Abstract
In chronic lymphocytic leukemia (CLL), the hypoxia-inducible factor 1 (HIF-1) regulates the response of tumor cells to hypoxia and their protective interactions with the leukemic microenvironment. In this study, we demonstrate that CLL cells from TP53-disrupted (TP53dis) patients have constitutively higher expression levels of the α-subunit of HIF-1 (HIF-1α) and increased HIF-1 transcriptional activity compared to the wild-type counterpart. In the TP53dis subset, HIF-1α upregulation is due to reduced expression of the HIF-1α ubiquitin ligase von Hippel-Lindau protein (pVHL). Hypoxia and stromal cells further enhance HIF-1α accumulation, independently of TP53 status. Hypoxia acts through the downmodulation of pVHL and the activation of the PI3K/AKT and RAS/ERK1-2 pathways, whereas stromal cells induce an increased activity of the RAS/ERK1-2, RHOA/RHOA kinase and PI3K/AKT pathways, without affecting pVHL expression. Interestingly, we observed that higher levels of HIF-1A mRNA correlate with a lower susceptibility of leukemic cells to spontaneous apoptosis, and associate with the fludarabine resistance that mainly characterizes TP53dis tumor cells. The HIF-1α inhibitor BAY87-2243 exerts cytotoxic effects toward leukemic cells, regardless of the TP53 status, and has anti-tumor activity in Em-TCL1 mice. BAY87-2243 also overcomes the constitutive fludarabine resistance of TP53dis leukemic cells and elicits a strongly synergistic cytotoxic effect in combination with ibrutinib, thus providing preclinical evidence to stimulate further investigation into use as a potential new drug in CLL.
Introduction
Chronic lymphocytic leukemia (CLL) patients with high-risk genomic features such as disruption of the TP53 gene [i.e. del(17p) and TP53 mutations] respond poorly to chemoimmunotherapy and frequently relapse.1–9 Significant advances have been made in the treatment of CLL following the introduction of Bruton tyrosine kinase (BTK) inhibitors.10 Ibrutinib, which is currently approved for the front-line treatment of CLL, induces long-lasting responses in the majority of patients, improving outcome with relatively limited toxicities.10 However, patients with disruption of the TP53 gene (TP53dis) treated with ibrutinib are still characterized by a poorer outcome.11
Hypoxia inducible factor 1 (HIF-1) is an essential regulator of cell adaptation to hypoxia and is often up-regulated in tumors due to intratumoral hypoxia or activation of oncogenic pathways.12,13 In tumors, HIF-1 fosters different tumor-promoting mechanisms, including metabolic adaptation, neoangiogenesis, cell survival and invasion.14
HIF-1 is a heterodimer, which consists of a constitutively expressed HIF-1β subunit and an inducible HIF-1α subunit. Besides its traditional regulation via proteasomal degradation, other signaling pathways, such as PI3K/AKT and RAS/ERK1-2, contribute to HIF-1α accumulation, via stability regulation or synthesis induction.12,15
HIF-1α is constitutively expressed in CLL cells compared to normal B cells due to microRNA-mediated down-regulation of the von Hippel-Lindau protein (pVHL),16 a ubiquitin ligase responsible for HIF-1α degradation.12 In addition, in CLL cells, HIF-1α is up-regulated by interactions with stromal cells (SC) and by exposure to hypoxic microenvironments, thus promoting the survival and propagation of leukemic cells, and their metabolic adaptation to the protective conditions of the tumor niche.17–20 We have already reported that HIF-1α is involved in drug resistance mechanisms in patients with unmutated (UM) immunoglobulin heavy chain variable region genes (IGHV).20 The TP53 gene encodes one of the best-studied tumor suppressor proteins, which is often mutated in cancer, thus promoting cell survival, proliferation and drug resistance.21 p53 may also play a pivotal role in the regulation of HIF-1α, since in conditions of prolonged hypoxia/anoxia, the protein accumulates and promotes HIF-1α destruction.22 In solid tumors, loss of TP53 function associates with constitutive elevated levels of HIF-1α.12,22,23
In this study, we found that HIF-1α is over-expressed in CLL cells from patients carrying TP53 aberrations, also elucidating the molecular mechanisms implicated in the constitutive (TP53-related) and inducible (hypoxia- and SC-induced) HIF-1α upregulation. In addition, we observed that the HIF-1α inhibitor BAY87-2243 exerts potent anti-tumor functions, overcoming the constitutive fludarabine resistance of TP53-disrupted CLL cells, and eliciting a strong synergistic cytotoxic effect in combination with ibrutinib.
Methods
Patients’ samples
A total of 102 patients with CLL, diagnosed according to the International Workshop on CLL-National Cancer Institute guidelines,24 were included in the study [40 TP53dis and 62 TP53-wild type (TP53wt) cases] (Online Supplementary Table S1). Healthy donors’ (HD, n=2) samples were provided by the local blood bank. Patients were untreated or off-therapy for at least 12 months before sampling of peripheral blood (PB) for the experiments. Samples were collected after patients’ informed consent in accordance with the Declaration of Helsinki and after approval by the local Institutional Review Board. PB mononuclear cells (PBMC) were isolated and characterized as detailed in the Online Supplementary Appendix.
Cell lines
The Burkitt’s lymphoma cell line, Séraphine, and the mantle cell lymphoma cell line, Granta-519, were kindly provided by T. Zenz. The TP53wt and the CRISPR/Cas9-mediated TP53 knockout version (TP53ko) of Granta-519 and Séraphine cell lines were used in the study. The M2-10B4 murine SC line (ATCC #CRL-1972) was also used. Cell lines were maintained as reported in the Online Supplementary Appendix.
Animals
C57BL/6 Em-TCL1 mice were maintained in specific pathogen-free animal facilities and treated in accordance with European Union and Institutional Animal Care and Use Committee (number 716) guidelines. Splenic cells (5×106) were injected intraperitoneally into syngeneic C57BL/6 mice, and experiments were performed with groups of 4-6 mice. Leukemic mice were treated when tumor cells reached 10% in PB. BAY87-2243 was administered at 4 mg/kg in ethanol/solutol/water solution once daily by oral gavage. Mice were sacrificed at the end of treatment.
Cell culture
In selected experiments, CLL cells were cultured in the presence or absence of M2-10B4 SC, and exposed to PD98059 (Sigma Aldrich, Milan), Y27632 (Sigma Aldrich) or LY249002 (Sellekchem, Houston, TX, USA) for 48 hours (h). CLL cells were exposed for 48 h to BAY87-2243 (Sellekchem); 2-Fluoroadenine-9-β-D-arabinofuranoside (F-ara-A, Sigma Aldrich); ibrutinib (Sellekchem) used alone or in combination, at the indicated concentrations. Culture conditions were normoxia or mild hypoxia (1% O2), 5% CO2 at 37°C.
Western blot
Full details can be found in the Online Supplementary Appendix together with the list of antibodies used for western blot (WB) analyses.
Quantitative real-time polymerase chain reaction
Full details of quantitative real-time polymerase chain reaction (qRT-PCR) experiments can be found in the Online Supplementary Appendix together with the list of primer sequences.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA, http://www.broad.mit.edu/gsea/index.jsp) was performed as previously described.25,26 Gene sets were assessed as significantly enriched in one of the phenotypes if the nominal P-value and the false discovery rate (FDR)-q value were <0.05.
RHOA and RAS, ERK1-2, AKT and RHOA kinase activity
The isoprenylated membrane-associated RAS or RHOA proteins and the non-isoprenylated cytosolic forms were detected as previously described.20 Details of measurement of ERK1-2, AKT and RHOA kinase activity are reported in the Online Supplementary Appendix.
Cell viability assay
Cell viability was evaluated by flow cytometry using Annexin-V/Propidium Iodide (Ann-V/PI) staining with the MEBCYTO-Apoptosis Kit (MBL Medical and Biological Laboratories, Naka-ku Nagoya).
Statistical analysis
GraphPad Prism (version 6.01, San Diego, CA, USA) was used to perform paired and unpaired t-test, and to calculate Pearson correlation coefficient. Results are expressed as mean±standard error of mean (SEM), unless otherwise specified. Statistical significance was defined as a P<0.05. Combination analysis was performed using Compusyn software; combinations were considered synergistic when the combination index (CI) was <1.
Results
HIF-1α is over-expressed in chronic lymphocytic leukemia cells from TP53dis patients and in TP53 knockout lymphoma cell lines
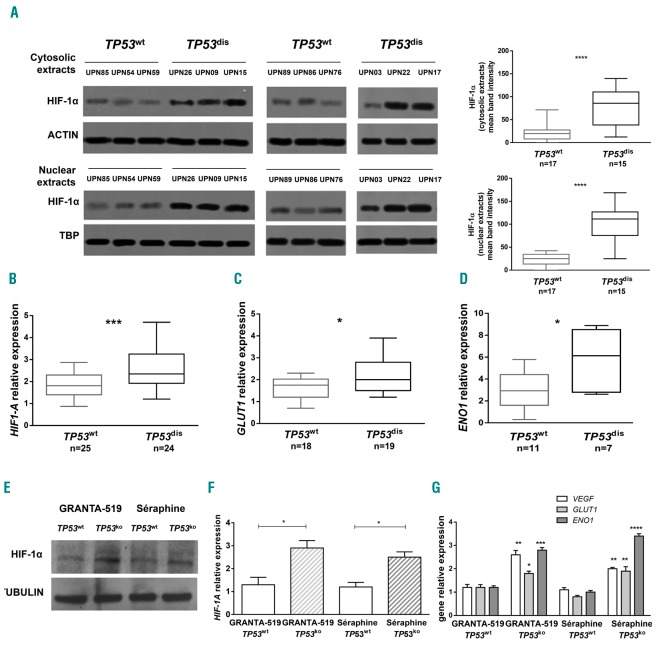
Expression levels of HIF-1α protein were comparatively evaluated in HD CD19+ cells, and in CLL cells isolated from TP53dis and TP53wt samples. As expected, HD CD19+ B cells did not express HIF-1α at the baseline normoxic conditions (data not shown). In contrast, leukemic cells from CLL patients exhibited detectable cytosolic and nuclear HIF-1α protein (Figure 1A). Interestingly, CLL cells from patients carrying TP53 abnormalities (TP53dis CLL cells) had significantly higher amounts of the cytosolic and nuclear fractions of HIF-1α subunit, as well as higher HIF-1A mRNA levels compared to CLL cells isolated from TP53wt cases (TP53wt CLL cells) (Figure 1A and B). We evaluated an enlarged cohort of cases and observed that the association between the expression of HIF-1α and the TP53 status was not influenced by the IGHV mutational status (Online Supplementary Figure S1). The transcriptional activity of HIF-1α was evaluated through the expression of selected target genes.13,15,27 We found a higher expression of GLUT1 and ENO1 in TP53dis CLL cells, compared to TP53wt samples (Figure 1C and D). To corroborate the finding of an association between HIF-1α expression and TP53 status we exploited cell line models. Interestingly, the expression of HIF-1α protein and mRNA was higher in TP53ko Granta-519 and Séraphine lymphoma cell lines, compared to the p53wt (Figure 1E and F). In line with this finding, expression of VEGF, GLUT1 and ENO1 was also significantly higher in TP53ko than in TP53wt Granta-519 and Séraphine cell lines (Figure 1G).
Figure 1.
HIF-1α is over-expressed and more active in primary cells isolated from patients with chronic lymphocytic leukemia (CLL) and in lymphoma cell lines carrying a TP53 disruption. The expression of HIF-1α and HIF-1α target genes was measured in TP53dis and TP53wt CLL cells and in lymphoma TP53wt and TP53ko cell lines. (A) Western blot (WB) analysis for HIF-1α protein expression in freshly isolated TP53dis and TP53wt CLL cells. A representative blot is shown with relative Unique Patient Numbers (UPN) and cumulative band intensity data obtained from the analysis of 17 TP53wt and 15 TP53dis CLL patients. Box plots represent median value and 25-75% percentiles, whiskers represent minimum and maximum values of band intensity for each group. (B-D) Real-time-polymerase chain reaction (RT-PCR) analysis of HIF-1A, and its target genes GLUT1 and ENO1 expression levels in TP53dis and TP53wt CLL cells. (E) WB analysis for HIF-1α protein expression in the TP53wt and TP53ko Granta-519 and Séraphine cell lines. Representative blot of three independent experiments is shown. (F) RT-PCR analysis of HIF-1A in the TP53wt and TP53ko Granta-519 and Séraphine cell lines. (G) RT-PCR analysis of VEGF, GLUT1 and ENO1 in the TP53wt and TP53ko Granta-519 and Séraphine cell lines showed a significantly higher expression level of all analyzed genes in TP53ko samples.(B, C and D) Box and whiskers plots represent median values and 25-75% percentiles; whiskers represent minimum and maximum values for each group. (F and G) Bar graphs represent mean results obtained from three experiments together with standard error of mean. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
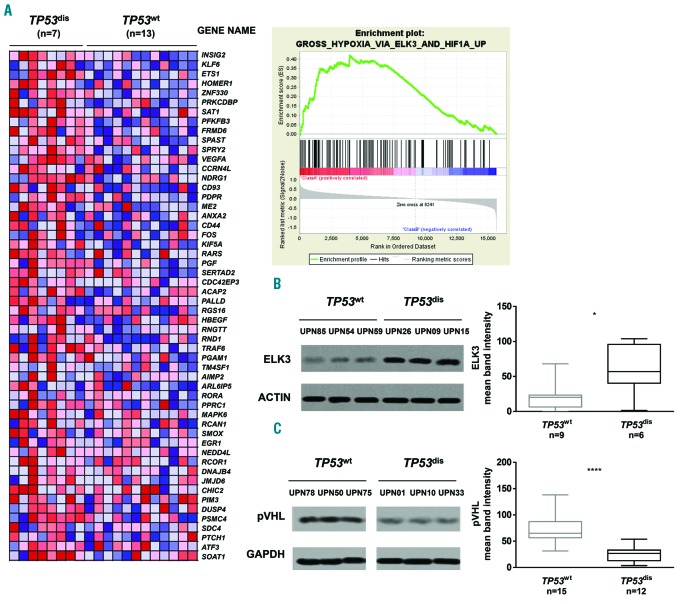
To further investigate the link between TP53 and HIF-1α, we performed GSEA on previously published microarray data from tumor cells isolated from seven TP53dis and 13 TP53wt cases (geocode GSE18971).28 Data of GSEA cases revealed that the TP53 abnormalities were associated with an upregulation of a number of genes belonging to the “GROSS_HYPOXIA_VIA_ELK3_AND_HIF1A_UP” gene set (Figure 2A). The protein ELK3 participates in the transcriptional response to hypoxia and controls the expression of several regulators of HIF-1α stability.29 Consistently, the baseline expression of ELK3 was higher in TP53dis compared to TP53wt CLL cells (Figure 2B).
Figure 2.
TP53dis chronic lymphocytic leukemia (CLL) cells show an upregulation of several genes involved in response to hypoxia and express reduced levels of von Hippel-Lindau protein (pVHL). (A) Gene set enrichment analysis (GSEA) on CLL cells from TP53dis and TP53wt patients. The plot of the Enrichment Score (ES) versus the gene list index and a portion of the corresponding heatmap highlighting the relative expression of gene members belonging to the “GROSS_HYPOXIA_VIA_ELK3_AND_HIF1A_UP” gene set are depicted. 54 out of 126 genes from the gene set were significantly up-regulated in the TP53dis cohort of patients compared to the TP53wt cohort. (B) Western blot (WB) for ELK3 baseline expression in TP53dis and TP53wt CLL cells. A representative blot is shown with relative Unique Patient Numbers (UPN) and cumulative band intensity data obtained from the analysis of 9 TP53wt and 6 TP53dis CLL patients. (C) WB for pVHL baseline expression in TP53dis and TP53wt CLL cells. A representative blot is shown with relative UPN and cumulative band intensity data obtained from the analysis of 15 TP53wt and 12 TP53dis CLL patients. Box plots represent median value and 25-75% percentiles; whiskers represent minimum and maximum values of band intensity for each group. *P<0.05; ****P<0.0001.
Given its role in HIF-1α regulation,12,30 we also compared pVHL expression in TP53dis and TP53wt samples. Notably, CLL cells from TP53dis patients had reduced amounts of pVHL compared to TP53wt patients, most likely being responsible for better stabilization of the HIF-1α protein and a repression of its proteasomal degradation in TP53dis cells (Figure 2C). As for HIF-1α expression, there were no differences between pVHL levels according to IGHV mutational status (Online Supplementary Figure S2). These data suggest that TP53 abnormalities lead to a reduced expression of pVHL and subsequently to an accumulation of HIF-1α protein.
Hypoxia and stromal cells further increase HIF-1α expression in chronic lymphocytic leukemia cells from TP53dis and TP53wt patients
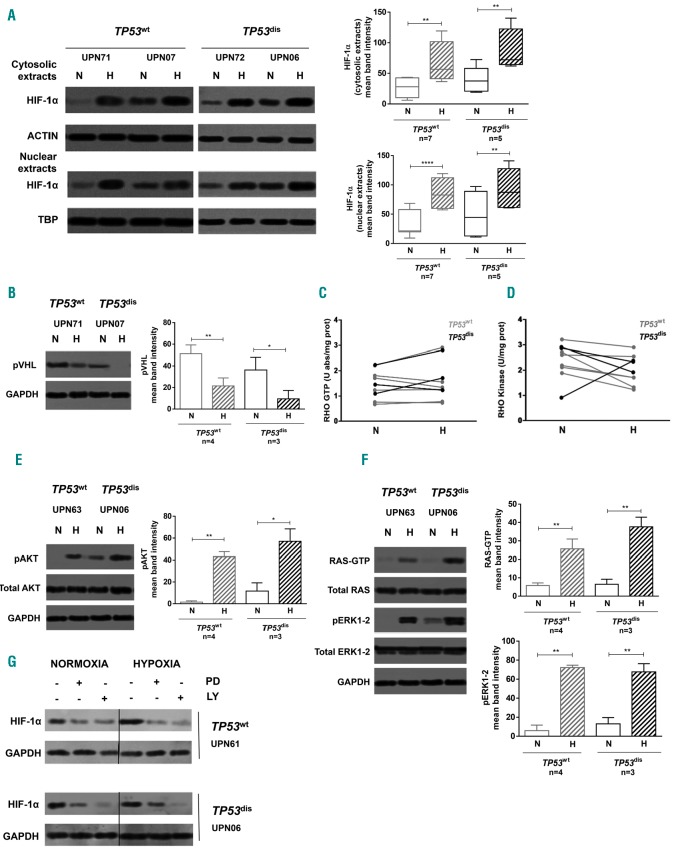
We next investigated whether microenvironmental signals, such as oxygen deprivation12 and the interactions with SC,20 had differential effects on HIF-1α according to the TP53 status of the leukemic cells, also in an attempt to better define the underlying molecular mechanisms. To this end, CLL cells were cultured for 48 h in condition of hypoxia or in the presence of SC. Of note, ex vivo culture partially abrogated the TP53-related differential expression of HIF-1α observed at the baseline in freshly isolated CLL cells. In hypoxia, we observed a marked upregulation of the cytosolic and nuclear fractions of HIF-1α protein, which was independent of TP53 status (Figure 3A), and was associated to a reduced expression of pVHL (Figure 3B), and to an activation of the PI3K/AKT and RAS/ERK1-2 pathways (Figure 3C-F). Consistently, we observed that blocking concentration of pharmacologic agents inhibiting ERK1-2 (PD98059) and PI3K (LY294002) effectively counteracted the hypoxia-induced HIF-1α upregulation, independently of TP53 status (Figure 3G).
Figure 3.
Hypoxia further increases HIF-1α expression in TP53dis and TP53wt chronic lymphocytic leukemia (CLL) cells via the PI3K/AKT and RAS/ERK1-2 signaling pathways. Primary CLL cells were cultured for 48 hours under normoxic and hypoxic conditions. (A and B) Western blot (WB) analyses detected a higher amount of cytosolic and nuclear HIF-1α and lower amount of von Hippel-Lindau protein (pVHL) in TP53dis and TP53wt CLL cells cultured in hypoxia (H) compared to normoxia (N). (C and D) Immuno-enzymatic measurement showed that RHOA-GTP and RHOA kinase activities were unaffected by hypoxia. (E and F) WB analyses for AKT, RAS and ERK1-2. TP53dis and TP53wt CLL cells had higher expression of the active form of AKT [p(Ser 473)AKT], RAS (RAS-GTP) and ERK1-2 [p(Thr202/Tyr204, Thr185/Tyr187]ERK1-2) in hypoxia compared to normoxia. (G) WB analyses for HIF-1α. The targeting of ERK1-2 with 10 μM PD98059 (PD) and of PI3K with 10 μM LY294002 (LY) reduced HIF-1α expression in CLL cells, both in hypoxia and normoxia, and independently from the TP53 status. (A) Results from two representative cases of seven TP53wt patients and two representative cases of five TP53dis patients. Representative blots are shown with relative Unique Patient Numbers (UPN) and cumulative band intensity data. Box plots represent median values and 25-75% percentiles; whiskers represent minimum and maximum values of band intensity for each group. (B, E and F) Results from one representative experiment in four TP53wt patients and one representative experiment in three TP53dis patients. Bar graphs represent mean values together with standard error of mean. (C and D) Multiple line graphs represent individual data values for the same sample in each condition. (G) Results from one representative experiment in three TP53wt patients and one representative experiment in three TP53dis patients. Vertical lines have been inserted to indicate repositioned gel lanes. *P<0.05; **P<0.01; ****P<0.0001.
In line with previous data,20 we observed a marked upregulation of the cytosolic and nuclear amounts of the HIF-1α when CLL cells were co-cultured with SC (Figure 4A). SC-induced HIF-1α elevation was not associated to a reduced pVHL expression in leukemic cells (Figure 4B), whereas we observed an increased activation of RHOA/RHOA kinase (Figure 4C and D), PI3K/AKT (Figure 4E), and RAS/ERK1-2 (Figure 4F) signaling pathways. As confirmation, we found that targeted inhibition of ERK1-2, PI3K and RHOA kinase by blocking concentrations of pharmacologic agents (i.e. PD98059, LY294002 and Y27632, respectively) effectively counteracted SC-induced HIF-1α upregulation (Figure 4G).
Figure 4.
Stromal cells (SC) increase HIF-1α expression in TP53dis and TP53wt chronic lymphocytic leukemia (CLL) cells via PI3K/AKT, RAS/ERK1-2 and RHOA/RHOA kinase signaling pathways. Primary CLL cells were cultured for 48 hours in the presence and in the absence of M2-10B4 SC. (A and B) Western blot (WB) analyses for HIF-1α and von Hippel-Lindau protein (pVHL). SC up-regulated the cytosolic and nuclear expression of HIF-1α but did not affect pVHL expression in TP53dis and TP53wt CLL cells. (C and D) Immuno-enzymatic measurement showed that the co-culture with SC increased RHOA-GTP and RHOA kinase activities in TP53dis and TP53wt CLL cells. (E and F) WB analyses for AKT, RAS and ERK1-2. Higher amount of the active form of AKT [p(Ser 473)AKT], RAS (RAS-GTP) and ERK1-2 [p(Thr202/Tyr204, Thr185/Tyr187)ERK1-2] were detectable in both TP53wt and TP53dis CLL cells cultured with SC. (G) WB analyses for HIF-1α. The targeting of ERK1-2 with 10 μM PD98059 (PD), of RHOA kinase with 10 μM Y27632 (Y276), and of PI3K with 10 μM LY294002 (LY) reduced HIF-1α expression in CLL cells, both in the presence and in the absence of SC, regardless of the TP53 status. (A) Results from two representative cases of ten TP53wt patients and two representative cases of four TP53dis patients. Representative blots are shown, together with Unique Patient Numbers (UPN) and cumulative band intensity data. Box plots represent median values and 25-75% percentiles; whiskers represent minimum and maximum values of band intensity for each group. (B and G) Results are from one representative experiment in three TP53wt patients and one representative experiment in three TP53dis patients. (C and D) Multiple line graphs represent individual data values for the same sample in each condition. (E and F) Results are from one representative experiment in five TP53wt patients and one representative experiment in four TP53dis patients. Bar graphs represent mean values together with standard error of mean. Vertical lines have been inserted to indicate repositioned gel lanes. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
The role of these pathways in modulating HIF-1α over-expression was corroborated by titration experiments showing that exposure of TP53dis and TP53wt CLL cells to increasing concentrations of PD98059, LY294002 and Y27632 induced a progressive reduction of the activity of the targeted kinases, which was associated to a dose-dependent decrease in HIF-1α levels (Online Supplementary Figure S3).
The selective HIF-1α inhibitor BAY87-2243 has anti-tumor activities in chronic lymphocytic leukemia
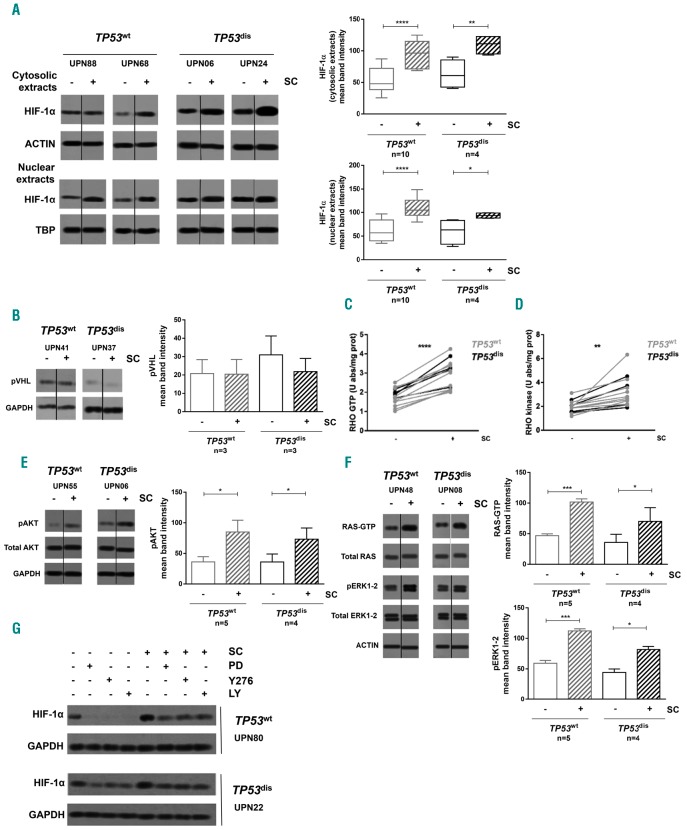
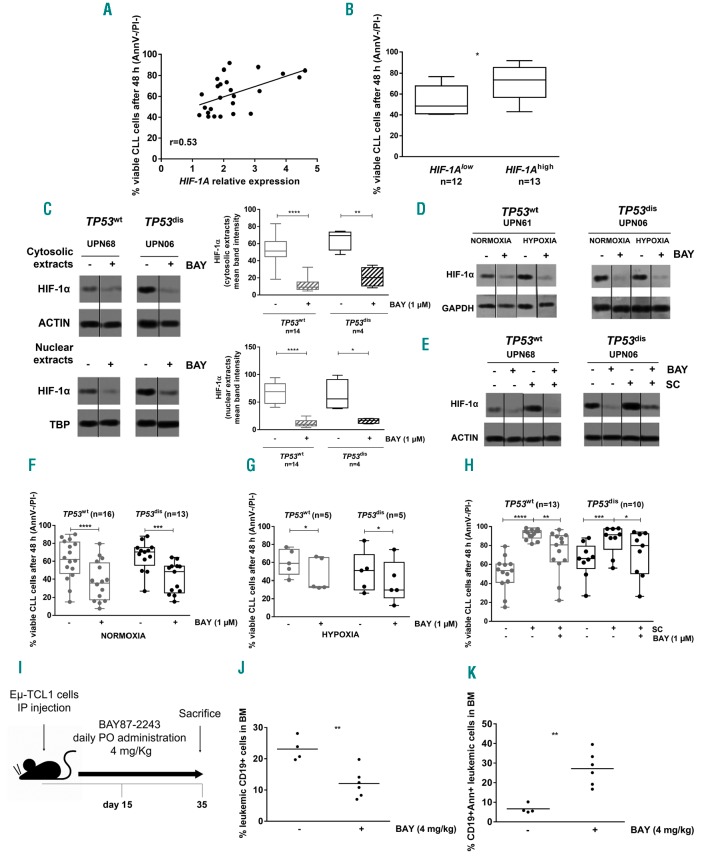
In line with the role of HIF-1α as a promoting factor for cell survival,12 we found a positive correlation between the baseline levels of HIF-1A mRNA and the 48-h viability of CLL cells during in vitro culture (Figure 5A). Consistently, the viability of leukemic cells isolated from samples characterized by baseline HIF-1A mRNA levels above the median value of the entire cohort (HIF-1Ahigh) was significantly higher than the viability of CLL cells displaying lower HIF-1A values (HIF-1Alow) (Figure 5B and Online Supplementary Figure S4). Based on these observations, and on previous data reporting HIF-1α as a potential therapeutic target in CLL,17 we evaluated the anti-tumor effect of BAY87-2243, a selective inhibitor of HIF-1α. First, we observed that BAY87-2243 effectively inhibited HIF-1α protein expression at the cytosolic and nuclear level, both in TP53dis and TP53wt CLL cells (Figure 5C), also counteracting the HIF-1α upregulation exerted by hypoxia and SC (Figure 5D and E). After 48 h, BAY87-2243 determined a strong cytotoxic effect toward leukemic cells isolated from both patient subsets (Figure 5F and Online Supplementary Figure S5). Of note, the downregulation of HIF-1α was also evident at 24-h exposure, when cell viability was still well preserved, thus confirming that it was determined by a targeted inhibitory effect rather than by a consequence of cell death (data not shown). BAY87-2243 exerted a cytotoxic effect also when TP53dis and TP53wt CLL cells were cultured for 48 h in the presence of extrinsic signals inducing a further upregulation of baseline levels of HIF-1α, such as hypoxia (Figure 5G and Online Supplementary Figure S6) and co-culture with SC (Figure 5H and Online Supplementary Figure S7).
Figure 5.
HIF-1α promotes chronic lymphocytic leukemia (CLL) cell survival and its inhibition exerts a direct cytotoxicity, also in the presence of HIF-1α inducing microenvironmental stimuli. (A) Correlation of HIF-1A gene expression and 48-hour (h) cell viability of CLL cells as determined by Annexin V/propidium iodide assay. (B) The median value of HIF-1A mRNA expression of a cohort of 25 CLL samples was selected as the cut-off to identify HIF-1Ahigh and HIF-1Alow CLL cells. HIF-1Ahigh showed significantly higher 48-h cell viability compared to HIF-1Alow CLL cells. (C) Western blot (WB) analyses for HIF-1α. The exposure to 1 μM BAY87-2243 (BAY) for 48 h reduced the cytosolic and nuclear amount of HIF-1α in CLL cells isolated from both TP53wt and TP53dis patient subsets. A representative blot is shown, together with Unique Patient Numbers (UPN) and cumulative band intensity data obtained from the analysis of 14 TP53wt and 4 TP53dis CLL patients. Box plots represent median value and 25-75% percentiles; whiskers represent minimum and maximum values of band intensity for each group. (D and E) Primary CLL cells were exposed to 1 μM BAY for 48 h under normoxic and hypoxic conditions, in the absence and in the presence of M2-10B4 stromal cells (SC), and evaluated for HIF-1α expression by WB. BAY87-2243 was able to lower HIF-1α expression in CLL cultured in normoxic and hypoxic conditions, and in the absence or in the presence of SC, independently of TP53 status. (F-H) Cell viability of TP53dis and TP53wt CLL cells exposed for 48 h to 1 μM BAY87-2243, under normoxia and hypoxia, or in co-culture with SC. The treatment with BAY87-2243 determined a significant decrease in the viability of TP53wt and TP53dis CLL cells, compared to untreated controls, both in normoxia and hypoxia. After exposure to SC, BAY87-2243 significantly reduced the viability of TP53dis and TP53wt CLL cells, not completely overcoming the SC protective effect. In blot representations, vertical lines have been inserted to indicate repositioned gel lanes. (I) Scheme of the in vivo experiment with mice transplanted with the EmTCL1-derived leukemia. (J) Percentage of leukemic cells (CD19+) in the bone marrow (BM) of mice transplanted with the Em-TCL1-derived leukemia, treated with BAY87-2243, and euthanized as in (I). (K) Percentage of AnnV+ leukemic cells (CD19+) in the BM of mice transplanted with Em-TCL1-derived leukemia, treated with BAY87-4432, and euthanized as in (I). (A) Data are represented by a scatter plot. (J and K) Data are represented as bee-swarm plots. (B) Box and whiskers plots represent median values and 25-75% percentiles; whiskers represent minimum and maximum values for each group. (D) Results are from one representative experiment in three TP53wt patients and one representative experiment in three TP53dis patients. (E) Results are from one representative experiment in seven TP53wt patients and one representative experiment in four TP53dis patients. (F, G and H) Box and whiskers plots represent median values and 25-75% percentiles; whiskers represent minimum and maximum values for each group, together with all points. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
To further corroborate these data and the ability of BAY87-2243 to exert effective anti-tumor functions in CLL, we used a murine model derived from the transfer of Em-TCL1 leukemic cells into syngeneic mice.17 In line with the results reported by Valsecchi et al.,17 showing that HIF-1α regulates the interaction of CLL cells with the bone marrow (BM) microenvironment, we observed that BAY87-2243 significantly reduced BM infiltration by leukemic cells, also inducing cytotoxicity in a consistent proportion of CLL cells (Figure 5I-K). The anti-tumor effect observed with BAY87-2243 in the BM was not evident in the PB and spleen compartments (data not shown), suggesting that, in a murine model of aggressive and rapidly growing CLL, HIF-1α may serve as a pro-survival factor, especially for the leukemia reservoir residing in the BM.
In conclusion, our data indicate that HIF-1α is a pro-survival factor in CLL, which can be effectively targeted by the pharmacologic agent BAY87-2243, a specific inhibitor with potent anti-tumor effects both in vitro and in vivo.
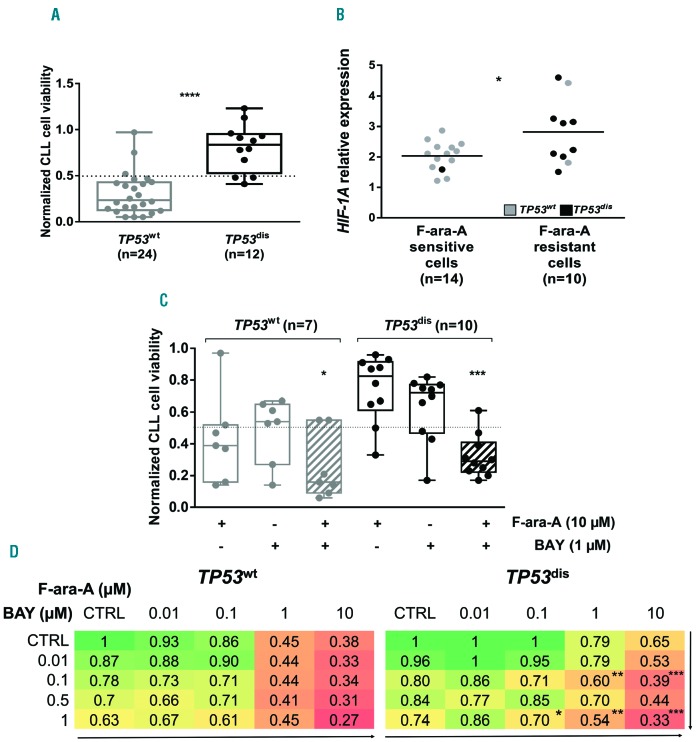
BAY87-2243 restores fludarabine sensitivity of TP53dis chronic lymphocytic leukemia cells and counteracts the protective effect of stromal cells
We next examined whether BAY87-2243 was also effective in overcoming the intrinsic resistance to fludarabine of CLL cells from TP53dis patients.9,31–33 As expected, in our cohort, the normalized cell viability after 48-h F-ara-A treatment was significantly higher in TP53dis compared to TP53wt CLL cells (Figure 6A). Consistently, we observed that CLL cells with a normalized cell viability ≥0.5, arbi trarily considered as fludarabine-resistant, were mostly TP53dis and had a significantly higher baseline expression of HIF-1A mRNA compared to fludarabine-sensitive cells (i.e. normalized cell viability <0.5) (Figure 6B). Interestingly, BAY87-2243 enhanced the cytotoxicity of fludarabine on TP53dis CLL cells, as shown by the significant impairment of cell viability observed after combined treatment with BAY87-2243 + fludarabine compared to each compound used as a single agent (Figure 6C). The cytotoxic effect exerted by the combination was strongly synergistic (CI=0.17) on TP53dis CLL cells and was also evident, although less remarkable, on TP53wt CLL cells (Figure 6C and Online Supplementary Figure S8). Interestingly, we observed that combinations consisting of lower concentrations of BAY87-2243 + fludarabine were capable of inducing significant reductions in cell viability compared to each drug used as a single agent, and this effect was particularly evident in the TP53dis CLL subset (Figure 6D). The cytotoxic activity of the combinations was also strongly synergistic, as shown by data on CI (Online Supplementary Figure S9). Notably, the significantly higher cytotoxicity of the combination BAY87-2243 + fludarabine was maintained even when both TP53dis and TP53wt CLL cells were cultured under hypoxia (Online Supplementary Figure S10A and B) or in the presence of SC (Online Supplementary Figure S10C and D).
Figure 6.
HIF-1A mRNA is over-expressed in fludarabine-resistant chronic lymphocytic leukemia (CLL) cells and HIF-1α inhibition is capable of restoring fludarabine sensitivity. (A) Normalized cell viability (i.e. the ratio between the percentage of AnnV/PI negative CLL cells cultured in the presence of F-ara-A and the percentage of AnnV/PI negative CLL cells left untreated) of TP53dis and TP53wt CLL cells exposed for 48 hours (h) to 10 μM F-ara-A. (B) A normalized cell viability of 0.5 was selected as the cut-off value to identify fludarabine-resistant (i.e. normalized cell viability ≥0.5) and fludarabine-sensitive (i.e. normalized cell viability <0.5) CLL cells. Fludarabine-resistant CLL cells showed significantly higher baseline levels of HIF-1A mRNA compared to fludarabine-sensitive. Eight of 10 (80%) fludarabine-resistant samples derived from TP53dis patients, and 13 of 14 (93%) fludarabine-sensitive samples derived from TP53wt patients. (C) Normalized cell viability of TP53dis and TP53wt CLL cells exposed for 48 h to 1 μM BAY87-2243 (BAY) and/or 10 μM F-ara-A. The combination BAY87-2243 + F-ara-A (striped pattern) determined a significant decrease in the viability of TP53wt and TP53dis CLL cells, compared to each compound used as single agent and to untreated controls. (D) Heatmaps showing normalized viability after 48-h treatment with BAY87-2243 + fludarabine as single agents or in combination, used at different concentrations. Asterisks indicate the combinations which determined a significant reduction in cell viability compared to each single agent, at the corresponding concentration. (A and C) Box plots represent median values and 25-75% percentiles; whiskers represent minimum and maximum values for each group, together with all points. (B) Data are represented as bee-swarm plot. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
These results indicate that BAY87-2243 and fludarabine synergistically eliminate primary CLL cells, and that their effect is maintained even in the presence of HIF-1α inducing factors that recapitulate the BM niche microenvironment.
The combination of BAY87-2243 and ibrutinib exerts a synergistic cytotoxic effect on chronic lymphocytic leukemia cells
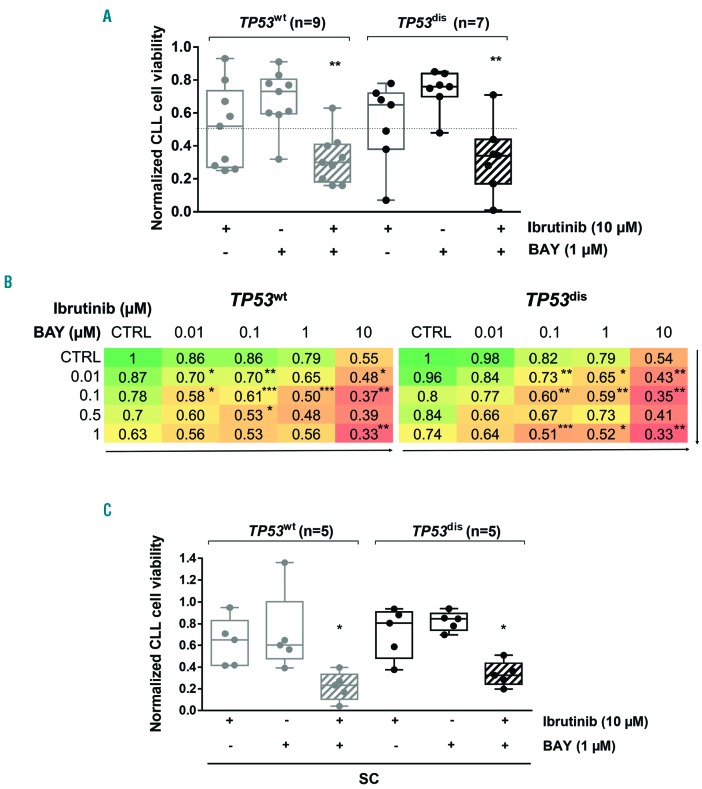
Previously reported data indicate that TP53-mutated CLL cells have a lower sensitivity to ibrutinib cytotoxicity in vitro34 and that ibrutinib-induced apoptosis is significantly reduced in conditions of hypoxia.19 Therefore, we hypothesized that the combination of an HIF-1α inhibitor and ibrutinib may represent a potentially attractive next step for patients carrying TP53 abnormalities, who are characterized by constitutively higher levels of HIF-1α. Our results show that the combination of BAY87-2243 + ibrutinib determined a significant impairment in the viability of TP53dis and TP53wt CLL cells compared to each compound used as a single agent, and was very strongly synergistic (Figure 7A and Online Supplementary Figure S11). Interestingly, similar effects were observed also with lower concentrations of both agents (Figure 7B and Online Supplementary Figure S12). Notably, the combination BAY87-2243 + ibrutinib exerted a significantly higher cytotoxic effect compared to each single compound even when CLL cells from both TP53dis and TP53wt samples were cultured in the presence of SC (Figure 7C).
Figure 7.
BAY87-2243 plus ibrutinib combinations exerts synergistic cytotoxic effects and is effective in impairing chronic lymphocytic leukemia (CLL) cell viability in the presence of stromal cells (SC). (A) Normalized 48-hour (h) viability of TP53dis and TP53wt CLL cells exposed to 1 μM BAY87-2243 (BAY) and 10 μM ibrutinib, used as single agents or in combination (striped pattern). Box plots represent median value and 25-75% percentiles; whiskers represent minimum and maximum values for each group, together with all points. (B) Heatmaps showing normalized viability after 48-h treatment with BAY87-2243 + ibrutinib as single agents or in combination, used at different concentrations. Asterisks indicate the combinations which determined a significant reduction in cell viability compared to each single agent, at the corresponding concentration. (C) Normalized 48-h viability of TP53dis and TP53wt CLL cells exposed to 1 μM BAY87-2243 and 10 μM ibrutinib, used as single agents or in combination (striped pattern), in co-culture with M2-10B4 SC. Box plots represent median value and 25-75% percentiles; whiskers represent minimum and maximum values for each group, together with all points. *P<0.05; **P<0.01; ***P<0.001.
Overall, these data demonstrate that BAY87-2243 exerts a compelling synergistic effect with ibrutinib, thus providing the rationale for future clinical translation.
Discussion
In this study, we investigated the expression and regulation of HIF-1α in TP53dis CLL cells and its potential role as a therapeutic target. We found that CLL cells carrying TP53 abnormalities express significantly higher baseline levels of HIF-1α and have increased HIF-1α transcriptional activity compared to TP53wt cells. Regardless of TP53 status, the resting levels of HIF-1α are susceptible to further upregulation by microenvironmental stimuli, such as hypoxia and SC. Our data show that HIF-1α is a suitable therapeutic target, the inhibition of which induces a strong cytotoxic effect, capable also of reversing the in vitro fludarabine resistance of TP53dis CLL cells and of exerting synergistic effects with ibrutinib.
Hypoxia has a detrimental role in the pathobiology of several solid and hematologic tumors.35,36 The identification of new potential targets in CLL is certainly important for high-risk patients, for whom there is still no effective cure, and, in addition, the development of new therapies might be effective in a broader setting of B-cell lymphoproliferative disorders.
It has been previously reported that HIF-1α levels vary considerably among CLL patients and that its overexpression is a predictor of a poor survival.17,37 In line with observations made in solid tumors, where p53 promotes the ubiquitination and proteasomal degradation of the HIF-1α subunit in hypoxia,12,22 we postulated that abnormalities of the TP53 gene might have an influence on the regulation of HIF-1α in CLL cells. To the best of our knowledge, this is the first study examining the differential expression and transcriptional activity of HIF-1α in patients with TP53-deficient CLL, also uncovering new mechanisms for HIF-1α modulation in leukemic cells. Our data show that the high-resting levels of HIF-1α detected in TP53dis samples associate both to an increased transcription, as shown by the higher HIF-1A mRNA levels, and to a decreased degradation, as shown by the higher baseline expression of ELK3 and by the lower pVHL amounts.
We next investigated the cellular pathways implicated in HIF-1α regulation mediated by extrinsic factors. Hypoxia-induced HIF-1α upregulation is not only due to a reduced pVHL-mediated protein degradation, which is a well-known mechanism in condition of oxygen deprivation, but also to an increased activation of RAS/ERK1-2 and PI3K/AKT signaling pathways. In contrast, pVHL expression in leukemic cells is not affected by SC, which instead activate the RAS/ERK1-2, RHOA/RHOA kinase and PI3K/AKT intracellular pathways, thus leading to HIF-1α overexpression. These results endorse recent data that implicate a number of paracrine factors in the transcriptional and translational regulation of HIF-1α in different tumor models.38
Previous data have suggested that HIF-1α targeting is a promising therapeutic strategy in CLL, potentially capable of synergizing with chemotherapeutic agents. The only HIF-1α inhibitor that has been preclinically tested in CLL is EZN-2208, a topoisomerase I inhibitor that has also been shown to down-modulate HIF-1α.17 We therefore tested BAY87-2243, a more selective HIF-1α inhibitor that has already shown in vivo anti-tumor efficacy in a lung tumor model, without any signs of toxicity.39 Interestingly, BAY87-2243 demonstrated a direct cytotoxicity towards leukemic cells isolated from CLL patients, and this effect was independent of TP53 status. As far as we know, this is the first evidence of the anti-tumor activity of BAY87-2243 in hematologic tumors, and particularly in CLL.
Patients with CLL and TP53 abnormalities are intrinsically resistant to fludarabine-based chemotherapy regimens.9,31–33,40,41 Our data demonstrate: i) higher baseline levels of HIF-1α in fludarabine-resistant CLL cells; and ii) a further upregulation of HIF-1α after exposure to hypoxia and SC. Given this, we investigated the ability of BAY87-2243 to overcome both intrinsic (i.e. TP53-related) and inducible (i.e. SC-induced) resistance to fludarabine. Our data show that BAY87-2243 potently synergizes with fludarabine in vitro, and that their combined cytotoxic effect was especially evident in TP53dis samples. Interestingly, HIF-1α inhibition was also effective in overcoming the TP53-independent fludarabine-resistance induced by extrinsic factors recapitulating the BM microenvironment.
Since HIF-1α critically regulates the interactions of CLL cells with the BM stroma,17 the cytotoxic effects exerted by BAY87-2243 in culture systems mimicking the tumor niche could, in part, be the result of a perturbation of molecular circuits triggered by microenvironmental stimuli that are implicated in cell survival and drug resistance. Data showing that hypoxia and SC induce HIF-1α overexpression support the hypothesis that, within the tumor niche, leukemic cells become highly dependent on its pro-survival effect. In line with this assumption, we found that treatment with BAY87-2243 induced a marked reduction in leukemic infiltration and a parallel increase in the proportion of apoptotic leukemic cells in the BM of a CLL transplantable model derived from the Em-TCL1 transgenic mice. Of note, the inhibition of HIF-1α may have beneficial effects also on the non-leukemic milieu. Recent data have shown that infiltration by CLL cells into BM could result in tissue-site hypoxia, causing: i) increased expression of HIF-1α in hematopoietic progenitors, which leads to an impaired hematopoiesis and a reduced output of innate immune cells into the blood; and ii) impaired functions of different immune cell subsets.19,42 Overall, this evidence endorses the concept of HIF-1α inhibition as a very promising therapeutic strategy in CLL.
In the era of new targeted treatments, ibrutinib has determined a dramatic change in the therapeutic landscape and has become the standard of care for the majority of CLL patients.43–45 However: i) ibrutinib is not suitable for all CLL patients and may have limited availability in several countries; ii) complete responses are infrequent, and indefinite drug administration is usually needed to maintain a clinical response; and iii) the development of ibrutinib resistance in CLL cells has been demonstrated.46,47 Even more importantly, TP53dis CLL patients show a suboptimal long-term response to ibrutinib,48 and TP53-mutated CLL cells have a lower sensitivity to ibrutinib cytotoxicity in vitro.34 Since our data show that TP53dis samples are characterized by higher levels and function of HIF-1α (which is a crucial target to overcome the constitutive and inducible drug resistance of CLL cells), we hypothesized that the combination of BAY87-2243 and ibrutinib might be an attractive approach for in vitro testing. We found that dual targeting of HIF-1α alongside BTK function produces a synergistic cytotoxic activity towards primary CLL cells, also in the presence of TP53 abnormalities; thus suggesting the possibility of improving ibrutinib efficacy through this novel therapeutic association.
Overall, our data indicate that HIF-1α is over-expressed in CLL cells, especially in the presence of TP53 aberrations, and that it is susceptible to further upregulation through microenvironmental stimuli. From the translational standpoint, the pharmacologic compound BAY87-2243, a selective inhibitor of HIF-1α, displays potent anti-tumor properties and warrants further pre-clinical evaluation in this disease setting, also in combination with other therapies. Indeed, on one hand, the synergism of BAY87-2243 and fludarabine may provide the rationale for future clinical application in countries with limited access to ibrutinib, particularly for the treatment of high-risk patients carrying TP53 abnormalities. On the other hand, BAY87-2243 coupled with ibrutinib may offer a rational combination to increase the proportion of minimal residual disease negative remissions, thus reducing the development of CLL clones with resistant mutations.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/4/1042
Funding
The authors would like to thank: Italian Association for Cancer Research (AIRC IG15232 and AIRC IG21408) (CR), (AIRC IG13119, AIRC IG16985, AIRC IG2174) (MM), (AIRC IG17622) (VGa), (AIRC 5×1000 project 21198, Metastatic disease: the key unmet need in oncology) (GG), (AIRC 5×1000 Special Programs MCO-10007 and 21198) (RF); Fondazione Neoplasie Sangue (Fo.Ne.Sa), Torino, Italy; University of Torino (local funds ex-60%) (MC); Ministero della Salute, Rome, Italy (Progetto Giovani Ricercatori GR-2011-02347441 [RB], GR-2009-1475467 [R.Bomben], and GR-2011-02351370 [MDB]). Fondazione Cassa di Risparmio di Torino (CRT) (VGr was recipient of a fellowship), Fondazione “Angela Bossolasco” Torino, Italy (VGr was recipient of the “Giorgio Bissolotti e Teresina Bosio” fellowship), the Italian Association for Cancer Research (AIRC, Ref 16343 VGr was recipient of the “Anna Nappa” fellowship and MT is currently the recipient of a fellowship from AIRC Ref 19653). Associazione Italiana contro le Leucemie, Linfomi e Mieloma (AIL) (CV was recipient of a fellowship). Pezcoller Foundation in collaboration with SIC (Società Italiana Cancerologia) (CV was a recipient of a "Fondazione Pezcoller - Ferruccio ed Elena Bernardi" fellowship).
References
- 1.Hallek M, Fischer K, Fingerle-Rowson G, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet Lond Engl. 2010;376(9747):1164–1174. [DOI] [PubMed] [Google Scholar]
- 2.Zenz T, Vollmer D, Trbusek M, et al. TP53 mutation profile in chronic lymphocytic leukemia: evidence for a disease specific profile from a comprehensive analysis of 268 mutations. Leukemia. 2010; 24(12): 2072–2079. [DOI] [PubMed] [Google Scholar]
- 3.Parikh SA. Chronic lymphocytic leukemia treatment algorithm 2018. Blood Cancer J. 2018;8(10):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutton L-A, Rosenquist R. Deciphering the molecular landscape in chronic lymphocytic leukemia: time frame of disease evolution. Haematologica. 2015;100(1):7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zenz T, Eichhorst B, Busch R, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28(29): 4473–4479. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez D, Martinez P, Wade R, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29(16):2223–2229. [DOI] [PubMed] [Google Scholar]
- 7.Campo E, Cymbalista F, Ghia P, et al. TP53 aberrations in chronic lymphocytic leukemia: an overview of the clinical implications of improved diagnostics. Haematologica. 2018;103(12):1956–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seiffert M, Dietrich S, Jethwa A, Glimm H, Lichter P, Zenz T. Exploiting biological diversity and genomic aberrations in chronic lymphocytic leukemia. Leuk Lymphoma. 2012; 53(6):1023–1031. [DOI] [PubMed] [Google Scholar]
- 9.Gaidano G, Rossi D. The mutational landscape of chronic lymphocytic leukemia and its impact on prognosis and treatment. Hematol Am Soc Hematol Educ Program. 2017;2017(1):329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson PA, Burger JA. Bruton’s tyrosine kinase inhibitors: first and second generation agents for patients with Chronic Lymphocytic Leukemia (CLL). Expert Opin Investig Drugs. 2018;27(1):31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien S, Furman RR, Coutre S, et al. Single-agent ibrutinib in treatment-naïve and relapsed/refractory chronic lymphocytic leukemia: a 5-year experience. Blood. 2018; 131(17):1910–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masoud GN, Li W. HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5(5):378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721. [DOI] [PubMed] [Google Scholar]
- 14.Singh D, Arora R, Kaur P, Singh B, Mannan R, Arora S. Overexpression of hypoxia-inducible factor and metabolic pathways: possible targets of cancer. Cell Biosci. 2017;762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semenza G. Signal transduction to hypoxia-inducible factor 1. Biochem Pharmacol. 2002; 64(5-6):993–998. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh AK, Shanafelt TD, Cimmino A, et al. Aberrant regulation of pVHL levels by microRNA promotes the HIF/VEGF axis in CLL B cells. Blood. 2009;113(22):5568–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valsecchi R, Coltella N, Belloni D, et al. HIF-1α regulates the interaction of chronic lym-phocytic leukemia cells with the tumor microenvironment. Blood. 2016; 127(16): 1987–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koczula KM, Ludwig C, Hayden R, et al. Metabolic plasticity in CLL: adaptation to the hypoxic niche. Leukemia. 2016;30(1):65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serra S, Vaisitti T, Audrito V, et al. Adenosine signaling mediates hypoxic responses in the chronic lymphocytic leukemia microenvironment. Blood Adv. 2016;1(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rigoni M, Riganti C, Vitale C, et al. Simvastatin and downstream inhibitors circumvent constitutive and stromal cell-induced resistance to doxorubicin in IGHV unmutated CLL cells. Oncotarget. 2015;6(30):29833–29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hientz K, Mohr A, Bhakta-Guha D, Efferth T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget. 2016;8(5):8921–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amelio I, Melino G. The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression. Trends Biochem Sci. 2015;40(8):425–434. [DOI] [PubMed] [Google Scholar]
- 23.Salnikow K, Costa M, Figg WD, Blagosklonny MV. Hyperinducibility of hypoxia-responsive genes without p53/p21-dependent checkpoint in aggressive prostate cancer. Cancer Res. 2000;60(20):5630–5634. [PubMed] [Google Scholar]
- 24.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008; 111(12):5446–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bomben R, Dal-Bo M, Benedetti D, et al. Expression of mutated IGHV3-23 genes in chronic lymphocytic leukemia identifies a disease subset with peculiar clinical and biological features. Clin Cancer Res. 2010; 16(2):620–628. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semenza GL, Jiang BH, Leung SW, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–32537. [DOI] [PubMed] [Google Scholar]
- 28.Dal Bo M, Pozzo F, Bomben R, et al. ARHG-DIA, a mutant TP53-associated Rho GDP dissociation inhibitor, is over-expressed in gene expression profiles of TP53 disrupted chronic lymphocytic leukaemia cells. Br J Haematol. 2013;161(4):596–599. [DOI] [PubMed] [Google Scholar]
- 29.Gross C, Dubois-Pot H, Wasylyk B. The ternary complex factor Net/Elk-3 participates in the transcriptional response to hypoxia and regulates HIF-1 alpha. Oncogene. 2008; 27(9):1333–1341. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Xin H, Eckert DT, Brown JA, Gnarra JR. Hypoxia and cell cycle regulation of the von Hippel-Lindau tumor suppressor. Oncogene. 2011;30(1):21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turgut B, Vural O, Pala FS, et al. 17p Deletion is associated with resistance of B-cell chronic lymphocytic leukemia cells to in vitro fludarabine-induced apoptosis. Leuk Lymphoma. 2007;48(2):311–320. [DOI] [PubMed] [Google Scholar]
- 32.Dietrich S, Oleś M, Lu J, et al. Drug-perturbation-based stratification of blood cancer. J Clin Invest. 2018;128(1):427–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadeu F, Delgado J, Royo C, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016; 127(17):2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guarini A, Peragine N, Messina M, et al. Unravelling the suboptimal response of TP53-mutated chronic lymphocytic leukaemia to ibrutinib. Br J Haematol. 2019;184(3):392–396. [DOI] [PubMed] [Google Scholar]
- 35.Kim J-Y, Lee J-Y. Targeting Tumor Adaption to Chronic Hypoxia: Implications for Drug Resistance, and How It Can Be Overcome. Int J Mol Sci. 2017;18(9):1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muz B, de la Puente P, Azab F, Luderer M, Azab AK. The role of hypoxia and exploitation of the hypoxic environment in hematologic malignancies. Mol Cancer Res. 2014;12(10):1347–1354. [DOI] [PubMed] [Google Scholar]
- 37.Kontos CK, Papageorgiou SG, Diamantopoulos MA, et al. mRNA overexpression of the hypoxia inducible factor 1 alpha subunit gene (HIF1A): An independent predictor of poor overall survival in chronic lymphocytic leukemia. Leuk Res. 2017; 53:65–73. [DOI] [PubMed] [Google Scholar]
- 38.Kuschel A, Simon P, Tug S. Functional regulation of HIF-1α under normoxia–is there more than post-translational regulation¿ J Cell Physiol. 2012;227(2):514–524. [DOI] [PubMed] [Google Scholar]
- 39.Ellinghaus P, Heisler I, Unterschemmann K, et al. BAY 87-2243, a highly potent and selective inhibitor of hypoxia-induced gene activation has antitumor activities by inhibition of mitochondrial complex I. Cancer Med. 2013;2(5):611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Döhner H, Fischer K, Bentz M, et al. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995;85(6):1580–1589. [PubMed] [Google Scholar]
- 41.Zenz T, Häbe S, Denzel T, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114(13):2589–2597. [DOI] [PubMed] [Google Scholar]
- 42.Manso BA, Zhang H, Mikkelson MG, et al. Bone marrow hematopoietic dysfunction in untreated chronic lymphocytic leukemia patients. Leukemia. 2019;33(3):638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eichhorst B, Robak T, Montserrat E, et al. appendix 6: Chronic lymphocytic leukaemia: eUpdate published online September 2016 (http://www.esmo.org/Guidelines/Haematological-Malignancies) Ann Oncol. 2016; 27(suppl 5):v143–v144. [DOI] [PubMed] [Google Scholar]
- 44.Eichhorst B, Robak T, Montserrat E, et al. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015; 26 Suppl 5:v78–84. [DOI] [PubMed] [Google Scholar]
- 45.NCCN Guidelines Insights: chronic lymphocytic leukemia/small lymphocytic leukemia, version 1.2017. http://www.jnccn.org/content/15/3/293.long (accessed February 11, 2018).
- 46.Woyach JA, Ruppert AS, Guinn D, et al. BTKC481S-mediated resistance to Ibrutinib in chronic lymphocytic leukemia. J Clin Oncol. 2017;35(13):1437–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones D, Woyach JA, Zhao W, et al. PLCG2 C2 domain mutations co-occur with BTK and PLCG2 resistance mutations in chronic lymphocytic leukemia undergoing ibrutinib treatment. Leukemia. 2017;31(7):1645–1647. [DOI] [PubMed] [Google Scholar]
- 48.Byrd JC, Furman RR, Coutre SE, et al. Three-year follow-up of treatment-naïve and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood. 2015; 125(16):2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]