Abstract
From the mid‐1960s onwards, it was believed that only two human coronavirus species infect humans: HCoV‐229E and HCoV‐OC43. Then, in 2003, a novel member of the coronavirus family was introduced into the human population: SARS‐CoV, causing an aggressive lung disease. Fortunately, this virus was soon expelled from the human population, but it quickly became clear that the human coronavirus group contains more members then previously assumed, with HCoV‐NL63 identified in 2004. Despite its recent discovery, ample results from HCoV‐NL63 research have been described. We present an overview of the publications on this novel coronavirus.
Keywords: human coronavirus, HCoV‐NL63, ACE2, croup, respiratory tract illness, Kawasaki disease
Introduction
Coronaviruses are positive strand RNA viruses with the largest viral genome among the RNA viruses (27–33 kb). The virus particles are enveloped and carry extended spike proteins on the membrane surface, providing the typical crown‐like structure (crown=corona) (Fig. 1). Coronaviruses are known to infect mammals and birds. Genotypically, the coronaviruses can be subdivided into three groups. The group III viruses are found exclusively in birds, whereas viruses from group I and II have mammals as their host.
Figure 1.
Negatively stained electron micrograph of HCoV‐NL63. Courtesy of Dr Bermingham and Dr Hoschler and the EM unit of the Health Protection Agency, Colindale, London.
The coronaviruses share a conserved organization of their (positive strand) RNA genome. The 5′ two‐thirds of the genome contains the large 1a and 1b ORFs, encoding the proteins necessary for RNA replication (the nonstructural proteins), whereas the 3′ one‐third contains the genes coding for the structural proteins: haemagglutinin esterase protein (only for group IIa), spike protein, envelope protein, membrane protein and nucleocapsid protein. Interspersed between the structural protein genes one can find accessory protein genes that differ in number and position for the various coronaviruses (Fig. 2).
Figure 2.

Genome organization of HCoV‐NL63 and other group I coronaviruses.
The coronaviruses can cause a variety of diseases in animals including gastroenteritis and respiratory tract disease. In humans, the coronaviruses (HCoV) are associated with respiratory tract illnesses, and its most villainous member is SARS‐CoV. This virus was identified in 2003 and causes severe acute respiratory syndrome (SARS), an often‐fatal lung disease in humans (2003, 2003, 2004). The SARS‐CoV probably originated from a wild animal reservoir, most likely bats (2005, 2005), and was transmitted in a zoonotic event to humans via infected civet cats that were marketed and prepared as food in China. The global spread of SARS in early 2003 caused at least 800 deaths and substantial morbidity. The epidemic was contained at the end of 2003 due to a highly effective global public health response, and SARS‐CoV is currently not circulating in humans. Nevertheless, at least four coronaviruses are continuously circulating in the human population, especially in young children. Two were discovered recently: HCoV‐NL63 and HCoV‐HKU1 (2004, 2005b); the other two, HCoV‐OC43 and HCoV‐229E, were identified in the mid‐1960s (Tyrrell & Bynoe, 1965; Hamre & Procknow, 1966). The latter viruses were tested for pathogenicity in human volunteers, which demonstrated that HCoV‐229E and HCoV‐OC43 cause the common cold (Bradburne et al., 1967). For the new viruses HCoV‐NL63 and HCoV‐HKU1 a human test system, but also an animal model system, is lacking. Nevertheless, since the discovery of HCoV‐NL63 in 2004, much knowledge has been gained on this virus. Several groups have studied the worldwide spread, the association with human disease and replication characteristics of HCoV‐NL63.
Identification of HCoV‐NL63
In January 2003, a 7‐month‐old child appeared in an Amsterdam hospital with coryza, conjunctivitis and fever. Chest radiography showed typical features of bronchiolitis and a nasopharyngeal aspirate specimen was collected 5 days after the onset of disease (sample NL63). Diagnostic tests for all known respiratory viruses were negative. The clinical sample was inoculated onto tertiary monkey kidney cells (tMK; Cynomolgus monkey) and a cytopathic effect was observed. The infectious agent could subsequently be passaged onto LLC‐MK2 cells (a monkey kidney cell line). Using the VIDISCA method (Virus Discovery cDNA‐AFLP; van der Hoek et al., 2004) a novel virus could be identified in the supernatant of the LLC‐MK2 culture. The initial PCR products showed sequence similarity to the genome sequence of members of the Coronaviridae family. The complete genome of the virus (named at that time HCoV‐NL63) showed that this virus was not a recombinant but rather a novel member of the group I coronaviruses (Fig. 3). HCoV‐NL63 forms a subcluster with HCoV‐229E, PEDV (porcine epidemic diarrhoea virus), and Bat‐CoV (Poon et al., 2005), which was assigned group Ib. The percentage nucleotide and amino acid identity among the Coronaviridae was determined for each gene and is listed in Table 1. For all genes except the M gene, the identity is the highest with HCoV‐229E.
Figure 3.
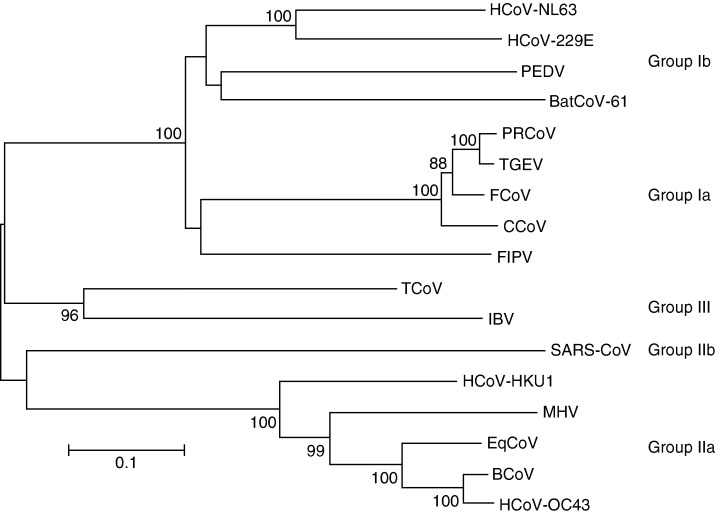
Phylogenetic analysis of coronaviruses. Analysis was carried out with the nucleotide sequences of a part of the spike gene (HCoV‐NL63 position 22854–24319) using the neighbour‐joining method of the mega program. Bootstrap resampling (1000 replicates) was employed to place approximate confidence limits on individual branches; bootstrap values above 80 are shown. The GenBank accession number of the sequences used in this phylogenetic analysis are HCoV‐229E: n; CCoV (canine coronavirus) n; FIPV (feline infectious peritonitis virus) n; PEDV (porcine epidemic diarrhea virus) n; TGEV (transmissible gastroenteritis virus): n; PRCoV (porcine respiratory coronavirus) n; MHV (mouse hepatitis virus) n; BCoV (bovine coronavirus) n; HCoV‐OC43: n; SARS‐CoV: n; IBV (avian infectious bronchitis virus) n; TCoV (turkey coronavirus) n; BatCoV strain61: n and HCoV‐HKU1: n.
Table 1.
Percentage nucleotide and amino acid identity of HCoV‐NL63 with other coronaviruses
| Group | Ib | Ia | IIa | IIb | III | ||
|---|---|---|---|---|---|---|---|
| Gene | HCoV‐229E | PEDV | TGEV | HCoV‐OC43 | HCoV‐ HKU1 | SARS‐CoV | IBV |
| 1a | 63/57 * | 58/50 | 53/38 | 48/23 | 49/23 | 47/23 | 46/22 |
| 1b | 76/81 | 71/77 | 69/71 | 59/52 | 62/53 | 59/55 | 60/52 |
| Spike | 56/55 | 55/44 | 53/42 | 47/22 | 48/22 | 46/21 | 43/23 |
| Envelope | 59/47 | 54/42 | 54/30 | 41/18 | 45/18 | 46/18 | 34/10 |
| Membrane | 64/61 | 65/65 | 49/44 | 49/33 | 51/33 | 45/29 | 49/24 |
| Nucleocapsid | 52/43 | 47/38 | 48/35 | 38/21 | 38/21 | 43/24 | 41/20 |
Percentage nucleotide identity/percentage amino acid identity.
A second research group in the Netherlands described essentially the same virus shortly after the first publication on HCoV‐NL63. Fouchier described a virus (which they named HCoV‐NL) in a Vero‐E6 cell culture supernatant (Fouchier et al., 2004). A nose swab sample was collected from an 8‐month‐old boy suffering from pneumonia in 1988. Virus from that sample was also cultivated first on tMK cells and subsequently passaged onto Vero cells. The similarity with the previously described HCoV‐NL63 strain was very high (98.8% at the nucleotide level) and it can be concluded that these two virus isolates represent the same species. Most differences between the two virus variants are clustered in the amino terminal region of the spike protein (Fig. 4).
Figure 4.

Nucleotide distance between HCoV‐NL63 isolate Amsterdam 1 and isolate NL. Each point plotted as the percentage genetic distance between the two isolates along the genome with a sliding window of 200 nt wide and a step size of 20 nt.
Almost 1 year later, a third group described the same human coronavirus (Esper et al., 2005b). Using universal coronavirus primers, patient samples were identified with coronaviruses that did not match at the nucleotide level with HCoV‐229E, HCoV‐OC43 or SARS‐CoV. These authors gave their virus the name ‘New Haven coronavirus’ (HCoV‐NH), although the partial sequences of their isolates clearly show that the novel coronaviruses identified in New Haven, CT, USA, are very similar to the isolates from the Netherlands (94–100% identical at nucleotide level) and thus represent the same species (van der Hoek & Berkhout, 2005).
Careful reading of the early coronavirus literature indicates that HCoV‐NL63, or the novel group II virus HCoV‐HKU1, may have been observed previously. In some of the earliest work on human coronaviruses during the mid‐1960s, viruses could be propagated in human embryonic tracheal organ culture that were not, or only distantly, related to HCoV‐229E or HCoV‐OC43 (1967, 1965). Unfortunately these strains [B814 (Tyrrell & Bynoe, 1965), HCoV‐OC16, HCoV‐OC37, and HCoV‐OC48 (McIntosh et al., 1967)] were lost for follow‐up studies, so it remains undetermined whether these viruses represent HCoV‐HKU1 or HCoV‐NL63, or yet undiscovered human coronaviruses. Additionally, in the beginning of the 1980s, several isolates of human coronaviruses were studied that were obtained by either cell culture or organ culture (HCoV strains AD, PA, LP, KI, PR, TO, RO, GI, and HO) (1980, 1981, 1984). Some of these isolates turned out to be serologically related to HCoV‐OC43, and therefore were termed group II isolates (strains RO, GI and HO), whereas others were serologically related to HCoV‐229E and thus were assigned as group I isolates (strains AD, PA, LP, KI, PR, and TO). Of these group I viruses, LP, KI, PR and TO could be cultured in MRC continuous cells (human), but strains AD and PA could only be cultured in human foetal tracheal and nasal organ cultures. In fact, this difference in culture properties reflects the difference in cell tropism of HCoV‐229E and HCoV‐NL63 (see ‘Cell tropism and receptor usage’) and thus raises the possibility that some of these early group I strains (for instance AD and PA) were in fact HCoV‐NL63 isolates. Unfortunately, this hypothesis cannot be tested because isolates AD and PA were also lost for further study. The earliest HCoV‐NL63 isolate remains the NL‐strain that was obtained from a patient in 1988, which was identified as HCoV‐NL63 in 2004 (Fouchier et al., 2004).
Infection with HCoV‐NL63
Worldwide spread
The first two studies on HCoV‐NL63 clearly indicated that infection by this virus is not a rare event (2004, 2004). Both reports presented several HCoV‐NL63 infected patients that suffered from upper or lower respiratory tract illnesses. Since then a number of reports have shown that the virus is not unique to the Netherlands, and that it can be found worldwide. In Australia, HCoV‐NL63 could be detected in 16 of 766 patients with acute respiratory disease (2.1%; Arden et al., 2005). A Japanese study on 118 children under the age of 2 that suffered from bronchiolitis and who were admitted to the hospital revealed three positive patients (2.5%; Ebihara et al., 2005a), and a second Japanese study presented five HCoV‐NL63 infections among 419 specimens collected from children with respiratory infections (1.2%) (Suzuki et al., 2005). Among 279 hospitalized children with upper and lower respiratory tract illness in Belgium, seven children were identified with HCoV‐NL63 (2.3%) (Moes et al., 2005). In France, 28 of 300 patients with upper and lower respiratory tract illness under the age of 20 harboured HCoV‐NL63 RNA (9.3%) (Vabret et al., 2005) and in a Swiss cohort of neonates, HCoV‐NL63 was identified in six of 82 cases (7%) during their first episode of lower respiratory tract illness (Kaiser et al., 2005). Two studies by a Canadian group reported 45 NL63‐positive patients of a total of 1765 specimens that were tested (2.5%) (2005a, 2005b). Seven of these Canadian patients were involved in an outbreak of acute respiratory‐tract infection in a personal care home for elderly. HCoV‐NL63 was also detected in Hong Kong in 15 of 587 (2.6%) children (Chiu et al., 2005). These Hong Kong children participated in a prospective clinical and virological study on children under the age of 18 with acute respiratory tract infection. The study group represented the Hong Kong population, so it could be estimated that HCoV‐NL63 infection causes more than 200 hospital admissions each year per 100 000 children under the age of 6. Among 949 samples of children with lower respiratory tract illness in Germany who participated in the PRI.DE study (Paediatric Respiratory Infection in Germany) HCoV‐NL63 was detected in 49 children (5.2%) (van der Hoek et al., 2005). On the whole, patients with HCoV‐NL63 range in age from 1 month to 100 years, but in all studies the proportion of positive specimens is the highest in the 0–5‐year group. HCoV‐NL63 infection is frequently observed in patients with an underlying disease, whether they are immunocompromised due to (chemo‐) therapy or HIV infection or have another medial condition such as chronic obstructive pulmonary disease (2005, 2005b, 2004, 2005). In conclusion, HCoV‐NL63 infection has been recognized all over the world. Infection of children, the elderly or weakened persons regularly requires hospitalization due to the severity of respiratory symptoms.
Seasonality of infection
HCoV‐OC43 and HCoV‐229E tend to be transmitted predominantly in the winter season in temperate climate countries (Hendley et al., 1972). The seasonality of HCoV‐NL63 in the Netherlands shows the same preference for the winter season. The studies in Belgium, Australia, Japan, Canada, Germany and France also report a winter predominance of this virus (2005, 2005a, 2005a, 2005, 2005, 2005). Only in Hong Kong did HCoV‐NL63 show a spring‐summer peak of activity, whereas in the same study HCoV‐OC43 was mostly found in the winter season (Chiu et al., 2005). This indicates that the seasonality of HCoV‐NL63 in tropical and subtropical regions may not be restricted to the winter season.
Genetic differences between isolates
RNA viruses have an enormous potential to generate genetic diversity due to their high mutation rates, which are estimated at approximately one mutation per genome per replication cycle (1999, 2004). As a consequence, RNA viruses are known to exist as a dynamic distribution of variants with a closely related, yet non‐identical genome centred on a master sequence, the so‐called quasispecies concept (Domingo 2002). These high mutation rates provide RNA viruses with the capacity to increase their level of genetic variability and thereby their ability to adapt quickly to a change in the environment. In addition to the high mutation rate, recombination events increase the level of genetic diversity. Coronavirus recombination is assumed to be mediated by a ‘copy choice mechanism’ with template switching by the polymerase during RNA‐dependent RNA synthesis (1998, 1985). In vivo recombination has been described between group I feline and canine coronaviruses, between strains of MHV (mouse hepatitis virus) and between strains of IBV (infectious bronchitis virus) (1998, 2001). Phylogenetic analysis of a small part of the 1a gene of HCoV‐NL63 shows diversity of the natural isolates and subsequent clustering in NL63 subgroups (2005, 2005a, 2005, 2004). The close clustering of isolates from the Netherlands, Belgium, Canada, Hong Kong, Korea and Australia indicates that this clustering does not correlate with the geographic origin (Fig. 5). Phylogenetic analysis of the spike gene also reveals the presence of subgroups (Moes et al., 2005). Interestingly, but not unexpectedly for coronaviruses, the clustering based on the 1a gene does not match with that of the spike gene, indicating that recombination has occurred between different HCoV‐NL63 isolates (Moes et al., 2005; Fig. 5). Sequencing of full‐length genomes of field isolates verifies the discordant clustering of 1a‐gene sequences and spike sequences and confirms that recombination is also common for HCoV‐NL63 (K. Pyrc, R. Dijkman, L.C. Deng, M.F. Jebbink, B. Berkhout & L. van der Hoek, unpublished results).
Figure 5.

Phylogenetic analysis of partial 1a and partial spike gene sequences of HCoV‐NL63 isolates. HCoV‐229E was used to root the tree for the 1a gene sequences. The neighbour‐joining method of the mega program was used with bootstrap resampling (1000 replicates) to place approximate confidence limits on individual branches; only bootstrap values above 80 are shown. GenBank numbers: n–n–n–n–n–n–n–n. CAN, Canada; Q, Australia; NL, the Netherlands; BE, Belgium; KR, Korea. NL63‐Amst1 is the prototype sequence of HCoV‐NL63. Red and blue indicate the strains that cluster for the 1a region and for which spike sequences are also available.
Besides variable regions, other parts of the HCoV‐NL63 genome are highly conserved among isolates. Especially the 5′ region of the 1b gene, which encodes the RNA‐dependent RNA polymerase, and the nucleocapsid protein gene show a low level of heterogeneity among natural isolates of HCoV‐NL63 (K. Pyrc, R. Dijkman, L.C. Deng, M.F. Jebbink, B. Berkhout & L. van der Hoek, unpublished results). These regions therefore present the best targets for the design of diagnostic primers and probes. Clinical samples will contain virus with its full‐length genome, but may also contain subgenomic mRNAs from infected cells. Considering that the sequence of the nucleocapsid gene is present in the genomic RNA and also in all subgenomic mRNAs (Pyrc et al., 2004), this region of the HCoV‐NL63 genome may represent the ideal target for the design of molecular‐based detection assays.
Combined infection with HCoV‐NL63 and a second respiratory virus
HCoV‐NL63 infections are often found in combination with a second respiratory virus, and the frequency of double infections can exceed 50% of all HCoV‐NL63 infections (van der Hoek et al., 2005). Some studies reported coinfection with influenza A virus H3N2 (Chiu et al., 2005), respiratory syncytial virus (2005, 2005b, 2005), parainfluenza‐3 (2005, 2005), or human metapneumovirus (2005, 2005b). Interestingly, the viral load of HCoV‐NL63 in patients with a coinfection is significantly lower than the load in singly infected HCoV‐NL63 patients (2005, 2005). The reduced viral load may be caused by direct competition for the same target cell in the respiratory tissues. Alternatively, the innate immune response triggered by one respiratory virus may inhibit replication of HCoV‐NL63. It is also likely that an initial HCoV‐NL63 infection may set the stage for a subsequent infection with another respiratory virus, and at the time that this second virus is causing symptoms, the HCoV‐NL63 infection may already be under the control of the immune system. Double infections are more often observed in hospitalized patients vs. outpatients (van der Hoek et al., 2005), suggesting that patients with a codetected respiratory virus have a more severe disease or a worse long‐term prognosis than patients with a single HCoV‐NL63 infection. However, further studies are required to confirm these trends.
Viral shedding
In the course of a coronavirus infection the virus can be shed for a long period. For instance, some cats infected with FIPV (feline infectious peritonitis virus) can continue to shed the virus in faeces for months (Herrewegh et al., 1997) and the duration of SARS‐CoV excretion in faeces is also up to a few months (Liu et al., 2004). The duration of SARS‐CoV excretion in sputa was on average a few weeks, but could for some persons extend to almost 2 months (Liu et al., 2004). For HCoV‐NL63 the situation does not seem very different. Three weeks after an initial HCoV‐NL63 infection the virus is still detectable in respiratory specimens in 50% of the infected children. At this time all children are free of symptoms (Kaiser et al., 2005).
Frequency of infection compared to other coronaviruses
A consensus‐coronavirus reverse transcriptase (RT)‐PCR assay that can amplify any member of the coronavirus family will provide a very useful diagnostic tool to screen for all known coronaviruses in a single assay. Besides quick screening for several pathogens in a clinical assay, the method also provides the opportunity to identify new coronaviruses. The consensus RT‐PCR assay designed in the polymerase gene as described by Stephensen et al. (1999), which was designed to amplify all known coronaviruses, might not be able to detect HCoV‐NL63 because of a mismatch in one of the consensus primers. Moes et al. (2005) recently optimized these consensus primers based on an alignment of the HCoV‐NL63 prototype sequence and 13 other coronavirus sequences. Even though the HCoV‐HKU1 sequence was not known at the time that this PCR was designed, alignment of the consensus primers confirms that these primers perfectly match with HCoV‐HKU1.
Moes et al. (2005) screened 309 respiratory samples from hospitalized children in Leuven, Belgium, and found human coronaviruses in 15 samples (5%, Table 2). Of these patients, seven were infected with HCoV‐NL63, seven with HCoV‐OC43, and a single person with HCoV‐229E. These results indicate that HCoV‐NL63 and HCoV‐OC43 are the two coronaviruses that cause most respiratory infections leading to hospitalization. Similar results were obtained in our laboratory for patients with respiratory disease who were diagnosed in the Public Health Laboratory at the Municipal Health Service of Amsterdam. Of the 63 respiratory samples that were tested for the three human coronaviruses (229E, OC43 and NL63) in the winter of 2002–2003, three samples were positive for HCoV‐NL63 (4.7%) and three for HCoV‐OC43 (4.7%); no HCoV‐229E was detected (S. Bruisten, M. van Zon, J. Spaargaren & L. van der Hoek, unpublished results). In Hong Kong, HCoV‐NL63 is the most frequently found coronavirus (2.6%), followed by HCoV‐OC43 (1.5%) and HCoV‐229E (0.3%) (Chiu et al., 2005). Vabret et al. obtained similar results in a screen for these three viruses in patients from Caen, France. HCoV‐NL63 (9.3%) was detected more frequently than HCoV‐OC43 (2.2%), and HCoV‐229E was not detected at all. Furthermore, HCoV‐NL63 was found most frequently in Swiss neonates with lower respiratory tract illness (7.3%), compared to HCoV‐OC43 (6.1%) and HCoV‐229E (3.7%) (Kaiser et al., 2005).
Table 2.
Frequency of infection by the various human coronaviruses
| Country | HCoV‐NL63 | HCoV‐229E | HCoV‐OC43 | Patient group | Method | Reference |
|---|---|---|---|---|---|---|
| Netherlands (n=63) * | 4.7 † | 0 | 4.7 | C A H O | RT‐PCR | Private communication ‡ |
| Belgium (n=309) | 2.3 | 0.3 | 2.3 | C H | RT‐PCR (pancorona RT‐PCR) | Moes et al. (2005) |
| Hong Kong (n=587) | 2.6 | 0.3 | 1.5 | C H | RT‐PCR (real time RT‐PCR) | Chiu et al. (2005) |
| France (n=300) | 9.3 | 0 | 2.2 | C H | RT‐PCR (multiplex RT‐PCR) | Vabret et al. (2005) |
| Switzerland (n=82) | 7.3 | 3.7 | 6.1 | C H O | RT‐PCR | Kaiser et al. (2005) |
Number of samples tested.
Percentage of the tested samples positive for the indicated virus.
S. Bruisten, M. van Zon, J. Spaargaren and L. van der Hoek (data not published).
C, children; A, adults; H, hospitalized patients; O, outpatients; HCoV, human coronavirus.
There are no reports yet that have screened for all four circulating human coronaviruses, including HCoV‐HKU1. The first study in Hong Kong identified two HCoV‐HKU1 infected adult patients among 400 (0.5%) patients with respiratory illnesses. A French study on HCoV‐HKU1 demonstrated six HCoV‐HKU1 positive samples among 135 hospitalized children (4.4%) (Vabret et al., 2006). In Australia, 3.1% of children with upper and lower respiratory tract illness were positive for HCoV‐HKU1 (Sloots et al., 2006). A second survey in Hong Kong demonstrated 10 additional HCoV‐HKU1 positive patients (2.4%) in adults with community‐acquired pneumonia (Woo et al., 2005c). This implies that infection with HCoV‐HKU1 is not unusual, that adults are infected relatively frequently and that HCoV‐HKU1 has also spread worldwide. It is also clear that more studies are required to confirm these trends.
Disease associated with infection
The first described cases of HCoV‐NL63 infection were young children with severe lower respiratory tract illnesses in hospital settings (2005, 2004, 2004). One of the HCoV‐NL63 infected elderly Canadian patients died 5 days after the onset of disease (Bastien et al., 2005a), showing that the severity of the respiratory disease can be substantial. All studies on the incidence of HCoV‐NL63 infection have used respiratory clinical material from patients suffering from respiratory tract illnesses, mostly lower respiratory tract illnesses. It is therefore not surprising that the symptoms associated with NL63‐infection represent respiratory diseases. Nevertheless, one can provide an initial overview of the symptoms related to HCoV‐NL63 infection by focusing on patients without a secondary infection. Table 3 summarizes the symptoms in patients with an HCoV‐NL63 infection. These results are derived only from studies that did not select for certain clinical symptoms, because selection for a respiratory disease (e.g. bronchiolitis or lower respiratory tract illness) will bias the overview towards these clinical symptoms. As mentioned above, a substantial number of patients suffered from lower respiratory tract illnesses that required hospitalization. On the other hand, some of the patients showed relatively mild symptoms like fever, cough, sore throat and rhinitis (Bastien et al., 2005a). For instance, only 16 of the 45 patients with HCoV‐NL63 in Canada had been hospitalized because of the respiratory illness, demonstrating that infection with HCoV‐NL63 does not always require hospitalization (2005a, 2005b). In the PRI.DE study, which investigated children with lower respiratory tract illness, HCoV‐NL63 infection was also more often detected in outpatients than hospitalized patients, confirming that HCoV‐NL63 generally causes less severe problems (van der Hoek et al., 2005). This suggests strongly that HCoV‐NL63, similar to HCoV‐229E and HCoV‐OC43, is a common cold virus (McIntosh, 1996) and that HCoV‐NL63, like HCoV‐229E and HCoV‐OC43, can cause a more severe clinical picture in young children, the elderly and immunocompromised persons (1974, 2004).
Table 3.
Clinical symptoms in HCoV‐NL63‐positive patients without a second respiratory infection *
| Country | Fever | Cough | Coryza/ rhinorrhoea | Pharyngitis/sore throat/ hoarseness | Bronchitis/ Bronchiolitis | Pneumonia | Croup | Underlying disease | Kawasaki disease | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Netherlands (n=4) | 50 † | 50 | 75 | ND | 0 | 0 | 0 | 100 | 0 | Fouchier et al. (2004) |
| Australia (n=10) | 70 | 70 | 60 | 50 | 50 | 0 | 10 | 30 | 0 | Arden et al. (2005) |
| Canada I (n=19) | 79 | 47 | 10 | 26 | 10 | 0 | 5 | 5 | 0 | Bastien et al. (2005a) |
| Belgium (n=5) | 80 | 60 | 20 | 20 | 60‡ | 0 | 0 | 80 | 0 | Moes et al. (2005) |
| Hong Kong (n=12) | 18 | 73 | 64 | 27 | 0 | 0 | 27 | 27 | 0 | Chiu et al. (2005) |
| France (n=18) § | 61 | ND | 39 | 22 | 33 | 5 | 0 | 5 | 0 | Vabret et al. (2005) |
| Canada II (n=24) | 30 | 58 | 42 | 8 | 46 | 4 | 4 | 69 | 0 | Bastien et al. (2005b) |
Negative for RSV PIV1‐3 influenza A and B and adenovirus. Some groups tested for additional pathogens: hMPV (2005, 2005a, 2005b, 2005, 2004), HCoV‐229E and HCoV‐OC43 (2005, 2004), PIV4 (2004, 2005) and rhinovirus (Fouchier et al., 2004).
Percentage of HCoV‐NL63 patients with indicated symptom.
Lower respiratory tract illness nonpneumonia.
Medical records were available for 18 of the 28 patients.
ND, not determined; HCoV, human coronavirus.
A link between HCoV‐NL63 and respiratory diseases was established in the German prospective population‐based study (PRI.DE) on lower respiratory tract illness in children less than 3 years of age (2004, 2004). That study demonstrated a strong association between laryngotracheitis (croup) and HCoV‐NL63 infection. Croup is an inflammation of the trachea and is characterized by a loud barking cough that usually worsens at night. Almost half of the HCoV‐NL63 positive patients were diagnosed with croup and a significantly higher fraction of samples from croup patients contained HCoV‐NL63 RNA (17.4%) compared to noncroup patients (4.2%, P<0.0001). However, in other studies, not many croup patients are among the HCoV‐NL63 positives (Table 3). Patient selection in these studies may explain this discrepancy. Croup patients are not often hospitalized, and even less frequently sampled for respiratory virus diagnostics. For this reason, croup patients are under‐represented in most studies that analyzed hospitalized RTI patients. The PRI.DE study and the Hong Kong study were the only population‐based studies, thus excluding a bias in patient selection. Both studies observed the high frequency of croup among the HCoV‐NL63 positive patients (43% and 27%, respectively) (2005, 2005).
Interestingly, HCoV‐NL63 has also been associated with Kawasaki disease (Esper et al., 2005a). Respiratory secretions from 11 children with Kawasaki disease were tested and eight samples were positive for HCoV‐NL63 (73%). HCoV‐NL63 was detected in only one subject of a control group of 22 patients matched for age with the Kawasaki disease patients who visited the hospital in the same period. These results published by Esper et al. are of obvious relevance. Kawasaki disease is one of the most common forms of childhood vasculitis (Burns & Glode, 2004). It presents with prolonged fever and a polymorphic exanthem, oropharyngeal erythema, and bilateral conjunctivitis. Scarlet fever and measles are the closest clinical mimics. A number of epidemiological and clinical observations suggested previously that an infectious agent might be the cause of Kawasaki disease. These observations include the acute self‐limiting nature of the febrile illness, the peak of incidence in patients under 18 months, the geographic clustering of outbreaks, and the seasonal predominance of the illness in late winter/early spring (Kuijpers et al., 1999). Coronary complications can be reduced significantly by the use of intravenous immunoglobulin therapy combined with oral aspirin (Burns, 2001). The link between HCoV‐NL63 and Kawasaki disease is fascinating; however, multiple groups recently questioned the association between Kawasaki disease and HCoV‐NL63. These groups screened for HCoV‐NL63 in respiratory material of Kawasaki disease patients, but none were able to confirm the findings by Esper et al. (2005, 2006, 2005b, 2005).
Sero‐prevalence
The ‘common cold’ coronaviruses are responsible for 10–30% of common cold cases each winter season (McIntosh, 1996). This means that during a lifetime virtually everybody will experience an infection with these viruses, and everybody will consequently carry antibodies against these viruses. Expression of a fragment of the HCoV‐NL63 spike protein – a part unique to HCoV‐NL63 – in Escherichia coli and subsequent analysis for antibody recognition revealed that this protein was recognized in all healthy adults who were tested (n=18) on Western blot. These sera were also capable of virus neutralization in cell culture infections, indicating that adults have neutralizing antibodies that are specific for HCoV‐NL63 (K. Pyrc and L. van der Hoek, unpublished results). This result was confirmed in quantitative infection assays using pseudotyped virus. Pseudotyping is a method that uses pseudovirions to investigate the interaction between the spike protein of HCoV‐NL63 and the target cell. The functional organization of coronavirus spike proteins is similar to that of the glycoprotein from retroviruses, and spike proteins can be incorporated into the membrane of retrovirus particles. These pseudovirions accurately mimic receptor engagement and membrane fusion of the coronavirus and can be used to study the capacity of sera to inhibit binding of the coronavirus spike to the target cell. Retroviral particles expressing the HCoV‐NL63 spike protein or the HCoV‐229E spike protein were compared in neutralization assays. Sera that showed specific reactivity against the NL63 spike on western blot and that neutralized the virus in cell culture infections also neutralized infection in the NL63‐pseudovirions assay (Hofmann et al., 2005). Surprisingly, a completely different result was obtained with the 229E‐pseudovirions. HCoV‐229E neutralization occurred only in a minority of the samples from adults (Hofmann et al., 2005). This result reinforces the idea that infection with HCoV‐NL63 is much more common than HCoV‐229E infection. Neutralizing NL63‐antibodies are, on average, acquired before the age of 8 years, but thus far only a limited number of sera from the different age groups have been tested (Hofmann et al., 2005). Screening a larger number of samples with an NL63‐specific‐ELISA will allow a more precise determination of the average age at which HCoV‐NL63 is acquired. An NL63‐ELISA will also prove valuable to critically test whether Kawasaki disease in young children is accompanied by seroconversion or an increase in HCoV‐NL63‐specific antibodies. Such results are needed to further document the putative role of HCoV‐NL63 as the causative agent of Kawasaki disease.
Entry of the target cell by HCoV‐NL63
Cell tropism and receptor usage
Coronaviruses bind to cellular receptors via the spike protein to mediate infection of specific target cells. The spike of HCoV‐NL63 is a class I fusion protein, similar to the influenza virus haemagglutinin and the HIV‐1 Env glycoprotein gp120/gp41. The amino‐terminal part of the spike protein (S1) contains the receptor‐binding domain and the carboxy‐terminal part (S2) contains a membrane‐spanning region, two heptad repeat regions (HR1 and HR2) and the fusion peptide. Before the discovery of HCoV‐NL63 it was generally thought that all group I coronaviruses use CD13 (also known as aminopeptidase N) as receptor, in view of the fact that it was described for HCoV‐229E, porcine coronaviruses TGEV (transmissible gastroenteritis virus) and PRCoV (porcine respiratory coronavirus), and the feline and canine coronaviruses (1992, 1996, 1992). Unexpectedly, HCoV‐NL63 is not able to use CD13 as receptor for cell entry (Hofmann et al., 2005). The cell types that support virus replication already suggested an unusual receptor usage of HCoV‐NL63. The virus replicates in monkey kidney cells (2004, 2004), whereas its closest relative, HCoV‐229E, does not. Intriguingly, SARS‐CoV is also able to replicate in monkey kidney cells. Being from a different coronavirus group (SARS‐CoV is from group IIb), this similarity in cell tropism is surprising. A more detailed analysis of the cell tropism revealed that certain human cells can also be infected by HCoV‐NL63, namely Huh‐7 cells: a human hepatocellular carcinoma cell line (Hofmann et al., 2005). This underscores the resemblance of HCoV‐NL63 and SARS‐CoV because Huh‐7 cells can also be infected by the latter coronavirus. The shared cell tropism is suggestive of a shared receptor usage by HCoV‐NL63 and SARS‐CoV. SARS‐CoV was unique among the coronaviruses in that it uses angiotensin converting enzyme 2 (ACE2) to bind to and to enter target cells (Li et al., 2003). ACE2 is a surface molecule that was not known previously as a receptor for coronaviruses. Subsequently, Hofmann et al. (2005) showed that HCoV‐NL63 is the only other coronavirus next to SARS‐CoV that uses ACE2 as entry receptor. This surface molecule is localized to the ciliated cells of human nasal and tracheobronchial airway epithelia, thus supporting the presence of virus infection in the upper airways (Sims et al., 2005).
The HCoV‐NL63 receptor ACE2 is a homologue of the ACE protein, and both are key enzymes of the renin–angiotensin system. ACE activates the renin–angiotensin system by cleaving angiotensin I into angiotensin II, whereas ACE2 negatively regulates this system by inactivating angiotensin II. The renin–angiotensin system has a crucial role in severe acute lung injury (Imai et al., 2005), with ACE2 playing a protective role in lung failure and its counterpart ACE promoting lung oedemas and impaired lung function during acute lung injury. Interestingly, infection with SARS‐CoV suppresses the expression of ACE2 protein and it has been hypothesized that the reduced level of ACE2 during SARS‐CoV infection is the main cause of the severe pneumonia and acute, often lethal, lung failure (Kuba et al., 2005). It seems important to compare the pathogenicity of SARS‐CoV and HCoV‐NL63 with respect to ACE2 binding and downregulation. The reduced pathogenicity of HCoV‐NL63 suggests that ACE2 binding by the virus is not the only factor that determines the severity of viral pathogenicity. Further research on this subject is warranted, especially on the modulation of the ACE2 expression levels during HCoV‐NL63 infection, to understand the difference in lung pathogenicity of SARS‐CoV and HCoV‐NL63.
pH‐dependent cell entry
The internalization of coronaviruses into the host cell occurs either by direct fusion with the plasma membrane or by endocytosis and subsequent fusion with the endosomal membrane. Viruses that use the latter entry route can be inhibited by lysosomotropic agents that lower the endosomal pH such as bafilomycin A, chloroquine and NH4Cl. Treatment of Huh‐7 cells with bafilomycin A or NH4Cl revealed that entry driven by NL63‐S protein depends on the low‐pH environment in intracellular vesicles (Hofmann et al., 2005). The same has been described for HCoV‐229E infection and SARS‐CoV‐S mediated infection (2001, 2004, 2004). There is also one study that reported the opposite result (Huang et al., 2005). In that report, NL63‐S mediated infection of ACE2 expressing‐HEK293T cells was not greatly influenced by NH4Cl. In the same system, SARS‐CoV‐S mediated entry showed strong pH dependence, similar to results described previously. The same study also demonstrated that cathepsin function is not essential for HCoV‐NL63 infection. Cathepsins are a diverse group of endosomal and lysosomal proteases with endo‐ and exopeptidase activity. One of these, cathepsin L, facilitates endosomal entry of SARS‐CoV (2005, 2005).
Transcription and replication of HCoV‐NL63
Unfortunately, an infectious cDNA clone of HCoV‐NL63 is currently not available. For this reason, basic research on the molecular properties of the virus and its replication cycle has been limited. Inspection of the HCoV‐NL63 genome and comparison with the genomes of other coronaviruses led to several predictions concerning its life cycle and replication strategies. The genome of HCoV‐NL63 contains important signals that are needed for efficient replication (Pyrc et al., 2005). Translation of the genomic RNA generates one large polyprotein encoded by the 1a gene, and a −1 ribosomal frameshift results in the production of the 1ab polyprotein. A potential pseudoknot RNA structure that could facilitate the frameshift for HCoV‐NL63 is located within the RNA‐dependent RNA polymerase gene. The predicted pseudoknot structure consists of a hairpin with a highly conserved 11‐base pair stem, and a loop of eight nucleotides that forms five base pairs with a sequence 167 nucleotides further downstream. The heptanucleotide UUUAAAC sequence is present just upstream of the pseudoknot and probably acts as the slippery sequence where the actual ribosomal frameshift occurs (Pyrc et al., 2005).
Coronaviruses employ posttranslational proteolytic processing as a key regulatory mechanism in the expression of their replicative proteins. The HCoV‐NL63 1a and 1ab polyproteins are potentially cleaved by viral proteases to facilitate the assembly of a multisubunit protein complex that is responsible for viral replication and transcription. The genome of HCoV‐NL63 is predicted to encode two proteinases in the 5′ region of the 1a polyprotein (Pyrc et al., 2005). First, a papain‐like proteinase (PLpro) is expressed by the nonstructural protein (nsp) 3 gene situated near the 5′ end of the genome. This putative papain‐like proteinase of HCoV‐NL63 consists of two domains – PL1pro and PL2pro– and both are expected to have catalytic activity. The enzyme is predicted to cleave the 1a/1b protein at three sites between nsp1|nsp2, nsp2|nsp3, and nsp3|nsp4, releasing the functional papain‐like proteinase protein (nsp3) molecule by auto‐cleavage (Table 4). Analysis of the predicted cleavage sites of PLpro indicates that this HCoV‐NL63 enzyme has the specificity to cut between two small amino acids with short uncharged side chains, similar to homologous enzymes in other coronaviruses (Lim & Liu, 1998).
Table 4.
Putative protein cleavage of the HCoV‐NL63 1a/1b polyprotein
| Protein | Cleavage site | Processing enzyme |
|---|---|---|
| nsp1 | FGHGAG|SVVFVD | PLpro |
| nsp2 | FTKLAG|GKISFS | PLpro |
| nsp3; PLpro | VAKQGA|GFKRTY | PLpro |
| nsp4 | YNSTLQ|SGLKKM | Mpro |
| nsp5; Mpro | YGVNLQ|SGKVIF | Mpro |
| nsp6 | KISTVQ|SKLTDL | Mpro |
| nsp7 | NSSTLQ|SVASSF | Mpro |
| nsp8 | RVVKLQ|NNEIMP | Mpro |
| nsp9 | ATIRLQ|AGKQTE | Mpro |
| nsp10 | DRTTIQ|SVDISY | Mpro |
| nsp12; RdRp | NSTILQ|AAGLCV | Mpro |
| nsp13; MBD NTPase Helicase | KHADLH|SSQVCG | Mpro |
| nsp14; ExoN | IETNLQ|SLENIA | Mpro |
| nsp15; NendoU | FYPQLQ|SAEWKC | Mpro |
| nsp16; 2′‐O‐MT | – | – |
HCoV, human coronavirus; nsp, nonstructural protein; PLpro, papain‐like proteinase; Mpro, main proteinase.
The nsp5 gene is predicted to encode the second proteinase of HCoV‐NL63. It contains a serine‐like proteinase domain and is designated the main proteinase (Mpro) to stress its important function. The putative NL63‐Mpro processes the majority of cleavage sites between nonstructural proteins in the 1a/1b polyprotein (Pinon et al., 1999). Analysis of the Mpro cleavage sites suggests that the substrate specificity of NL63‐Mpro is different from that of the other coronaviruses. The cleavage site between nsp13 and nsp14 contains a histidine residue at position P1 instead of the standard glutamine that is present in other coronavirus species (Table 4). The histidine at P1 is present in all HCoV‐NL63 isolates sequenced thus far (Amsterdam 1, Amsterdam 57, Amsterdam 496 and NL). Intriguingly, the recently discovered HCoV‐HKU1 virus seems to share this unusual cleavage site (Woo et al., 2005a).
HCoV‐NL63 genome replication generates several subgenomic mRNAs for the spike, ORF3, envelope, membrane and nucleocapsid protein genes (Pyrc et al., 2004). All subgenomic mRNAs have an identical 5′ part, the untranslated leader of 72 nt. Creation of subgenomic mRNA requires pausing at a transcription regulatory sequence immediately upstream of the ORF during negative strand RNA synthesis. Subsequently, the nascent RNA chain switches in a strand transfer event to a transcription regulatory sequence located in the 5′ untranslated region of the genome such that a common leader sequence is added to each subgenomic mRNA. The transcription regulatory sequence motif of HCoV‐NL63 is AACUAAA (Pyrc et al., 2004). This transcription regulatory sequence core sequence is conserved in all subgenomic mRNA, except for the envelope protein gene that has the suboptimal transcription regulatory sequence core AACUAUA (Pyrc et al., 2004). The subgenomic mRNA for the nucleocapsid protein is the most abundant and there is an inverse correlation between the distance of a gene to the 3′ untranslated region and its RNA expression level. The envelope protein gene forms a notable exception for which the amount of the subgenomic mRNA is lower than expected, which may be due to the suboptimal transcription regulatory sequence motif (Pyrc et al., 2004).
Antiviral agents
An effective antiviral treatment may be required for HCoV‐NL63‐infected patients that end up in the intensive care unit due to acute respiratory disease. Several inhibitors are known to reduce replication of at least some coronaviruses including HCoV‐NL63 (2006a, 1987). These inhibitors act at various steps of the coronavirus replication cycle, e.g. receptor binding, membrane fusion, transcription and posttranslational processing.
An interesting HCoV‐NL63 inhibitor is intravenous immunoglobulin (Pyrc et al., 2006a), which is already approved as an intravenously delivered drug by the Food and Drug Administration. Intravenous immunoglobulin has been used successfully to treat several diseases, mostly primary immune deficiencies and autoimmune neuromuscular disorders, but also respiratory diseases (e.g. RSV; Hemming et al., 1987) and Kawasaki disease (Stiehm et al., 1987).
Inhibition of viral replication can also occur at the level of fusion of the viral and cellular membranes. The spike of HCoV‐NL63 contains two heptad repeat regions, HR1 and HR2, situated in the S2 part of the spike protein close to the transmembrane domain. After binding of virus to the receptor, a conformational change leads to the formation of a six‐helix bundle containing three HR1s and three HR2s and subsequent exposure of the fusion peptide mediates membrane fusion between the virus and the host cell. For retroviruses, paramyxoviruses and coronaviruses (2004, 2006a), peptides derived from the HR2 domain can inhibit virus infection, most likely by interacting with HR1. The peptide thus blocks formation of the natural HR1–HR2 interaction, prevents membrane fusion and as a consequence reduces infection.
Another novel means to inhibit replication is RNA interference (RNAi; Haasnoot et al., 2003). Pyrc et al. (2006a) explored the antiviral potential of small interfering RNA (siRNA) targeting HCoV‐NL63. The inhibitory properties of two siRNAs targeting conserved sequences within the spike protein gene were analyzed in cell culture infections. Transfection of a relatively low amount of siRNA into HCoV‐NL63‐susceptible cells made them resistant to virus infection.
HCoV‐NL63 can also be inhibited at the transcriptional level by pyrimidine nucleoside analogues: β‐d‐N4‐hydroxycytidine and 6‐azauridine (Pyrc et al., 2006a). As yet, however, the exact mechanism by which these agents inhibit HCoV‐NL63 transcription is unclear. Finally, protease inhibitors act at the level of posttranslational processing. The Mpro of coronaviruses has a highly conserved substrate‐recognition pocket, thus providing the opportunity to design broad‐spectrum antiviral drugs against several coronavirus species (Yang et al., 2005). Yang et al. designed main protease inhibitors and measured the inhibitory capacity in an in vitro protease activity assay. One potent inhibitor, N3, showed wide‐spectrum inhibition of various Mpro enzymes, including the one encoded by HCoV‐NL63.
Conclusions
HCoV‐NL63 was first described in 2004, but it soon became clear that this virus is not an emerging virus in humans. The isolate from 1988 proves this, but also the literature from the mid 1960s and 1980s indicates that human coronaviruses other than HCoV‐229E and HCoV‐OC43 circulated in those days. The symptoms observed during infection with HCoV‐NL63 resemble those observed in children infected with HCoV‐229E and HCoV‐OC43: upper and lower respiratory tract illnesses. However, the high frequency of croup is typical for HCoV‐NL63 infection. The first publications on HCoV‐NL63 were from two Dutch research groups, but infection with this virus is not restricted to the Netherlands. Numerous groups have reported HCoV‐NL63 infections worldwide. The virus was detected in 1–10% of the screened patient groups and a peak is observed in the winter season in most countries. Only in Hong Kong has a spring–summer seasonality been described, which may be caused by the subtropical climate.
The HCoV‐NL63 genome sequence is most similar to HCoV‐229E, and has a low similarity to the group II coronaviruses HCoV‐OC43, HCoV‐HKU1 and SARS‐CoV. Given the evolutionary distance with the latter virus, it is striking that HCoV‐NL63 and SARS‐CoV share the same receptor for target cell entry. This receptor, ACE2, is a member of the renin–angiotensin system, but it is also involved in the protection against lung damage. The influence of HCoV‐NL63 infection on ACE2 expression is of interest as it has been postulated that the down‐regulation of ACE2 during SARS‐CoV infection is a major factor in the pathogenicity of SARS.
The identification of NL63‐variants with distinct genetic markers underscores that HCoV‐NL63 was not recently introduced into the human population. The clustering of the NL63‐variants is not based on geographical distribution, as the different subtypes appear to coexist. Phylogenetic analysis of the 1a and spike genes of field isolates supplies evidence for the occurrence of recombination. The shared receptor usage by SARS‐CoV provides the opportunity for double infection of the same cell. Consequently, there is a risk of recombination between these two viruses, with the possibility that more pathogenic variants of HCoV‐NL63 can evolve. Several antiviral agents are in development that potentially inhibit HCoV‐NL63 infection, and these can be used to develop an effective antiviral treatment.
Finally, there is an urgent need for an infectious cDNA clone and an appropriate animal model system to study the virus in more detail. Those tools would provide the possibility to investigate pathogenicity markers of the virus, and to compare the differences between SARS‐CoV and HCoV‐NL63 infections.
Acknowledgements
L.vd.H. is supported by VIDI grant 016.066.318 from Netherlands Organization for Scientific Research (NWO), The Netherlands.
Editor: Arie van der Ende
References
- Arden KE, Nissen MD, Sloots TP & Mackay IM (2005) New human coronavirus HCoV‐NL63 associated with severe lower respiratory tract disease in Australia. J Med Virol 75: 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N, Anderson K, Hart L, Van Caeseele P, Brandt K, Milley D, Hatchette T, Weiss EC & Li Y (2005a) Human coronavirus NL63 infection in Canada. J Infect Dis 191: 503–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N, Robinson JL, Tse A, Lee BE, Hart L & Li Y (2005b) Human coronavirus NL‐63 infections in children: a 1-year study. J Clin Microbiol 43: 4567–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belay ED, Erdman DD, Anderson LJ, Peret TC, Schrag SJ, Fields BS, Burns JC & Schonberger LB (2005) Kawasaki disease and human coronavirus. J Infect Dis 192: 352–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau DM & Holmes KV (2001) Human coronavirus HCoV‐229E enters susceptible cells via the endocytic pathway. Adv Exp Med Biol 494: 193–198. [DOI] [PubMed] [Google Scholar]
- Bosch BJ, Martina BE, Van Der Zee R, Lepault J, Haijema BJ, Versluis C, Heck AJ, De Groot R, Osterhaus AD & Rottier PJ (2004) Severe acute respiratory syndrome coronavirus (SARS‐CoV) infection inhibition using spike protein heptad repeat‐derived peptides. Proc Natl Acad Sci USA 101: 8455–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradburne AF, Bynoe ML & Tyrrell DA (1967) Effects of a ‘new’ human respiratory virus in volunteers. Br Med J 3: 767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC (2001) Kawasaki disease. Adv Pediatr 48: 157–177. [PubMed] [Google Scholar]
- Burns JC & Glode MP (2004) Kawasaki syndrome. Lancet 364: 533–544. [DOI] [PubMed] [Google Scholar]
- Chang LY, Chiang BL, Kao CL, Wu MH, Chen PJ, Berkhout B, Yang HC & Huang LM (2006) Lack of association between infection with a novel human coronavirus (HCoV) HCoV‐NH and Kawasaki Disease in Taiwan. J Infect Dis 193: 283–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SS, Chan KH, Chu KW, Kwan SW, Guan Y, Poon LL & Peiris JS (2005) Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong China. Clin Infect Dis 40: 1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B, Gelfi J, L'Haridon R, Vogel LK, Sjostrom H, Noren O & Laude H (1992) Aminopeptidase N is a major receptor for the entero‐pathogenic coronavirus TGEV. Nature 357: 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E (2002) Quasispecies theory in virology. J Virol 76: 463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW & Holland JJ (1999) Mutation rates among RNA viruses. Proc Natl Acad Sci USA 96: 13910–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Gunther S, Preiser W, et al (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348: 1967–1976. [DOI] [PubMed] [Google Scholar]
- Ebihara T, Endo R, Ma X, Ishiguro N & Kikuta H (2005a) Detection of human coronavirus NL63 in young children with bronchiolitis. J Med Virol 75: 463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara T, Endo R, Ma X, Ishiguro N & Kikuta H (2005b) Lack of association between New Haven coronavirus and Kawasaki disease. J Infect Dis 192: 351–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F, Shapiro ED, Weibel C, Ferguson D, Landry ML & Kahn JS (2005a) Association between a novel human coronavirus and Kawasaki Disease. J Infect Dis 191: 499–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F, Weibel C, Ferguson D, Landry ML & Kahn JS (2005b) Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J Infect Dis 191: 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J, Ihorst G, Rieger CH, et al (2004) Prospective population‐based study of viral lower respiratory tract infections in children under 3 years of age (the PRI DE study). Eur J Pediatr 163: 709–716. [DOI] [PubMed] [Google Scholar]
- Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, De Jong JC, Simon JH & Osterhaus AD (2004) A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA 101: 6212–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot PCJ, Cupac D & Berkhout B (2003) Inhibition of virus replication by RNA interference. J Biomed Sci 10: 607–616. [DOI] [PubMed] [Google Scholar]
- Hamre D & Procknow JJ (1966) A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med 121: 190–193. [DOI] [PubMed] [Google Scholar]
- Hemming VG, Rodriguez W, Kim HW, Brandt CD, Parrott RH, Burch B, Prince GA, Baron PA, Fink RJ & Reaman G (1987) Intravenous immunoglobulin treatment of respiratory syncytial virus infections in infants and young children. Antimicrob Agents Chemother 31: 1882–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendley JO, Fishburne HB & Gwaltney JM Jr (1972) Coronavirus infections in working adults. Eight‐year study with 229 E and OC 43. Am Rev Respir Dis 105: 805–811. [DOI] [PubMed] [Google Scholar]
- Herrewegh AA, Mahler M, Hedrich HJ, Haagmans BL, Egberink HF, Horzinek MC, Rottier PJ & De Groot RJ (1997) Persistence and evolution of feline coronavirus in a closed cat‐breeding colony. Virology 234: 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh AA, Smeenk I, Horzinek MC, Rottier PJ & De Groot RJ (1998) Feline coronavirus type II strains 79‐1683 and 79‐1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol 72: 4508–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Hattermann K, Marzi A, Gramberg T, Geier M, Krumbiegel M, Kuate S, Uberla K, Niedrig M & Pohlmann S (2004) S protein of severe acute respiratory syndrome‐associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J Virol 78: 6134–6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Pyrc K, Van Der Hoek L, Geier M, Berkhout B & Pohlmann S (2005) Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA 102: 7988–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang IC, Bosch BJ, Li F, et al (2005) SARS coronavirus but not human coronavirus NL63 utilizes cathepsin L to infect ACE2‐expressing cells. J Biol Chem 281: 3198–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Kuba K, Rao S, et al (2005) Angiotensin‐converting enzyme 2 protects from severe acute lung failure. Nature 436: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser L, Regamey N, Roiha H, Deffernez C & Frey U (2005) Human coronavirus NL63 associated with lower respiratory tract symptoms in early life. Pediatr Infect Dis J 24: 1015–1017. [DOI] [PubMed] [Google Scholar]
- Konig B, Konig W, Arnold R, Werchau H, Ihorst G & Forster J (2004) Prospective study of human metapneumovirus infection in children less than 3 years of age. J Clin Microbiol 42: 4632–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek TG, Erdman D, Goldsmith CS, et al (2003) A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348: 1953–1966. [DOI] [PubMed] [Google Scholar]
- Kuba K, Imai Y, Rao S, et al (2005) A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus‐induced lung injury. Nat Med 11: 875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers TW, Wiegman A, Van Lier RA, Roos MT, Wertheim‐van Dillen PM, Pinedo S & Ottenkamp J (1999) Kawasaki disease: a maturational defect in immune responsiveness. J Infect Dis 180: 1869–1877. [DOI] [PubMed] [Google Scholar]
- Lai MMC & Holmes KV (2001) Coronaviridae: the viruses and their replication. Fields Virology (Knipe DM. & Howley PM, eds), pp. 1163–1186. Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- Lai MM, Baric RS, Makino S, Keck JG, Egbert J, Leibowitz JL & Stohlman SA (1985) Recombination between nonsegmented RNA genomes of murine coronaviruses. J Virol 56: 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson HE, Reed SE & Tyrrell DA (1980) Isolation of rhinoviruses and coronaviruses from 38 colds in adults. J Med Virol 5: 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH & Yuen KY (2005) Severe acute respiratory syndrome coronavirus‐like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA 102: 14040–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moore MJ, Vasilieva N, et al (2003) Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426: 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu M, et al (2005) Bats are natural reservoirs of SARS‐like coronaviruses. Science 310: 676–679. [DOI] [PubMed] [Google Scholar]
- Lim KP & Liu DX (1998) Characterization of the two overlapping papain‐like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C‐terminal cleavage site of an 87‐kDa protein. Virology 245: 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tang F, Fontanet A, et al (2004) Long‐term SARS coronavirus excretion from patient cohort China. Emerg Infect Dis 10: 1841–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnaughton MR, Madge MH & Reed SE (1981) Two antigenic groups of human coronaviruses detected by using enzyme‐linked immunosorbent assay. Infect Immun 33: 734–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K (1996) Coronaviruses. Fields virology (Fields BN, Knipe DM. & Howley PM, et al, eds), 1095 pp. Lippincott‐Raven Publishers, Philadelphia. [Google Scholar]
- McIntosh K, Dees JH, Becker WB, Kapikian AZ & Chanock RM (1967) Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci USA 57: 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K, Chao RK, Krause HE, Wasil R, Mocega HE & Mufson MA (1974) Coronavirus infection in acute lower respiratory tract disease of infants. J Infect Dis 130: 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes E, Vijgen L, Keyaerts E, Zlateva K, Li S, Maes P, Pyrc K, Berkhout B, Van Der Hoek L & Van Ranst M (2005) A novel pancoronavirus RT‐PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect Dis 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Holmes EC & Gonzalez‐Candelas F (2004) The population genetics and evolutionary epidemiology of RNA viruses. Nat Rev Microbiol 2: 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhaus AD, Fouchier RA & Kuiken T (2004) The aetiology of SARS: Koch's postulates fulfilled. Philos Trans R Soc London B Biol Sci 359: 1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon JD, Teng H & Weiss SR (1999) Further requirements for cleavage by the murine coronavirus 3C‐like proteinase: identification of a cleavage site within ORF1b. Virology 263: 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon LL, Chu DK, Chan KH, et al (2005) Identification of a novel coronavirus in bats. J Virol 79: 2001–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K, Jebbink MF, Berkhout B & Van Der Hoek L (2004) Genome structure and transcriptional regulation of human coronavirus NL63. Virol J 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K, Berkhout B & Van Der Hoek L (2005) Molecular characterization of human coronavirus NL63 Recent Research Developments in Infection & Immunity (Pandelai SG, ed.), pp. 25–48. Transworld Research Network, Kerala, India. [Google Scholar]
- Pyrc K, Bosch BJ, Berkhout B, Jebbink MF, Dijkman R, Rottier P & Van Der Hoek L (2006a) Inhibition of HCoV‐NL63 infection at early stages of the replication cycle. Antimicrob Agents Chemother 50: 2000–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SE (1984) The behaviour of recent isolates of human respiratory coronavirus in vitro and in volunteers: evidence of heterogeneity among 229E-related strains. J Med Virol 13: 179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu C, Shike H, Baker SC, et al (2005) Human coronavirus NL63 is not detected in the respiratory tracts of children with acute Kawasaki Disease. J Infect Dis 192: 1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ & Bates P (2004) Characterization of severe acute respiratory syndrome‐associated coronavirus (SARS‐CoV) spike glycoprotein‐mediated viral entry. Proc Natl Acad Sci USA 101: 4240–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL & Bates P (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA 102: 11876–11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims AC, Baric RS, Yount B, Burkett SE, Collins PL & Pickles RJ (2005) Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol 79: 15511–15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD & Mackay IM (2006) Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol 35: 99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephensen CB, Casebolt DB & Gangopadhyay NN (1999) Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res 60: 181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiehm ER, Ashida E, Kim KS, Winston DJ, Haas A & Gale RP (1987) Intravenous immunoglobulins as therapeutic agents. Ann Intern Med 107: 367–382. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Okamoto M, Ohmi A, Watanabe O, Miyabayashi S & Nishimura H (2005) Detection of human coronavirus‐NL63 in children in Japan. Pediatr Infect Dis J 24: 645–646. [DOI] [PubMed] [Google Scholar]
- Tresnan DB, Levis R & Holmes KV (1996) Feline aminopeptidase N serves as a receptor for feline canine porcine and human coronaviruses in serogroup I. J Virol 70: 8669–8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell DAJ & Bynoe ML (1965) Cultivation of novel type of common‐cold virus in organ cultures. Br Med J 1: 1467–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A, Mourez T, Dina J, Van Der Hoek L, Gouarin S, Petitjean J, Brouard J & Freymuth F (2005) Human coronavirus NL63 France. Emerg Infect Dis 11: 1225–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A, Dina J, Gouarin S, Petitjean J, Corbet S & Freymuth F (2006) Detection of the new human coronavirus HKU1: a report of 6 cases. Clin Infect Dis 42: 634–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Hoek L & Berkhout B (2005) Questions concerning the New Haven coronavirus. J Infect Dis 192: 350–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Hoek L, Pyrc K, Jebbink MF, Vermeulen‐Oost W, Berkhout RJ, Wolthers KC, Wertheim‐van Dillen PM, Kaandorp J, Spaargaren J & Berkhout B (2004) Identification of a new human coronavirus. Nat Med 10: 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Hoek L, Sure K, Ihorst G, Stang A, Pyrc K, Jebbink MF, Petersen G, Forster J, Berkhout B & Uberla K (2005) Croup is associated with the novel coronavirus NL63. PLoS Med 2: e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elden LJ, Van Loon AM, Van Alphen F, Hendriksen KA, Hoepelman AI, Van Kraaij MG, Oosterheert JJ, Schipper P, Schuurman R & Nijhuis M (2004) Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real‐time reverse‐transcriptase polymerase chain reaction. J Infect Dis 189: 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Huang Y, Lau SK, Tsoi HW & Yuen KY (2005a) In silico analysis of ORF1ab in coronavirus HKU1 genome reveals a unique putative cleavage site of coronavirus HKU1 3C‐like protease. Microbiol Immunol 49: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Chu CM, et al (2005b) Characterization and complete genome sequence of a novel coronavirus HKU1 from patients with pneumonia. J Virol 79: 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Tsoi HW, et al (2005c) Clinical and molecular epidemiological features of coronavirus HKU1‐associated community‐acquired pneumonia. J Infect Dis 192: 1898–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Xie W, Xue X, et al (2005) Design of wide‐spectrum inhibitors targeting coronavirus main proteases. PLoS Biol 3: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT & Holmes KV (1992) Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357: 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]


