scholar]