Abstract
Foreign nucleic acids, the essential signature molecules of invading pathogens that act as danger signals for host cells, are detected by endosomal nucleic acid-sensing toll-like receptors (TLRs) 3, 7, 8, 9, and 13. These TLRs have evolved to recognize ‘non-self’ nucleic acids within endosomal compartments and rapidly initiate innate immune responses to ensure host protection through induction of type I interferons, inflammatory cytokines, chemokines, and co-stimulatory molecules and maturation of immune cells. In this review, we highlight our understanding of the recognition of pathogen-associated nucleic acids and activation of corresponding signaling pathways through endosomal nucleic acid-sensing TLRs 3, 7, 8, 9, and 13 for an enormous diversity of pathogens, with particular emphasis on their compartmentalization, intracellular trafficking, proteolytic cleavage, autophagy, and regulatory programs.
Keywords: innate immunity, endosomal nucleic acid-sensing toll-like receptor, danger signal, compartmentalization, signaling pathway
Introduction
As a universal and ancient form of host defense against invading pathogens, the innate immune system has developed a wide variety of germ-line encoded host sensors known as pattern-recognition receptors (PRRs). PRRs recognize danger signals that are essential, structure-conserved, and specific signature molecules of a diverse array of pathogens, which are called pathogen-associated molecular patterns (PAMPs), and danger/damage-associated molecular patterns that are molecules released from intracellular stores of damaged or stressed host cells. This recognition triggers the secretion of a large amount of inflammatory cytokines and antiviral cytokines, such as type I interferons (IFNs), for effective host defense by eradicating infectious pathogens and for the development of long-lasting pathogen-specific adaptive immunity through B and T cells [1–3].
Toll-like receptors (TLRs) are evolutionarily conserved germ-line encoded PRRs thought to be the primary sensors of infective pathogens. As an important family of type I transmembrane (TM) glycoprotein receptors, all TLRs are composed of an ectodomain (ECD) containing multiple leucine-rich repeats (LRRs) directly involved in the recognition of PAMPs, a TM domain required for the subcellular localization of TLRs, and an intracellular domain with a conserved cytoplasmic signaling region called the Toll/IL-1 receptor (TIR), which is required for transduction of downstream signaling [1–4].
A total of 10 TLRs in humans and 12 TLRs in mice have been identified to date. TLRs 1, 2, 4, 5, 6, and 10 are primarily expressed on the surface of immune cells and mainly recognize bacterial and fungal cell wall components and viral envelope proteins, as well as protozoal components [1,2,5]. In contrast, the nucleic acid-sensing TLRs, which include TLRs 3, 7, 8, 9, and 13 (only expressed in mice), are completely localized within the endosomal compartments of immune cells and recognize double stranded RNA (dsRNA), single stranded RNA (ssRNA) and DNA derived from viruses, bacteria, fungi, and parasites, respectively. Upon recognition of respective foreign nucleic acids, signal transduction through these receptors is initiated by the recruitment of myeloid differentiation factor 88 (MyD88) or the TIR domain-containing adapter molecule (TRIF) (also known as the TIR domain containing adapter molecule 1, TICAM1) that results in the activation of IFN-regulatory factor 3 (IRF3), IRF7, activator protein 1 (AP-1), and nuclear transcription-κB (NF-κB) and transcription of inflammatory cytokines, type I IFNs, chemokines, and antimicrobial peptides and innate immune response genes (Table 1) [1–5]. However, recognition of ‘self’-nucleic acids by the nucleic acid-sensing TLRs contributes to the pathology associated with several autoimmune or autoinflammatory diseases under certain pathological conditions. In this review, we discuss the recent progress in studies of pathogen-associated nucleic acid recognition and signaling pathways within endosomal compartments, and the individual functions of TLRs 3, 7, 8, 9, and 13 in innate immunity. In addition, we discuss the compartmentalization, intracellular trafficking, proteolytic cleavage, autophagy, and regulatory programs of these TLRs in innate immunity.
Table 1.
Summary of the signaling pathways of TLR3, 7, 8, 9, and 13
| Type | Subcellular localization | Natural and synthetic ligand | Adaptor | Transcription factor | Effector cytokines | References |
|---|---|---|---|---|---|---|
| TLR3 | Endosomes | dsRNAs, siRNAs, poly(I:C) | TRIF | NF-κB, AP-1, IRF3 | Inflammatory cytokines, type I interferons | [1,2,5–7] |
| TLR7 | Endosomes | ssRNAs, ORNs, siRNAs, imidazoquinolines | MyD88, PACSIN1 | NF-κB, AP-1, IRF3/7 | Inflammatory cytokines, type I interferons | [1,2,5,7–11] |
| TLR8 | Endosomes | ssRNAs, ORNs, siRNAs, poly(A)/T, DNA, imidazoquinolines | MyD88 | NF-κB, AP-1, IRF3/7 | Inflammatory cytokines, type I interferons | [1,2,5,7,9,12] |
| TLR9 | Endosomes | CpG DNAs, CpG ODNs | MyD88, PACSIN1 | NF-κB, AP-1, IRF3/7 | Inflammatory cytokines, type I interferons | [1,2,5,7–11] |
| TLR13 | Endosomes | 23S rRNA | MyD88 | NF-κB, AP-1, IRF3/7 | Inflammatory cytokines, type I interferons | [13–15] |
Distribution, Subcellular Localization and Regulation of TLRs 3, 7, 8, 9, and 13
Distribution of TLRs 3, 7, 8, 9, and 13
The expression of TLRs 3, 7, 8, 9, and 13 is distinct in different cell types of human and murine origins. TLR3 is observed in the human myeloid DCs (mDCs), macrophages, natural killer cells, and B cells [6,16]. TLR3 mRNA has also been detected in epithelial cells of many different organs including airway, uterine, corneal, vaginal, biliary, and intestinal organs because the mucous membranes of these organs act as efficient innate barriers to infection from the respiratory tract, and gastrointestinal and urogenital systems [6,16]. Surprisingly, higher expression level of TLR3 is also observed in neurons, astrocytes, and microglia, suggesting a specific function in response to encephalitogenic viruses of the central nervous system [6,17,18]. TLRs 7 and 9 are predominantly expressed in plasmacytoid DCs (pDCs) in humans and are involved in strong IFN-α induction, but TLR8 tends to be detected in human monocytes and mDCs, and predisposes the induction of inflammatory cytokines [1,19–23]. In mice, TLR7 is highly expressed in pDCs, while TLR8 is expressed in all four splenic DC subsets including CD4+ DCs, CD8+ DCs, CD4− CD8− DCs, and pDCs. The precise function of murine TLR8 remains to be elucidated [1,19–23]. Expression of TLR9 is detected in murine B cells, pDCs, monocytes/macrophages, and conventional DCs (cDCs) [1,19–23]. Unlike it in mouse, TLR9 expression in human is restricted to pDCs and B cells [1,19–23], reflecting possible species-specific differences in the expression of TLRs 7, 8, and 9 between mouse and human. Interestingly, a recent report showed that virus-induced signaling adaptor (VISA) regulates B cell expression of TLR7 and CD23 [24], suggesting an unexpected link between TLRs and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs). In addition, TLR13 is a novel functional TLR that is predominantly expressed in cDCs and macrophages [13,14].
Compartmentalization and regulation of TLRs 3, 7, 8, 9, and 13
TLRs 3, 7, 8, 9, and 13 are localized in intracellular compartments, such as endosomes, lysosomes, multivesicular bodies, and endoplasmic reticulum (ER) [1,2,5,25,26]. However, they are only activated within acidified endosomal compartments, because TLRs 3-, 7-, 8-, 9-, and 13-induced responses to foreign nucleic acids are suppressed by some endosomal acidification inhibitors, such as chloroquine, ammonium chloride or bafilomycin A1 [1,2,5,25,26]. Upon stimulation, nucleic acid-sensing TLRs are translocated directly from the ER to endosomes via the conventional secretory pathway through the Golgi [25–27]. After pathogens are internalized, the endosomal nucleic acid-sensing TLRs enable them to recognize nucleic acids delivered to the endosomal compartments [14,25–27]. By contrast, cellular nucleic acids presented outside endosomes are rapidly degraded by nucleases and are not detected by endosomal nucleic acid-sensing TLRs. Therefore, the compartmentalization of these receptors appears to be a key regulatory mechanism to modulate ligand binding and to distinguish ‘self’ nucleic acids from ‘non-self’ nucleic acids [1,2,5,25–28]. Accordingly, the compartmentalization of nucleic acid-sensing TLRs is regulated by multiple regulatory mechanisms, including trafficking, proteolysis and autophagy, and involves the TM region of these TLRs. Failure to maintain a physical separation between ‘self’ nucleic acids and endosomal nucleic acid-sensing TLRs is frequently associated with autoimmune disorders [1,2,5,25–27,29–31].
Importance of the TM region of TLRs 3, 7, 8, 9, and 13
As described above, the TM domain of TLRs mediates their subcellular localization. Domain analysis using chimeric nucleic acid-sensing TLRs has revealed that the endosomal TLR3 is determined by the linker region between its TM and TIR domains, whereas the endosomal localization of TLRs 7, 8, and 9 is determined by their TM domains [28,31,32]. Interestingly, a tyrosine-based internalization motif in the cytosolic tail of human TLR9 or the TM α-helix of mouse TLR9 is required to target it to the endosomal compartments, but mutation of structural motifs of TLR9 results in its redistribution to the cell surface [33]. Surprisingly, cell-surface expression of a mutant form of TLR9 (TLR9TM-MUT) in TLR9-deficient myeloid cells resulted in more MyD88 recruitment to TLR9 in response to cytidine-phosphate-guanosine DNA (CpG-DNA) or self-DNA than that caused by expression of wild-type TLR9, even after inhibition of proteolytic processing [34].
Trafficking regulation of TLRs 3, 7, 8, 9, and 13
Trafficking and processing of nucleic acid-sensing TLRs in intracellular compartments are tightly regulated by a series of regulatory molecules to prevent the ‘self’ nucleic acids recognition. The heat shock protein glycoprotein 96 (gp96, also known as grp94) not only is required for folding and mobilization of cell surface TLR, but also regulates the intracellular trafficking of nucleic acid-sensing TLRs [35,36]. In the absence of gp96, TLR7 and TLR9 remain in the ER and gp96-deficient macrophages fail to respond to agonists of TLR7 and TLR9 [35,36]. Another chaperone proteins known as protein associated with TLR4 (PRAT4A, also known as Cnpy3) is required for trafficking of nucleic acid-sensing TLRs from the ER to endosomes and of cell-surface TLRs from the ER to the plasma membrane. Interestingly, PRAT4A associates with and translocates TLRs 7 and 9 rather than TLR3 from the ER to the endosomes, indicating that trafficking of TLRs 3, 7, and 9 is differentially regulated [37,38]. Therefore, gp96 and PRAT4A are critical for both TLR9 egress from the ER and for the conformational stability and function of these receptors in the endosomal compartments. In addition to gp96 and PRAT4A, regulatory molecules solute carrier family 15 member 4 (Slc15a4), adapter-related protein complex-3 (AP-3), hermansky-Pudlak syndrome proteins of the biogenesis of lysosome-related organelle complex (BLOC)-1 and BLOC-2 groups, phospholipid scramblase 1 (PLSCR1), and hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) are essential for TLR7 and/or TLR9-mediated specialized membrane trafficking in pDCs [39–41].
In addition, Unc93b1, an ER resident protein with 12 membrane-spanning domains, acts as a chaperone in association with TLRs 3, 7, 8, 9, and 13, and controls the translocation of these TLRs from the ER to endosomal compartments [42–44]. Unc93b1 is also required for cytokine production, up-regulation of co-stimulatory molecules, and efficient cross-presentation of exogenous antigens via MHC class I and class II molecules [42–44]. Although the precise mechanism by which Unc93b1 controls each TLR trafficking is yet to be elucidated, it is known that Unc93b1 is essential for the activation of TLRs 3, 7, 8, 9, and 13, and efficient translocation of each TLR from the ER to endosomes [42–44]. Unc93b1 with an H412R mutation cannot exit the ER and overexpression of Unc93b1 increases trafficking of TLRs 3, 7, 8, and 9 to endosomes [45]. Furthermore, TLRs 7 and 9, but not TLR3, compete with each other for association with Unc93b1. Under normal circumstances, Unc93b1 physically associates with TLR9, and overexpression of TLR9 inhibits TLR7 signal transduction, resulting in a weaker TLR7 response to RNA. However, a D34A mutation in Unc93b1 has been found to up-regulate ligand-induced trafficking of TLR7 and down-regulate that of TLR9, leading to systemic lethal inflammation [46–49]. This indicates that physical association between UNC93B and nucleic acid-sensing TLRs is essential and, specifically, Unc93B1 controls homeostatic TLR7 activation by balancing trafficking of TLR9 and TLR7 to protect host from the initiation of autoimmune diseases. Thus, the compartmentalization of nucleic acid-sensing TLRs is regulated by ER-resident molecules, such as gp96, PRAT4A, and Unc93b1.
Proteolytic regulation of TLRs 3, 7, 8, 9, and 13
Proteolytic cleavage of the ECDs of nucleic acid-sensing TLRs is important for the binding of DNA and/or RNA, recruitment of MyD88 or TRIF, and initiation of signal transduction within endosomes (Fig. 1). Three research groups have independently shown that TLR9 ECD is proteolytically cleaved and removed at its flexible loop between LRR 14 and 15 by multiple acid-dependent proteases within endosomal compartment, generating the resultant half ECD, as well as the TM and the cytosolic TIR domain. The TIR domain forms a functional signaling complex with MyD88 to directly mediate DNA recognition and initiate signal transduction [50–52]. Surprisingly, full-length TLR9 is found only in the ER, whereas the cleaved, functional TLR9 is restricted to within endosomal compartment [50–52]. Proteases involved in TLR9 cleavage include cathepsins, such as cathepsin B, cathepsin S, cathepsin L, cathepsin H, and cathepsin K, and asparagine endopeptidase (AEP) [53–55]. However, the structural basis for recognition of CpG DNA by functional cleavage of TLR9 remains unclear so far. Interestingly, specific deletion of LRR in TLR9 renders it unresponsive to CpG DNA, mutations in the positively charged N-terminal region of TLR9 cannot affect its binding toCpG DNA, indicating the importance of multiple LRRs in TLR9 activation [55–57].
Figure 1.
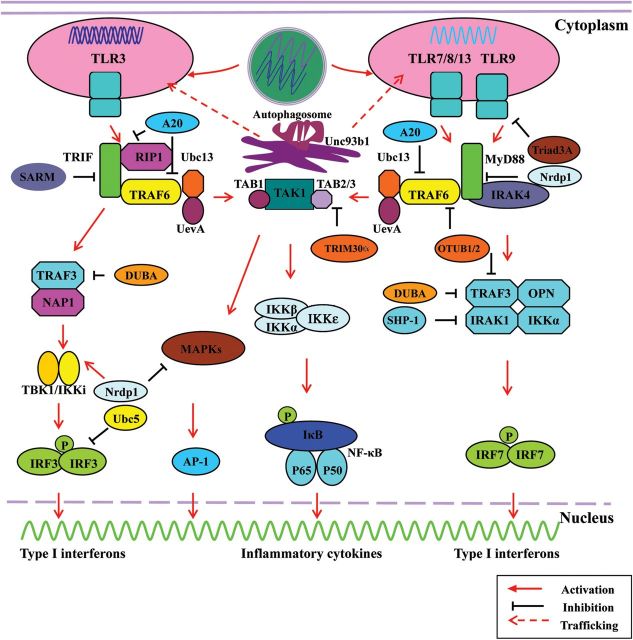
Overview of nucleic acid-sensing TLRs 3, 7, 8, 9, and 13 signaling pathways, compartmentalization, trafficking, and regulation mechanism Diagram showing the stimulation of TLR3, 7, 8, 9, and 13 by dsRNA, ssRNA, and CpG DNA, leading to the translocation of TLR3, 7, 8, 9, and 13 from the ER to endosomal compartments and activation of the MyD88- and TRIF-dependent signaling pathways, respectively. Upon stimulation, TLR3 recruits TRIF, which in turn recruits a set of adaptor molecules lead to the formation of a complex and the activation of IRF3, NF-κB, and AP-1. However, TLRs 7, 8, 9, and 13 interact with the TIR domain of MyD88 to recruit a set of adaptor molecules lead to the formation of a complex and the activation of IRF7, NF-κB, and AP-1. To achieve a balanced output, TLR3, 7, 8, 9, and 13 signaling pathways are positively and negatively regulated by modulatory molecules, such as A20, deubiquitinating enzyme A, SARM, SHP-2, Nrdp1, and Ubc5.
Recently, it has been shown that compartmentalized proteolytic cleavage of nucleic acid-sensing TLRs is a multistep process. The majority of the TLR9 ECD is to be removed and can be performed by AEP or multiple cathepsins, and then a trimming event mediated solely by cathepsin is required for optimal receptor signaling of functional TLR9 [55]. In addition, similar processing of TLR3 and TLR7 also occurs in murine RAW264.7 macrophages and/or human retinal epithelial cell line RPE1, implying that receptor proteolysis is a general regulatory strategy for restricting the capacity of all nucleic acid-sensing TLRs to signals only from the endosomal compartments where they sense nucleic acids [55,58,59].
Autophagy in regulation of TLRs 3, 7, 8, 9, and 13
Autophagy is an evolutionary and fundamental cellular homeostatic process involved in cell survival during starvation and in maintaining quality control of intracellular organelles [30,31,60–63]. The importance of autophagy in the regulation of innate immune recognition of pathogens in phagosomes or autophagosomes has been revealed by a number of recent studies. Autophagy is important for the regulation of nucleic acid-sensing TLRs, especially TLR7 [30,31,60–63]. A subset of nucleic acid-sensing TLR ligands, including synthetic nucleic acid-analogs, bacteria, viruses, fungi, and protozoa, can induce autophagy. Synthetic nucleic acid-analogs and pathogens are able to induce the formation of phagolysosomes or autolysosomes via fusion of double-membrane vesicles (phagosomes or autophagosomes) with late endosomes or lysosomes, which are highly acidifying environments containing abundant degradation enzymes. The ligands are then delivered to endosomal compartments where recognition by nucleic acid-sensing TLRs occurs in pDCs, macrophage cells, and B cells [30,31,60–63]. Although host ssRNAs can also be taken up by macrophages or pDCs during phagocytosis of necrotic cells/infected cells, they rarely reach the endosomal compartments. Several essential components of the autophagic machinery, such as autophagy-related proteins (ATGs) and Beclin, are required for phagolysosome or autolysosome formation, endosomal maturation, and activation of signaling pathways downstream of nucleic acid-sensing TLRs after ligand stimulation [30,31,64].
It has been reported that the engagement of TLR7 induces autophagy and promotes the elimination of Bacillus Calmette-Guerin (BCG) in autolysosomes [65,66]. In addition to this role in bacteria, the role of autophagy in defense against viruses in pDCs has been demonstrated, especially in the innate recognition of viral replication products, such as those of vesicular stomatitis virus (VSV) [60], Sendai virus (SeV) [60], herpes simplex virus (HSV) [60], influenza A virus (IAV), Dengue virus (DENV) [67], Poliovirus [68], coronaviruses [69], Coxsackie B virus (CBV) [70], foot and mouth disease virus (FMDV) [71], and hepatitis C virus (HCV) [72]. Two different ligands of TLR7, ssRNA and imiquimod, are able to induce the formation of autophagosomes via the TLR7-MyD88-dependent signaling pathway in macrophages, characterized by microtubule-associated light chain 3-green fluorescent protein (LC3-GFP) puncta formation for the elimination of BCG, and both Atg5 and Beclin are required for the induction of autophagy [65]. In the case of IAV, the virus internalizes into endosomes to be recognized by TLR7, but VSV and SeV replication intermediates formed during virus infection are required for recognition, and autophagosomes are necessary for host cells to respond to VSV and SeV infection via TLR7 [60]. In addition, pDCs take advantage of an autophagic process in which ‘self’ nucleic acids are degraded inside double-membrane vesicles called autophagosomes [60]. In the absence of ATG5 in pDCs, type I IFNs cannot be induced in response to CpG-DNA and HSV-1 infection [60]. It suggests that cytoplasmic virions are engulfed by autophagosomes, which then fuse with lysosomes. Autophagy may therefore control either the endosomal maturation required for CpG-DNA sensing. Alternatively, autophagy may regulate the TLR9-MyD88-IRF7 signaling pathways in pDCs to induce type I IFNs.
In addition, polyinosine-polycytidylic acid [poly(I:C)] can induce autophagy formation, as evidenced by GFP-LC3 puncta formation and increased proteolysis of long-lived proteins in murine RAW264.7 macrophages, and by LC3-II conversion in BMMs [56]. However, further studies are needed to ascertain if TLR3 is involved in induction of autophagy, as poly(I:C) can also activate RIG-I and melanoma differentiation-associated protein 5 (MDA-5). Although significant advances have been made in the molecular machinery underlying autophagy, our understanding of the relationship or interaction between TLRs and autophagy, and whether TLR ligand-induced autophagy is a general phenomenon that promotes trafficking of pathogenic nucleic acids to endosomal nucleic acid-sensing TLRs remained limited. Further studies are needed to elucidate the molecular mechanism by which TLR signaling activates autophagy.
Recognition of Foreign Nucleic Acids by TLRs 3, 7, 8, 9, and 13
Recognition of dsRNA by TLR3
TLR3 is one of the endosomal nucleic acid-sensing TLRs responsible for the recognition of synthetic RNA analogs poly(I:C) and polyinosine [poly(I)], as well as viral dsRNA (Fig. 1 and Table 2). TLR3 was originally identified as a receptor that recognizes poly(I:C). Subsequently, numerous reports have shown that TLR3 also acts as a sensor in the host response to the genomes of dsRNA viruses such as reovirus [73], or to the replication intermediates formed during infection by ssRNA viruses, such as respiratory syncytial virus (RSV) [74], West Nile virus (WNV) [75], DENV [76], IAV [77], encephalomyocarditis virus (EMCV) [78], lymphocytic choriomeningitis virus (LCMV) [79], Semliki Forest virus (SFV) [80], Coxsackievirus group B serotype 3 (CVB3) [81], poliovirus [82], Punta Toro virus (PTV) [83], Epstein-Barr virus (EBV) [84], HCV [85], and Kaposi's sarcoma-associated herpes virus (KSHV) [86]. In addition, TLR3 also recognizes viral dsRNAs found in the products of transcription intermediates of DNA viruses, including vaccinia virus (VACV) [87], HSV [88], and mouse cytomegalovirus (MCMV) [89] (Table 2).
Table 2.
List of pathogens recognized by TLR3, 7, 8, 9, and 13
| Receptors | Pathogens | References |
|---|---|---|
| TLR3 | Viruses | |
| Reovirus | [73] | |
| Respiratory syncytial virus | [74] | |
| West Nile virus | [75] | |
| Dengue virus | [76] | |
| Influenza A virus | [77] | |
| Encephalomyocarditis virus | [78] | |
| Lymphocytic choriomeningitis virus | [79] | |
| Semliki forest virus | [80] | |
| Coxsackievirus group B serotype 3 | [81] | |
| Poliovirus | [82] | |
| Punta Toro virus | [83] | |
| Epstein-Barr virus | [84] | |
| Hepatitis C virus | [85] | |
| Kaposi's sarcoma-associated herpesvirus | [86] | |
| Vaccinia virus | [87] | |
| Herpes simplex virus | [88] | |
| Mouse cytomegalovirus | [89] | |
| Protozoa | ||
| Schistosoma mansoni | [90] | |
| Fungus | ||
| Aspergillus fumigatus | [91] | |
| TLR7 and/or TLR8 | Viruses | |
| Vaccinia virus | [12] | |
| Modified VV Ankara | [12] | |
| Human immunodeficiency virus | [92] | |
| Influenza A virus | [93] | |
| Sendai virus | [94] | |
| Vesicular stomatitis virus | [94] | |
| Coxsacki B virus | [95] | |
| Human parechovirus 1 | [96] | |
| Foot and mouth disease virus | [97] | |
| Newcastle disease virus | [98] | |
| Dengue virus | [99] | |
| Hepatitis C virus | [100] | |
| Mouse hepatitis virus | [101] | |
| Pneumonia virus of mice | [102] | |
| Mouse mammary tumor virus | [103] | |
| Murine leukemia virus | [103] | |
| Bacteria | ||
| Borrelia burgdorferi | [104] | |
| Helicobacter pylori | [105] | |
| Mycobacterium bovis | [104] | |
| Escherichia coli | [104] | |
| Group B Streptococcus | [106] | |
| Protozoa | ||
| Trypanosoma cruzi | [107] | |
| Fungus | ||
| Candida spp. | [108] | |
| Saccharomyces cerevisiae | [109] | |
| TLR9 | Viruses | |
| Human papillomavirus | [2] | |
| Herpes simplex virus 1/2 | [110,111] | |
| Mouse cytomegalovirus | [112] | |
| Adenovirus | [113] | |
| Baculovirus | [114] | |
| Epstein-Barr virus | [115] | |
| Human herpesvirus type 6B | [116] | |
| Varicella-zoster virus | [117] | |
| Murine gammaherpesvirus 68 | [118] | |
| Ectromelia virus | [119] | |
| Bacteria | ||
| Salmonella typhimurium | [120] | |
| Streptococcus suis | [121] | |
| Mycobacterium tuberculosis | [122] | |
| Streptococcus pneumoniae | [123] | |
| M1T1 group A Streptococcus | [124,125] | |
| Propionibacterium acnes | [126] | |
| Protozoa | ||
| Trypanosoma cruzi | [127] | |
| Trypanosoma brucei | [128] | |
| Leishmania major | [129] | |
| Plasmodium falciparum | [130,131] | |
| Fungus | ||
| Aspergillus fumigatus | [132] | |
| Candida albicans | [109] | |
| Saccharomyces cerevisiae | [109] | |
| Malassezia furfur | [109,133] | |
| Cryptococcus neoformans | [109,133] | |
| TLR13 | Viruses | |
| Vesicular stomatitis virus | [13] | |
| Bacteria | ||
| Streptococcus aureus | [14,134] | |
| Streptococcus pyogenes | [135] |
TLR3 is involved in the recognition of small interfering RNA (siRNA) in a sequence-independent manner, resulting in the production of IL-12 and IFN-γ [136]. Interestingly, RNA-binding capsid proteins (CPs) from two positive-strand RNA viruses, Ross River virus (RRV) and Brome mosaic virus (BMV), and the hepadnavirus hepatitis B virus (HBV) were demonstrated to be potent enhancers of TLR3 signaling by poly(I:C) or viral dsRNAs [137]. Deletion of an arginine-rich RNA-binding domain from the HBV CP abolished its enhancement effect on TLR3 signaling [137], demonstrating that several viral RNA-binding proteins can enhance the dsRNA-dependent innate immune response initiated by TLR3. In addition, anti-microbial peptide LL37, the only known human member of the cathelicidin family of antimicrobial peptides, enhances poly(I:C) or rhinovirus infection-induced TLR3 signaling, and enables the recognition of viral dsRNAs by TLR3 in a human bronchial epithelial cell line (BEAS2B) and human peripheral blood mononuclear cells (PBMCs) [138].
The physiological function of TLR3 in antiviral immunity was studied in TLR3-/- mice, showing that TLR3-/- mice are more susceptible to reovirus, LCMV, MCMV, and VSV infections, compared with wild-type mice [73,79,89]. However, TLR3 may support viral growth or pathogenesis rather than protect mice from infection by viruses. For example, TLR3-/- mice were found to have stronger resistance to WNV infection, as WNV could disrupt the blood–brain barrier leading to the enhancement of virus entry into the brain via TLR3-mediated induction of peripheral inflammatory cytokines [75]. TLR3 has a synergetic function with RIG-I and MDA5 in the restriction of DENV in cultured human cells [76]. Additionally, studies of wild-type mice with IAV infection have shown that TLR3 is essential for mediating inflammatory response to host morbidity and lethality [77]. In addition, TLR3 also senses dsRNA derived from protozoa and fungi, such as Schistosoma mansoni, and Aspergillus fumigatus [90,91]. Taken together, the various known functions of TLR3 reflect the complexity of its roles in the regulation of host immune response.
Recognition of ssRNA by TLRs 7, 8, and 13
TLRs 7 and 8 are responsible for the recognition of ssRNA from the genomes of ssRNA viruses, and specific bacteria, synthetic ssRNA, imidazoquinolines, guanine analogs and siRNA (Fig. 1 and Table 2). Human TLRs 7 and 8, and mouse TLR7 recognize imidazoquinolines and initiate immune response, whereas mouse TLR8 is thought not to be activated by imidazoquinolines or ssRNA due to a five amino-acid deletion in the ECD [139,140]. Nevertheless, mouse TLR8 could be activated by poly(dT)17 oligodeoxyribonucleotides (ODNs) combined with the 3M-002, leading to the induction of TNF-α [141], suggesting that mouse TLR8 may be functionally active in the detection of DNA viruses. In support of this, a previous study showed that vaccinia virus (VACV) as well asvaccinia viral poly(A)/T-rich DNA could activate pDCs in a TLR8-dependent manner [12], suggesting that mouse TLR8 may regulate innate immunity against VACV infection. Interestingly, mouse TLR8 was found to inhibit TLR7-sensing of 3M-001 in HEK293 cells, and mouse TLR8−/− DCs showed increased responses to various TLR7 ligands and NF-κB activation, indicating that TLR8 may directly modulate TLR7 function [142]. In addition, guanine analogs, such as 7-allyl-8-oxoguanosine (loxoribine) and gardiquimod, and pyrimidine analog bropirimine can induce the production of various cytokines via TLR7 in humans and mice [139,140], suggesting the possibility of exploiting imidazoquinolines and nucleoside analogs as adjuvants for therapy, vaccination, and anti-tumor response in humans and animals.
TLRs 7 and 8 also recognize guanosine (G)/uridine (U)-rich synthetic ssRNA derived from ssRNA viruses. Adenosines (A)/U or G/U-rich synthetic ssRNA (RNA40) derived from the U5 region of human immunodeficiency virus (HIV) and polyU RNA of IAV are sensed by TLR7 in mice and by TLR7 and TLR8 in humans [92,93]. More specifically, GU-rich synthetic ssRNAs may be responsible for TLR7-mediated IFN-α production, and AU-rich synthetic ssRNA for the induction of inflammatory cytokines in a TLR8-dependent manner [92,93,143]. In addition to HIV and IAV, GU-rich ssRNAs derived from a variety of viruses, such as SeV [94], VSV [94], CBV [95], human parechovirus 1 [96], FMDV [97], Newcastle disease virus (NDV) [98], DENV [99], HCV [100], and mouse hepatitis virus (MHV) [101], have been identified as natural ligands for TLR7 and/or TLR8 (Table 2). TLR7 could also recognize mouse pneumonia virus, mouse mammary tumor virus, and murine leukemia virus (MuLV) via the TLR7-MyD88-dependent signaling pathway [102,103]. Interestingly, the dsRNA bluetongue virus induces the production of a significant amount of type I IFNs and inflammatory cytokines in pDCs via a MyD88-dependent but TLR7/8-independent signaling pathway [144]. pDCs do not express TLR3 and do not use the RIG-1/MDA5 signaling pathway, however, it is still not known how pDCs respond to dsRNA virus infection [144]. These data indicate that dsRNA viruses may induce the secretion of type I IFNs and inflammatory cytokines in pDCs via a novel TLR-independent and MyD88-dependent pathway.
Modifications of ssRNAs, such as 5-methylcytosine (m5C), N6-methyladenosine (m6A), 5-methyluridine (m5U), 2′-O-methyl groups, 2′-thiolated uridine (s2U) or pseudouridine, are commonly found in endogenous RNAs of the host [30,145]. These modified ssRNAs are usually not recognized by TLRs 7 and 8. However, as viral RNA also contains these modifications in infected cells and purified ‘self’ mRNA or DNA can induce TLR7-and TLR9-mediated signaling [30,145,146], it suggests that the structure, sequence or modification of RNA/DNA may be a minor factor in discriminating ‘self’ and ‘non-self’ by TLRs 7, 8, and 9, and any endogenous RNA/DNA sequence may result in activation of these TLRs, but the compartmentalization of nucleic acid-sensing TLRs and nucleic acid-based regulatory programs are keys for distinguishing ‘self’ from foreign nucleic acids. Surprisingly, CD14 is dispensable for viral uptake but is required for triggering MyD88-dependent induction of inflammatory cytokines via TLRs 7, 8, and 9 by stimulatory ssRNA/CpG DNA and various types of VSV [147,148]. This suggests that CD14 not only promotes the selective uptake of nucleic acids, but also acts as a co-receptor for endosomal TLR activation.
TLR7 and TLR8 also recognize dissociated ssRNA derived from certain siRNA in endosomes [149], and detect Borrelia burgdorferi, Mycobacterium bovis, Helicobacter pylori and RNAs from Escherichia coli and group B Streptococcus (GBS) in the endosomes by phagolysosomes or autolysosomes, but cannot detect other bacteria such as Listeria monocytogenes and Group A Streptococcus (GAS) (Table 2) [104–106]. Although Trypanosoma cruzi is likely to be sensed by TLR7, the recognition of parasite RNA mediated by TLRs 7 and 8 is much less documented [107]. Recent studies also showed that IFN-α/β is induced by Candida spp. and Saccharomyces cerevisiae RNA/DNA by a mechanism that involves endosomal recognition of fungal RNA and DNA by TLR7 and TLR9, respectively, and absolutely requires MyD88 [108,109].
A novel endosomal nucleic acid-sensing TLRs called TLR13 appears to be able to recognize VSV and induce the production of type I IFNs through the activation of NF-κB and IRF7 in a MyD88- and transforming growth factor-activated protein kinase 1 (TAK1)-dependent manner. However, it is currently unknown if ssRNA derived from VSV can be acted as a ligand for TLR13 [13]. Recent studies have identified a conserved 23S rRNA sequence ‘CGGAAAGACC’ derived from Streptococcus aureus as a natural ligand for TLR13 [14,134]. Interestingly, the rRNA sequence recognized by TLR13 is bound by certain antibiotics, and bacterial strains that have evolved to resist these antibiotics cannot be detected by TLR13 [14,134]. We speculate that widespread ancient antibiotic resistance has subverted TLR13-driven antibacterial immune resistance, and may explain why TLR13 expression has been lost in certain mammalian species (including humans). The function of TLR13 might be replaced by an RNA-sensing PRR in humans that can still recognize erythromycin resistance-forming RNA modifications. In addition, TLR13 has also been shown to activate NF-κB in response to bacterial RNA or Streptococcus pyogenes in an RNA-specific manner [135].
Recognition of CpG DNA by TLR9
TLR9 induces the production of both inflammatory cytokines and type I IFNs by recognizing unmethylated 2′-deoxyribo CpG motif-containing DNA from bacteria, viruses, protozoa, fungi and synthetic ODNs (Fig. 1 and Table 2). In the host, recognition of methylated CpG motifs by TLR9 is severely suppressed and immune response is normally not triggered.
Although TLR9 recognizes bacterial unmethylated CpG DNA [150], the functions of TLR9 in fighting bacterial infections have not been elucidated in detail. Collaboration of several TLRs may be the key factor in host resistance to Mycobacterium tuberculosis because TLR2-/--TLR4-/--TLR9-/- mice showed markedly enhanced susceptibility to Salmonella typhimurium, Streptococcus suis and M. tuberculosis infections [120–122]. TLR9 activation by pneumococcal DNA was observed only in cells with live bacterial infection [123]. A recent study reported that TLR9 was important in host defense against M1T1 GAS infections by increasing macrophage hypoxia-inducible factor-1α levels, and oxidative burst and nitric oxide production in response to GAS, contributing to GAS clearance in vivo in both localized cutaneous and systemic infection models [124]. On the other hand, the virulence factor DNase Sda1 of M1T1 GAS has the ability to degrade bacterial DNA and suppress TLR9-dependent INF-α and TNF-α induction [125]. Thus, this is a novel mechanism of bacterial innate immune evasion based on autodegradation of CpG-rich DNA by a bacterial DNase. TLR9 was also shown to be critical for the induction of type I IFNs signaling and the phosphorylation of signal transducers and activators of transcription 1 in DCs in response to S. aureus, illustrating an additional mechanism through which S. aureus induces the innate immune signaling during infection [126]. One possibility is that bacterial DNA may be delivered to endosomal compartments where acidic conditions (pH = 5.5 to 6.2) lead to the degradation of dsDNA containing multiple CpG motif sequences which are recognized by TLR9. This suggests again that the compartmentalization is a very important element in TLR9 (as well as TLRs 3, 7, 8, and 13) activation.
CpG motif-containing DNAs from human papillomavirus [2], HSV-1/2 [110,111], MCMV [112], adenovirus (ADV) [113], baculovirus [114], EBV [115], human herpesvirus type 6B (HHV-6B) [116], varicella-zoster virus (VZV) [117], murine gammaherpesvirus 68 (MHV-68) [118], and ectromelia virus (ECTV) are also sensed by TLR9 and induce the secretion of robust type I IFNs and inflammatory cytokines in pDCs [119] (Table 2). TLR9 was shown to be involved in VZV-induced IFN-α production, which could be inhibited by inhibitory CpG ODN in pDCs [117]. TLR9-mediated IFN-α response to HSV-1/2 was cell-type specific and limited to pDCs, because TLR9 could recognize live, or heat- or UV-inactivated HSV-1/2 and induced secretion of high levels of IFN-α in pDCs, whereas macrophages produced IFN-α upon HSV infection via a TLR9-independent mechanism [110,111].
TLR9 also senses CpG DNA derived from protozoa and fungi (Table 2), including genomic DNA derived from T. cruzi, Trypanosoma brucei, and Leishmania major [127–129]. Unmethylated CpG motifs in A. fumigatus DNA stimulate mouse bone marrow-derived DCs (BMDCs) and human pDCs to secrete inflammatory cytokines in a TLR9-dependent manner [132], contributing to the establishment of anti-fungi immune response states. In addition to A. fumigatus, Candida albicans, S. cerevisiae, Malassezia furfur, and Cryptococcus neoformans DNAs are capable of triggering TLR9 recruitment, demonstrating that TLR9 is compartmentalized selectively to fungal phagosomes and negatively modulates macrophage antifungal effector functions [108,109,133]. Interestingly, hemozoin, a crystalline metabolite of hemoglobin produced by Plasmodium falciparum, acts as an inducer of TLR9 and potently activates macrophages and DCs to produce inflammatory cytokines and chemokines [130]. Notably, TLR9 recognizes P. falciparum DNA contained in purified hemozoin, which appears to be a carrier that transports P. falciparum DNA to endosomes [131], consistent with the conclusion that ligand delivery is an essential condition for intracellular TLR9 recognition. However, the role of hemozoin in TLR9 activation needs to be more fully elucidated in future studies.
Signaling Networks of TLRs 3, 7, 8, 9, and 13
TRIF signaling pathways of TLR3
TLR3 participates in the TRIF-dependent pathway (Fig. 1). In this case, TRIF directly interacts with the TIR domain of TLR3 to recruit a set of adaptor molecules, including TNF receptor-associated factor 6 (TRAF6), TNF receptor-associated death domain, Pellino1, and receptor interacting protein 1 (RIP1) for the activation of TAK1 [1,2,7,8,151], which in turn leads to the activation of IRF3, mitogen-activated protein kinases (MAPKs), and NF-κB. On the other hand, interactions of TLR3-TRIF with TRAF3, non-classical NAK-associated protein 1 (NAP1), TBK1, and IKKi (also called IKKɛ) lead to the formation of a complex and the activation of IRF3, which then translocates to the nucleus to induce the expression of IFN-β and IFN-inducible genes [1–3,7,8,151].
MyD88-signaling pathways of TLRs 7, 8, 9, and 13
TLRs 7, 8, 9, and 13 induce the production of inflammatory cytokines and type I IFNs through a MyD88-dependent signaling pathway (Fig. 1 and Table 1). MyD88 is a specific adaptor molecule containing a TIR domain in its C-terminal region and a dead domain in its N terminal region. Upon stimulation, TLRs 7, 8, and 9 interact with the TIR domain of MyD88 to recruit signaling molecules IL-1 receptor-associated kinases 4/1/2 (IRAK4/1/2) and interact with TRAF3/6, Ubc13, and Uev1A, forming a signaling complex [1–3,7–9,151]. This complex recruits TAK1 and TAK1-binding proteins 1/2/3 (TAB1/2/3), and is activated by the K63-linked polyubiquitin chain of TRAF6, phosphorylated IKKβ, and MAP kinase kinase 6 (MKK6). This phenomenon results in the activation of a classical IKK complex, consisting of IKK1/α, IKK2/β, and IKK3/γ (also called NF-κB essential modulator, NEMO), which leads to IκB degradation and thereby contributes to the activation of NF-κB and MAPK signaling pathways to induce secretion of inflammatory cytokines [1–3,7–9,151–153].
In pDCs, the TLR7 and TLR9 signaling pathways are unique in that they both require MyD88 for the induction of type I IFNs. TLR7 and TLR9 recruit MyD88 and IRAK4, which then interact with TRAF6, TRAF3, IRAK1, IKKα, osteopontin (OPN), and IRF7 to form a super-molecular complex [1,2,7,8,151]. Ultimately, IRF7 is phosphorylated by IRAK1 and/or IKKα and translocated to the nucleus to induce the transcription of type I IFNs [1,2,7,8,151]. In addition, a phosphoinositol 3-OH kinase (PI3K) mammalian target of the rapamycin (mTOR)-p70S6K pathway is beneficial for the induction of type I IFNs in MyD88-dependent activation of IRF7 in pDCs [154]. IRF8 has an essential function in the induction of type I IFNs and inflammatory cytokines through TLR9 in pDCs and cDCs [155]. However, in cDCs, secretion of IFN-β and inflammatory cytokines in a TLR7- and/or TLR9-MyD88-dependent manner is mediated by IRF1 and IRF5 [10,11]. Interestingly, protein kinase C and casein kinase substrate in neurons (PACSIN) 1 is specifically expressed in pDCs and represents a pDC-specific adaptor molecule that plays an important role in TLR7- and TLR9-mediated type I IFN responses in vitro and in vivo [8].
TLR13, another intracellular nucleic acid-sensing TLR, not only activates a MyD88- and TAK1-dependent TLR signaling pathway to activate NF-κB but also induces type I IFNs through IRF7, a characteristic similar to that of TLRs 7, 8, and 9 [13–15]. However, the mechanism by which TLR13 mediates recognition of foreign nucleic acids, its associated signaling pathway and its regulation are poorly understood.
Regulation of TLRs 3, 7, 8, 9, and 13 signaling pathways
The TLR-signaling pathways are regulated by positive and negative modulatory mechanisms to achieve a balanced output (Fig. 1 and Table 3). In general, phosphorylation/dephosphorylation, ubiquitination/deubiquitination, sumoylation/desumoylation, acetylation/deacetylation, and competitive effects of negative regulators are the principal regulatory mechanisms that directly or indirectly regulate the activation of signaling pathways associated with TLRs 3, 7, 8, 9, and 13.
Table 3.
List of regulatory molecules required for TLR3, 7, 8, 9, and 13 signaling pathways
| Regulators |
Targets | Regulatory mechanisms | Effectors | References | |
|---|---|---|---|---|---|
| Positive regulation | TRAF6 | TRAF6, cIAP1/2, IRAKs | Ubiquitination of targets | Activation of TAK1, TBK1 and IKKi | [156] |
| Ubc5 | IRF3 | Ubiquitination of IRF3 | Activation of IRF3 | [157] | |
| MARCH5 | TANK | Polyubiquitination of TANK | Activation of IRF7 | [158] | |
| Viperin | IRAK1, TRAF6 | Polyubiquitination of IRAK1 | Activation of IRF7 | [159] | |
| SUMO | TANK | SUMOylation of TANK | Controls IKKɛ-TBK1 interactions | [160] | |
| Nrdp1 | TBK1 | Ubiquitination of TBK1 | Activation of TBK1 and IRF3 | [161] | |
| miR-181b-1 | CYLD | Inhibits CYLD production | Activation of NF-kB | [162–164] | |
| Negative regulation | Triad3A | TLR9 | Ubiquitination and proteolytic degradation of TLR9 | Inhibition TLR9 activation | [156,165] |
| ST2L | MyD88 | Sequestration of MyD88 | Inhibition TLR9 activation | [7,156] | |
| SIGIRR | TRAF6, IRAK | Inhibition of TRAF6 and IRAK | Inhibition TLR9 activation | [7,156] | |
| PI3 | p38, JNK, NF-κB | Inhibition NF-κB and AP-1 activation | Inhibition of cytokine production | [7,156] | |
| TRIM30α | TAB2, TAB3 | Lysosome-related degradation of TAB2 and TAB3 | Inhibition NF-κB activation | [166] | |
| Nrdp1 | MyD88 | Proteosomal degradation of MyD88 | Inhibition IRAK1, TAK1, IKKβ, MAPKs, and NF-κB activation | [161] | |
| A20 | RIP1, TRAFs, TRIF, cIAP1/2, and IKK complex | Ubiquitination or deubiquitination of targets, and disruption of protein–protein interaction | Inhibition NF-κB and IRF3 activation | [167–169] | |
| DUBA | TBK1 | Ubiquitination of TBK1 | Inhibition of TRAF3 and IRFs | [170] | |
| CYLD | TBK1 | Ubiquitination of TBK1 | Inhibition of TRAF3 and IRFs | [171] | |
| OTUB1, OTUB2 | TRAF3/6 | Ubiquitination of TRAF3/6 | Inhibition of NF-κB and IRF3 activation | [172] | |
| TANK | TRAF6 | Ubiquitination of TRAF6 | Inhibition of NF-κB and AP-1 activation | [173] | |
| Atg16L1 | TRIF | Surpression of TRIF-dependent pathway | Suppression of caspase-1 activation | [174] | |
| SHP-2 | TBK1 | Phosphorylation of TBK1 | Inhibition of cytokine production | [175] | |
| SHP-1 | IRAK1, IRAK2 | IRAK1 and IRAK2 | Inhibition of cytokine production | [176] | |
| CD2AP | Cbl | Ubiquitination of Cbl | Inhibition of TLR9-mediated type 1 IFN production | [177] | |
| sTLR9 | TLR9 | Sequestration of TLR9 | Inhibition of TLR9-mediated signaling activation | [178] | |
| IRAKM | IkBa, IRAK-1 | Phosphorylation of IkBa, Inhibition of IRAK-1 | Inhibition of cytokine production | [179] | |
| MyD88s | MyD88 | Sequestration of MyD88 | Inhibition of MyD88-mediated signaling activation | [180] | |
| SARM | TRIF | Sequestration of TRIF | Inhibition of TRIF-mediated signaling activation | [181] | |
| IRF4 | IRF5 | Sequestration of IRF5 | Inhibition of TLR signaling activation | [182] | |
| miR-146 | TRAF6, IRAK1/2 | down-regulates IRAK1/2 and TRAF6 | Inhibition of the MyD88/NF-kB signaling activation | [183] | |
| miR-155 | IKKi, IKKβ, TAB2, and MyD88 | Inhibition of IKKi and IKKβ | Inhibition of NF-κB activation | [184] | |
| miR-9 | p105, p50 | Inhibition of p50/p52 mature | Modulation of the NF-kB pathway | [162–164] | |
| miR-199a, miR-214 | IKKβ | down-regulates IKKβ | Inhibition of NF-kB activity | [162–164] | |
| miR-223, miR-15, miR-16 | IKKα, NIK, TRAF2, and p52 | Inhibition of targets | Inhibition of noncanonical NF-kB pathway | [162–164] | |
DUBA, deubiquitinating enzyme A; CYLD, cyclindomatosis.
Positive regulation of TLRs 3, 7, 8, 9, and 13 signaling pathways
TRAF3 and TRAF6, which act as E3 ubiquitin ligases in the formation of K63-linked ubiquitin chain by catalyzing itself sequentially, are important for the activation of downstream signaling molecules, such as TAK1, TBK1 and IKKi [156]. An E2 ubiquitin ligase, Ubc5, is required for IRF3 activation by catalyzing K63-type ubiquitin chain formation [157]. Degradation of TRAF3 is responsible for the activation of MAPKs and NF-κB, and the production of inflammatory cytokines through proteasome-dependent mechanisms [156]. Mitochondrial protein MARCH5, a novel E3 ubiquitin ligase, catalyzes K63-linked poly-ubiquitination of TANK and positively modulates TLR7 signaling by attenuating TANK and inhibiting TRAF6 [158]. Viperin, an IFN-inducible antiviral protein, interacts with IRAK1 and TRAF6 to recruit them to lipid bodies and facilitates K63-linked ubiquitination of IRAK1 to induce nuclear translocation of transcription factor IRF7, thereby promoting TLR7- and TLR9-mediated type I IFN production in pDCs [159]. Furthermore, modification of TANK by the small ubiquitin-related modifier (SUMO) is triggered by the kinase activity of IKKɛ and TBK1 after stimulation of TLR7 ligands, which in turn weakens the interaction with IKKɛ and thus relieves the negative effect of TANK on signal propagation [160].
Negative regulation of TLRs 3, 7, 8, 9, and 13 signaling pathways
Although activation of TLR signal transduction is required for hosts to eliminate invading pathogens, excessive activation of TLR-signaling pathways may disrupt immune homeostasis, resulting in autoimmune, chronic inflammatory and infectious diseases. Therefore, TLR-associated signaling pathways and immune functions must be under strict negative regulation to maintain immune balance. Several negative regulators involved in suppressing TLR-signaling pathways at multiple levels, including splice variants of adaptors or their related proteins, ubiquitin ligases, deubiquitinases, transcriptional regulators, tyrosine phosphatases, kinases, signaling proteins, microRNAs, and even viral proteins, have been described [156,161–185].
Several RING finger proteins have been identified to be negative regulators of TLRs 3, 7, 8, 9, and 13 signaling pathways. The first one is Triad3A, a RING finger protein functioning as an E3 ubiquitin ligase, can enhance ubiquitination and proteolytic degradation of TLR9 [156,165]. The second RING protein, tripartite motif protein 30α (TRIM30α), functions as a negative regulator of TLR-mediated NF-κB activation by targeting TAB2 and TAB3 degradation in a ubiquitin-proteasome-independent pathway [166]. The third RING protein, neuregulin receptor degradation protein-1 (Nrdp1), functions as a RING protein-containing E3 ligase and plays a different function in regulating TLR signaling. It enhances the TRIF-dependent activation of TBK1 and IRF3 via K63-linked ubiquitination of TBK1 [161]. In contrast, MyD88-dependent activation of IRAK1, TAK1, IKKβ, MAPKs, and NF-κB are inhibited by Nrdp1-mediated degradation of MyD88 via K48-linked ubiquitination [161].
A number of deubiquitinating enzymes also play important roles in negative regulation of TLRs 3, 7, 8, 9, and 13 signaling pathways. A20, a cytoplasmic zinc finger protein, is one of the deubiquitin enzymes (DUBs) of the ovarian tumor (OTU) family. As a deubiquitinating enzyme A20 can remove K63-polyubiquitin chains from TRAF6 to turn off the NF-κB signaling. A20 also has a unique E3 ligase activity that allows it to catalyze K48-polyubiquitination to degrade RIP1 in a proteasome-dependent manner after cleavage of K63-polyubiquitin chains to restrict NF-κB activation [167]. Another important function of A20 is to disrupt the interaction of E3 ligases (TRAF6, TRAF2, TRAF3, IKK complex, and cIAP1/2) with E2 conjugating enzymes (Ubc13 and Ubc5hc), which results in effective downregulation of the NF-κB-signaling pathway [168]. Furthermore, A20 has also been reported to negatively regulate IFN-β transcription by inhibiting IRF3 activation [169]. Two other important deubiquitinating enzymes, deubiquitinating enzyme A and cyclindomatosis, are involved in the negative regulation of IFN-β production by removing a K63-polyubiquitin chain from TBK1 and inhibiting the activation of TRAF3 and IRFs [170,171].
Two OTU family members OTUB1 and OTUB2, originally identified as proteases, suppress IFN-β promoter activity through cleavage of the K63-polyubiquitin chain from TRAF3 and inhibit NF-κB signaling via deubiquitination of TRAF6 [172]. TANK may also act as a negative regulator of TRAF6 ubiquitination in macrophages and B cells [173], whereas Atg16L1 negatively regulates TRIF-dependent pathways that lead to intestinal inflammation suppression and caspase-1 activation [174]. Src homology 2 domain-containing protein tyrosine phosphatase (SHP)-2 inhibits cytokine production by suppressing the phosphorylation of TBK1 [175], while SHP-1 inhibits TLR-mediated production of inflammatory cytokines by suppressing the function of IRAK1 and IRAK2 [176]. Interestingly, CD2-associated adaptor protein (CD2AP) has the ability to positively regulate BDCA2/FcɛR1γ signaling via forming a complex with SHIP1 to inhibit the E3 ubiquitin ligase Cbl and TLR9-mediated type I IFNs production [177].
Several splice variants are also involved in negative regulation of the signaling pathways associated with endosomal nucleic acid-sensing TLRs by competing with various adapters and transcription factors for binding sites, such as soluble decoy TLRs (such as sTLR9), IRAKM, MyD88 short (MyD88s), Armadillo motif-containing protein (SARM) and IRF4 [178–182]. In addition, a range of miRNAs, including miR-9, miR-124, miR-155, miR-218, miR-15, miR-16, miR-223, miR-199a, miR-520h, miR-301a, and miR-181b-1, may also act as important negative regulators of those pathways, as they are involved in the down-regulation of signaling proteins related to innate immune response [162–164,183,184]. Thus, a complex network involving multiple regulatory molecules and pathways are likely important for the recognition of endosomal nucleic acid-sensing TLRs, which is essential for preventing serious inflammatory disorders and autoimmune diseases. In addition, nucleic acid-sensing TLR signaling, especially some important adapters such as NF-κB, TRIF, and IRFs, was negatively regulated by viruses (such as VACV, HCV, and HIV and herpesviruses, polyomaviruses, ascoviruses, and adenoviruses), which encoded a number of proteins and miRNAs to inhibit the production of type I IFNs and inflammatory cytokine [186,187].
Concluding Remarks
Over the past decade, significant progress has been made in our understanding of nucleic acid recognition. In particular, the functions of endosomal nucleic acid-sensing TLRs 3, 7, 8, 9, and 13 in innate immunity, especially their restricted expression/compartmentalization, ligand specificity, trafficking, proteolysis, autophagy, signaling, and regulation have been studied in detail. Several studies provided new insights into the compartmentalization of endosomal nucleic acid-sensing TLRs based on ligand recognition and revealed a series of regulatory mechanisms that are key for discrimination between ‘self’ and ‘non-self’. As previously mentioned, ‘self’-derived nucleic acids are properly degraded by extracellular and endosomal nucleases before they can be sensed by nucleic acid-sensing TLRs within endosomal compartments under normal conditions. In addition, compartmentalization of nucleic acid-sensing TLRs is important for avoiding contact with ‘self’-nucleic acids. Furthermore, intracellular trafficking and proteolytic maturation of nucleic acid-sensing TLRs, mediated in part through their TM domains, is important for preventing inappropriate recognition of ‘self’-nucleic acids by leakage of these receptors to the cell surface.
As discussed above, Unc93b1 acts as a chaperone for the endosomal nucleic acid-sensing TLRs 3, 7, 8, 9, and 13, and controls the translocation of these TLRs from the ER to endosomal compartments. Interestingly, another member of the TLR family, a protein-recognizing receptor TLR11, was also recently found to be expressed in the ER along with UNC93B1, TLR11 regulates the activation of DCs in response to protozoan parasite Toxoplasma gondii profilin (PFTG) [188], suggesting that in addition to nucleic acid-sensing TLRs, UNC93B1 also regulates at least one protein-sensing TLR in intracellular compartments. Although this knowledge is needed for our understanding of nucleic acid-sensing TLRs defense against protozoan parasites, further studies are needed to dissect whether TLR11 is trafficked from the ER to endosomal compartments by UNC93B1 to detect foreign nucleic acids, and the molecular mechanism of TLR11 activation by PFTG and T. gondii or others protozoan parasites. A surprising finding from recent experiments is that VACV and its poly(A)/T-rich DNA motifs are potent inducers of pDC-derived IFN-α in a TLR9-independent, and exclusively TLR8-dependent pathway [12]. Although it is not clear why the recognition of VACV/modified VACV Ankara (MVA) and ECTV in pDC is mediated through different TLRs and whether TLR8-mediated recognition of VACV by non-pDCs is important in VACV control in humans, these result may reflect differences between VACV/MVA and ECTV in terms of their genetic compositions and endosomal trafficking pathways, and that different pathogens have evolved to adopt different mechanisms to effectively activate the innate immune system.
It has been known for years that ‘self’-nucleic acids that are modified or inappropriately localized can be recognized as ‘non-self’ to induce autoimmune response. On the one hand, ‘self’-derived nucleic acids form nucleic acid–protein immune complexes (ICs) with a variety of endogenous proteins such as autoantibodies, anti-microbial peptides, small ribonucleoprotein (snRNPs), and high mobility group box 1 (HMGB-1), they may become resistant to nucleases and reach endosomal nucleic acid-sensing TLRs, leading to autoimmune reaction [1–3,189–190]. On the other hand, imbalance between positive and negative regulators of the TLR-associated may disrupt immune homeostasis, resulting in autoimmune diseases. Interestingly, intracellular trafficking of TLR7 and TLR9 from the ER to endosomes is also induced by LPS, which may increase the possibility of autoimmune diseases [186]. Furthermore, autophagy is not only important for the degradation of intracellular pathogens, cellular proteins, and organelles, but also for the regulation of nucleic acid-sensing TLR-mediated recognition of pathogens in multiple ways. However, autophagy can be also exploited by a diverse array of pathogens, which interfere with autophagy-mediated degradation of intracellular pathogens and substrate recognition by nucleic acid-sensing TLR, and suppress or escape the host's antiviral innate immune response. However, a detailed understanding of how such pathogens achieve these feats will need to be elucidated in future studies.
In addition to nucleic acid-sensing TLRs, other nucleic acid-sensing PRRs, such as RLRs, nucleotide-binding oligomerization domain-like receptors (NLRs), absent in melanoma 2 (AIM2)-like receptors (ALRs), and DNA recognition receptors (cytosolic sensors for DNA), also provide defense against a wide range of pathogens [1–3]. However, very little is known about possible crosstalk between nucleic acid-sensing TLRs and these PRRs. Therefore, future studies should aim to improve our understanding of innate immune systems and immunobiology, including host-pathogen interactions, crosstalk between immune networks and their influence on the development of adaptive immunity. Such studies will likely also reveal information essential for the identification and development of novel anti-pathogen targets, therapeutic strategies, and better adjuvant for vaccines.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (No. 30871884) and the National High Technology Research and Development Program of China (No. 2011AA10A211).
References
- 1.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Ann Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 3.Hansena JD, Vojtechb LN, Laingc KJ. Sensing disease and danger: a survey of vertebrate PRRs and their origins. Dev Comp Immunol. 2011;35:886–897. doi: 10.1016/j.dci.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Kang JY, Lee JO. Structural biology of the toll-like receptor family. Annu Rev Biochem. 2011;80:917–941. doi: 10.1146/annurev-biochem-052909-141507. [DOI] [PubMed] [Google Scholar]
- 5.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumoto M, Oshiumi H, Seya T. Antiviral responses induced by the TLR3 pathway. Rev Med Virol. 2011;21:67–77. doi: 10.1002/rmv.680. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M, Takeda K. Current views of Toll-like receptor signaling pathways. Gastroent Res Pract. 2010;2010:240365. doi: 10.1155/2010/240365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esashi E, Bao M, Wang YH, Cao W, Liu YJ. PACSIN1 regulates the TLR7/9-mediated type I interferon response in plasmacytoid dendritic cells. Eur J Immunol. 2012;42:573–579. doi: 10.1002/eji.201142045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–9. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Negishi H, Fujita Y, Yanai H, Sakaguchi S, Ouyang X, Shinohara M, Takayanagi H, et al. Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc Natl Acad Sci USA. 2006;103:15136–15141. doi: 10.1073/pnas.0607181103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang Foreman HC, Van Scoy S, Cheng TF, Reich NC. Activation of interferon regulatory factor 5 by site specific phosphorylation. PLoS One. 2012;7:e33098. doi: 10.1371/journal.pone.0033098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez J, Huang X, Yang Y. Toll-like receptor 8-mediated activation of murine plasmacytoid dendritic cells by vaccinia viral DNA. Proc Natl Acad Sci USA. 2010;107:6442–6447. doi: 10.1073/pnas.0913291107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Z, Cai Z, Sanchez A, Zhang T, Wen S, Wang J, Yang JH, et al. A novel Toll-like receptor that recognizes vesicular stomatitis virus. J Biol Chem. 2011;286:4517–4524. doi: 10.1074/jbc.M110.159590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oldenburg M, Krüger A, Ferstl R, Kaufmann A, Nees G, Sigmund A, Bathke B, et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012;337:1111–1115. doi: 10.1126/science.1220363. [DOI] [PubMed] [Google Scholar]
- 15.Shi Z, Cai Z, Wen S, Chen C, Gendron C, Sanchez A, Patterson K, et al. Transcriptional regulation of the novel Toll-like receptor Tlr13. J Biol Chem. 2009;284:20540–20547. doi: 10.1074/jbc.M109.022541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dieudonné A, Torres D, Blanchard S, Taront S, Jeannin P, Delneste Y, Pichavant M, et al. Scavenger receptors in human airway epithelial cells: Role in response to double-stranded RNA. PLoS One. 2012;7:e41952. doi: 10.1371/journal.pone.0041952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagyoszi P, Wilhelm I, Farkas AE, Fazakas C, Dung NT, Haskó J, Krizbai IA. Expression and regulation of toll-like receptors in cerebral endothelial cells. Neurochem Int. 2010;57:556–64. doi: 10.1016/j.neuint.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Kaul D, Habbel P, Derkow K, Krüger C, Franzoni E, Wulczyn FG, Bereswill S, et al. Expression of Toll-like receptors in the developing brain. PLoS One. 2012;7:e37767. doi: 10.1371/journal.pone.0037767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexopoulou L, Desnues B, Demaria O. Toll-like receptor 8: the awkward TLR8. Med Sci (Paris) 2012;28:96–102. doi: 10.1051/medsci/2012281023. [DOI] [PubMed] [Google Scholar]
- 20.Vincent IE, Zannetti C, Lucifora J, Norder H, Protzer U, Hainaut P, Zoulim F, et al. Hepatitis B virus impairs TLR9 expression and function in plasmacytoid dendritic cells. PLoS One. 2011;6:e26315. doi: 10.1371/journal.pone.0026315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards AD, Diebold SS, Slack EM, Tomizawa H, Hemmi H, Kaisho T, Akira S, et al. Toll-like receptor expression in murine DC subsets: Lack of TLR7 expression by CD8 alpha+ DC correlates with unresponsiveness to imidazoquinolines. Eur J Immunol. 2003;33:827–833. doi: 10.1002/eji.200323797. [DOI] [PubMed] [Google Scholar]
- 23.Poth JM, Coch C, Busch N, Boehm O, Schlee M, Janke M, Zillinger T, et al. Monocyte-mediated inhibition of TLR9-dependent IFN-α induction in plasmacytoid dendritic cells questions bacterial DNA as the active ingredient of bacterial lysates. J Immunol. 2010;185:7367–7373. doi: 10.4049/jimmunol.1001798. [DOI] [PubMed] [Google Scholar]
- 24.Xu LG, Jin L, Zhang BC, Akerlund LJ, Shu HB, Cambier JC. VISA is required for B cell expression of TLR7. J Immunol. 2012;188:248–258. doi: 10.4049/jimmunol.1100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaturvedi A, Pierce SK. How location governs toll-like receptor signaling. Traffic. 2009;10:621–628. doi: 10.1111/j.1600-0854.2009.00899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chockalingam A, Brooks JC, Cameron JL, Blum LK, Leifer CA. TLR9 traffics through the Golgi complex to localize to endolysosomes and respond to CpG DNA. Immunol Cell Biol. 2009;87:209–217. doi: 10.1038/icb.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 28.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 29.Lee CC, Avalos AM, Ploegh HL. Accessory molecules for Toll-like receptors and their function. Nat Rev Immunol. 2012;12:168–179. doi: 10.1038/nri3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Into T, Inomata M, Takayama E, Takigawa T. Autophagy in regulation of Toll-like receptor signaling. Cell Signal. 2012;24:1150–1162. doi: 10.1016/j.cellsig.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh S, Miyake K. Regulatory molecules required for nucleotide-sensing toll-like receptors. Immunol Rev. 2009;227:32–43. doi: 10.1111/j.1600-065X.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- 32.Nishiya T, Kajita E, Miwa S, Defranco A. TLR3 and TLR7 are targeted to the same intracellular compartments by distinct regulatory elements. J Biol Chem. 2005;280:37107–37117. doi: 10.1074/jbc.M504951200. [DOI] [PubMed] [Google Scholar]
- 33.Leifer CA, Brooks JC, Hoelzer K, Lopez J, Kennedy MN, Mazzoni A, Segal DM. Cytoplasmic targeting motifs control localization of toll-like receptor 9. J Biol Chem. 2006;281:35585–35592. doi: 10.1074/jbc.M607511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouchess ML, Arpaia N, Souza G, Barbalat R, Ewald SE, Lau L, Barton GM. Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation. Immunity. 2011;35:721–732. doi: 10.1016/j.immuni.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrançois L, Li Z, et al. Heat shock protein gp96 is a master chaperones for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007a;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks JC, Sun W, Chiosis G, Leifer CA. Heat shock protein gp96 regulates Toll-like receptor 9 proteolytic processing and conformational stability. Biochem Biophys Res Commun. 2012;421:780–784. doi: 10.1016/j.bbrc.2012.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiyokawa T, Akashi-Takamura S, Shibata T, Matsumoto F, Nishitani C, Kuroki Y, Seto Y, et al. A single base mutation in the PRAT4A gene reveals differential interaction of PRAT4A with Toll-like receptors. Int Immunol. 2008;20:1407–1415. doi: 10.1093/intimm/dxn098. [DOI] [PubMed] [Google Scholar]
- 38.Liu B, Yang Y, Qiu ZJ, Staron M, Hong F, Li Y, Wu S, et al. Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat Commun. 2010;1:1–10. doi: 10.1038/ncomms1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blasius AL, Arnold CN, Georgel P, Rutschmann S, Xia Y, Lin P, Ross C, et al. Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107:19973–19978. doi: 10.1073/pnas.1014051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang CY, Engel A, Opaluch AM, Ramos I, Maestre AM, Secundino I, De Jesus PD, et al. Cofactors required for TLR7- and TLR9-dependent innate immune responses. Cell Host Microbe. 2012;11:306–318. doi: 10.1016/j.chom.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chevrier N, Mertins P, Artyomov MN, Shalek AK, Iannacone M, Ciaccio MF, Gat-Viks I, et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. 2011;147:853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 44.Fukui R, Saitoh S, Matsumoto F, Kozuka-Hata H, Oyama M, Tabeta K, Beutler B, et al. Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA-but against RNA-sensing. J Exp Med. 2009;206:1339–1350. doi: 10.1084/jem.20082316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 46.Itoh H, Tatematsu M, Watanabe A, Iwano K, Funami K, Seya T, Matsumoto M. UNC93B1 physically associates with human TLR8 and regulates TLR8-mediated signaling. PLoS One. 2012;6:e28500. doi: 10.1371/journal.pone.0028500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukui R, Saitoh S, Kanno A, Onji M, Shibata T, Ito A, Onji M, et al. Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity. 2011;35:69–81. doi: 10.1016/j.immuni.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 48.Crane MJ, Gaddi PJ, Salazar-Mather TP. UNC93B1 mediates innate inflammation and antiviral defense in the liver during acute murine cytomegalovirus infection. PLoS One. 2012;7:e39161. doi: 10.1371/journal.pone.0039161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nickerson KM, Christensen SR, Shupe J, Kashgarian M, Kim D, Elkon K, Shlomchik MJ. TLR9 regulates TLR7- and MyD88-dependent autoantibody production and disease in a murine model of lupus. J Immunol. 2010;184:1840–1848. doi: 10.4049/jimmunol.0902592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, Lennon-Duménil AM, et al. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 53.Asagiri M, Hirai T, Kunigami T, Kamano S, Gober HJ, Okamoto K, Nishikawa K, et al. Cathepsin K-dependent toll-like receptor 9 signaling revealed in experimental arthritis. Science. 2008;319:624–627. doi: 10.1126/science.1150110. [DOI] [PubMed] [Google Scholar]
- 54.Matsumoto F, Saitoh S, Fukui R, Kobayashi T, Tanimura N, Konno K, Kusumoto Y, et al. Cathepsins are required for Toll-like receptor 9 responses. Biochem Biophys Res Commun. 2008;367:693–699. doi: 10.1016/j.bbrc.2007.12.130. [DOI] [PubMed] [Google Scholar]
- 55.Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208:643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peter ME, Kubarenko AV, Weber AN, Dalpke AH. Identification of an N-terminal recognition site in TLR9 that contributes to CpG-DNA-mediated receptor activation. J Immunol. 2009;182:7690–7697. doi: 10.4049/jimmunol.0900819. [DOI] [PubMed] [Google Scholar]
- 57.Kasperkovitz PV, Cardenas ML, Vyas JM. TLR9 is actively recruited to Aspergillus fumigatus phagosomes and requires the N-terminal proteolytic cleavage domain for proper intracellular trafficking. J Immunol. 2010;185:7614–7622. doi: 10.4049/jimmunol.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Cattaneo A, Gobert FX, Müller M, Toscano F, Flores M, Lescure A, Del Nery E, et al. Cleavage of Toll-like receptor 3 by cathepsins B and H is essential for signaling. Proc Natl Acad Sci USA. 2012;109:9053–9058. doi: 10.1073/pnas.1115091109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qi R, Singh D, Kao CC. Proteolytic processing regulates Toll-like receptor 3 stability and endosomal localization. J Biol Chem. 2012;287:32617–3229. doi: 10.1074/jbc.M112.387803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 61.Delgado M, Singh S, De Haro S, Master S, Ponpuak M, Dinkins C, Ornatowski W, et al. Autophagy and pattern recognition receptors in innate immunity. Immunol Rev. 2009;227:189–202. doi: 10.1111/j.1600-065X.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oh JE, Lee HK. Autophagy in innate recognition of pathogens and adaptive immunity. Yonsei Med J. 2012;53:241–247. doi: 10.3349/ymj.2012.53.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 64.Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010;189:925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delgado MA, Deretic V. Toll-like receptors in control of immunological autophagy. Cell Death Differ. 2009;16:976–983. doi: 10.1038/cdd.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaitseva E, Yang ST, Melikov K, Pourmal S, Chernomordik LV. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog. 2010;6:e1001131. doi: 10.1371/journal.ppat.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81:12543–12553. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, Bernasconi R, et al. Coronaviruses Hijack the LC3-I-positive edemosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taylor MP, Kirkegaard K. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy. 2008;4:286–289. doi: 10.4161/auto.5377. [DOI] [PubMed] [Google Scholar]
- 72.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 74.Audry M, Ciancanelli M, Yang K, Cobat A, Chang HH, Sancho-Shimizu V, Lorenzo L, et al. NEMO is a key component of NF-κB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J Allergy Clin Immunol. 2011;128:610–617. doi: 10.1016/j.jaci.2011.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10:1366–1373. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- 76.Nasirudeen AMA, Wong HH, Thien PL, Xu S, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Tropl Dis. 2011;5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le Goffic R, Balloy V, Lagranderie M, Alexopoulou L, Escriou N, Flavell R, Chignard M, et al. Detrimental contribution of the Toll-like receptor (TLR) 3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2006;2:e53. doi: 10.1371/journal.ppat.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hardarson HS, Baker JS, Yang Z, Purevjav E, Huang CH, Alexopoulou L, Li N, et al. Toll-like receptor 3 is an essential component of the innate stress response in virus-induced cardiac injury. Am J Physiol Heart Circ Physiol. 2007;292:H251–H258. doi: 10.1152/ajpheart.00398.2006. [DOI] [PubMed] [Google Scholar]
- 79.Edelmann KH, Richardson-Burns S, Alexopoulou L, Tyler KL, Flavell RA, Oldstone MB. Does Toll-like receptor 3 play a biological role in virus infections? Virology. 2004;322:231–238. doi: 10.1016/j.virol.2004.01.033. [DOI] [PubMed] [Google Scholar]
- 80.Schulz O, Diebold SS, Chen M, Näslund TI, Nolte MA, Alexopoulou L, Azuma YT, et al. Toll-like receptor 3 promotes cross-priming to virus- infected cells. Nature. 2005;433:887–892. doi: 10.1038/nature03326. [DOI] [PubMed] [Google Scholar]
- 81.Negishi H, Osawa T, Ogami K, Ouyang X, Sakaguchi S, Koshiba R, Yanai H, et al. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc Natl Acad Sci USA. 2008;105:20446–20451. doi: 10.1073/pnas.0810372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oshiumi H, Matsumoto M, Seya T. TICAM-1/TRIF, a TLR3 adaptor, is essential for protection against poliovirus infection. Int Immunol. 2010;22:i19. [Google Scholar]
- 83.Gowen BB, Hoopes JD, Wong MH, Jung KH, Isakson KC, Alexopoulou L, Flavell RA, et al. TLR3 deletion limits mortality and disease severity due to phlebovirus infection. J Immunol. 2006;177:6301–6307. doi: 10.4049/jimmunol.177.9.6301. [DOI] [PubMed] [Google Scholar]
- 84.Iwakiri D, Zhou L, Samanta M, Matsumoto M, Ebihara T, Seya T, Imai S, et al. Epstein-Barrvirus (EBV)- encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J Exp Med. 2009;206:2091–2099. doi: 10.1084/jem.20081761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li K, Li NL, Wei D, Pfeffer SR, Fan M, Pfeffer LM. Activation of chemokine and inflammatory cytokine response in hepatitis C virus-infected hepatocytes depends on Toll-like receptor 3 sensing of hepatitis C virus double-stranded RNA intermediates. Hepatology. 2012;55:666–675. doi: 10.1002/hep.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.West J, Damania B. Upregulation of the TLR3 pathway by kaposi's sarcoma-associated herpesvirus during primary infection. J Virol. 2008;82:5440–5449. doi: 10.1128/JVI.02590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hutchens M, Luker KE, Sottile P, Sonstein J, Lukacs NW, Núñez G, Curtis JL, et al. TLR3 increases disease morbidity and mortality from vaccinia infection. J Immunol. 2008;180:483–491. doi: 10.4049/jimmunol.180.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, Segal D, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 89.Tabeta K, Georgel P, Janssen E, Du X, Hoebe K, Crozat K, Mudd S, et al. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc Natl Acad Sci USA. 2004;101:3516–3522. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aksoy E, Zouain CS, Vanhoutte F. Double stranded RNAs from the helminth parasite activates TLR3 in dendritic cells. J Biol Chem. 2005;280:277–283. doi: 10.1074/jbc.M411223200. [DOI] [PubMed] [Google Scholar]
- 91.Carvalho A, De Luca A, Bozza S, Cunha C, D'Angelo C, Moretti S, Perruccio K, et al. TLR3 essentially promotes protective class I-restricted memory CD8+T-cell responses to Aspergillus fumigatus in hematopoietic transplanted patients. Blood. 2012;119:967–977. doi: 10.1182/blood-2011-06-362582. [DOI] [PubMed] [Google Scholar]
- 92.Diebold SS, Kaisho T, Hemmi H, Akira S, Reise SC. Innate antiviral responses by means of TLR7- mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 93.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1481–1482. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 94.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Triantafilou K, Orthopoulos G, Vakakis E, Ahmed MA, Golenbock DT, Lepper PM, Triantafilou M. Human cardiac inflammatory responses triggered by Coxsackie B viruses are mainly Toll-like receptor (TLR) 8-dependent. Cell Microbiol. 2005;7:1117–1126. doi: 10.1111/j.1462-5822.2005.00537.x. [DOI] [PubMed] [Google Scholar]
- 96.Triantafilou K, Vakakis E, Orthopoulos G, Ahmed MA, Schumann C, Lepper PM, Triantafilou M. TLR8 and TLR7 are involved in the host's immune response to human parechovirus 1. Eur J Immunol. 2005;35:2416–2423. doi: 10.1002/eji.200526149. [DOI] [PubMed] [Google Scholar]
- 97.Zhang YL, Guo YJ, Sun SH, Su X, Cai K, Wang D, Chen H, et al. Molecular cloning and functional characterization of porcine toll-like receptor 7 involved in recognition of single-stranded RNA virus/ssRNA. Mol Immunol. 2008;45:1184–1190. doi: 10.1016/j.molimm.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 98.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, et al. Cell type specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 99.Wang JP, Liu P, Latz E, Golenbock DT, Finberg RW, Libraty DH. Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. J Immunol. 2006;177:7114–7121. doi: 10.4049/jimmunol.177.10.7114. [DOI] [PubMed] [Google Scholar]
- 100.Zhang YL, Guo YJ, Li B, Sun SH. Hepatitis C virus single-stranded RNA induces innate immunity via Toll-like receptor 7. J Hepatol. 2009;51:29–38. doi: 10.1016/j.jhep.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 101.Cervantes-Barragan L, Zust R, Weber F, Spiegel M, Lang KS, Akira S, Thiel V, et al. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kane M, Case LK, Wang C, Yurkovetskiy L, Dikiy S, Golovkina TV. Innate immune sensing of retroviral infection via Toll-like receptor 7 occurs upon viral entry. Immunity. 2011;35:135–145. doi: 10.1016/j.immuni.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Davidson S, Kaiko G, Loh Z, Lalwani A, Zhang V, Spann K, Foo SY, et al. Plasmacytoid dendritic cells promote host defense against acute pneumovirus infection via the TLR7-MyD88-dependent signaling pathway. J Immunol. 2011;186:5938–5948. doi: 10.4049/jimmunol.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cervantes JL, Weinerman B, Basole C, Salazar JC. TLR8: the forgotten relative revindicated. Cell Mol Immunol. 2012;9:434–438. doi: 10.1038/cmi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cervantes JL, Dunham-Ems SM, La Vake CJ, Petzke MM, Sahay B, Sellati TJ, Radolf JD, et al. Phagosomal signaling by Borrelia burgdorferi in human monocytes involves Toll-like receptor (TLR) 2 and TLR8 cooperativity and TLR8-mediated induction of IFN-beta. Proc Natl Acad Sci USA. 2011;108:3683–3688. doi: 10.1073/pnas.1013776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, Teti G, et al. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat Immunol. 2009;10:587–594. doi: 10.1038/ni.1733. [DOI] [PubMed] [Google Scholar]
- 107.Caetano BC, Carmo BB, Melo MB, Cerny A, dos Santos SL, Bartholomeu DC, Golenbock DT, et al. Requirement of UNC93B1 reveals a critical role for TLR7 in host resistance to primary infection with Trypanosoma cruzi. J Immunol. 2011;187:1903–1911. doi: 10.4049/jimmunol.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bourgeois C, Majer O, Frohner IE, Lesiak-Markowicz I, Hildering KS, Glaser W, Stockinger S, et al. Conventional dendritic cells mount a type I IFN response against Candida spp. requiring novel phagosomal TLR7-mediated IFN-beta signaling. J Immunol. 2011;186:3104–3112. doi: 10.4049/jimmunol.1002599. [DOI] [PubMed] [Google Scholar]
- 109.Biondo C, Signorino G, Costa A, Midiri A, Gerace E, Galbo R, Bellantoni A, et al. Recognition of yeast nucleic acids triggers a host-protective type I interferon response. Eur J Immunol. 2011;41:1969–1979. doi: 10.1002/eji.201141490. [DOI] [PubMed] [Google Scholar]
- 110.Hochrein H, Schlatter B, O'Keeffe M, Wagner C, Schmitz F, Schiemann M, Bauer S, et al. Herpes simplex virus type-1 induces IFN-alpha production via Toll-like receptor 9-dependent and-independent pathways. Proc Natl Acad Sci USA. 2004;101:11416–11421. doi: 10.1073/pnas.0403555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Delale T, Paquin A, Asselin-Paturel C, Dalod M, Brizard G, Bates EE, Kastner P, et al. MyD88-dependent and-independent murine cytomegalovirus sensing for IFN-alpha release and initiation of immune responses in vivo. J Immunol. 2005;175:672367–32. doi: 10.4049/jimmunol.175.10.6723. [DOI] [PubMed] [Google Scholar]
- 113.Zhu J, Huang X, Yang Y. Innate immune response to adenoviral vectors is mediated by both Toll-like receptor-dependent and-independent pathways. J Virol. 2007;81:3170–3180. doi: 10.1128/JVI.02192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Abe T, Hemmi H, Miyamoto H, Moriishi K, Tamura S, Takaku H, Akira S, et al. Involvement of the Toll-like receptor 9 signaling pathway in the induction of innate immunity by baculovirus. J Virol. 2005;79:2847–2858. doi: 10.1128/JVI.79.5.2847-2858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zauner L, Nadal D. Understanding TLR9 action in Epstein-Barr virus infection. Front Biosci. 2012;17:1219–1231. doi: 10.2741/3982. [DOI] [PubMed] [Google Scholar]
- 116.NordstrÖm I, Eriksson K. HHV-6B induces IFN-lambda1 responses in cord plasmacytoid dendritic cells through TLR9. PLoS one. 2012;7:e38683. doi: 10.1371/journal.pone.0038683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu HR, Huang HC, Kuo HC, Sheen JM, Ou CY, Hsu TY, Yang KD. IFN-α production by human mononuclear cells infected with varicella-zoster virus through TLR9-dependent and -independent pathways. Cell Mol Immunol. 2011;8:181–188. doi: 10.1038/cmi.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guggemoos S, Hangel D, Hamm S, Heit A, Bauer S, Adler H. TLR9 contributes to antiviral immunity during gammaherpesvirus infection. J Immunol. 2008;180:438–443. doi: 10.4049/jimmunol.180.1.438. [DOI] [PubMed] [Google Scholar]
- 119.Samuelsson C, Hausmann J, Lauterbach H, Schmidt M, Akira S, Wagner H, Chaplin P, et al. Survival of lethal poxvirus infection in mice depends on TLR9, and therapeutic vaccination provides protection. J Clin Invest. 2008;118:1776–1784. doi: 10.1172/JCI33940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Arpaia N, Godec J, Lau L, Sivick KE, McLaughlin LM, Jones MB, Dracheva T, et al. TLR signaling is required for Salmonella typhimurium virulence. Cell. 2011;144:675–688. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lecours MP, Segura M, Fittipaldi N, Rivest S, Gottschalk M. Immune receptors involved in Streptococcus suis recognition by dendritic cells. PLoS One. 2012;7:e44746. doi: 10.1371/journal.pone.0044746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Saiga H, Shimada Y, Takeda K. Innate immune effectors in mycobacterial infection. Clin Dev Immunol. 2011;2011:347594. doi: 10.1155/2011/347594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Albiger B, Dahlberg S, Sandgren A, Wartha F, Beiter K, Katsuragi H, Akira S, et al. Toll-like receptor 9 acts at an early stage in host defence against pneumococcal infection. Cell Microbiol. 2007;9:633–644. doi: 10.1111/j.1462-5822.2006.00814.x. [DOI] [PubMed] [Google Scholar]
- 124.Zinkernagela AS, Hruzc P, Uchiyamaa S, von Köckritz-Blickwedea M, Schuepbachd RA, Hayashic T, Carsonc DA, et al. Importance of Toll-like receptor 9 in host defense against M1T1 group A Streptococcus infections. J Innate Immun. 2012;4:213–218. doi: 10.1159/000329550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Uchiyama S, Andreoni F, Schuepbach RA, Nizet V, Zinkernagel AS. DNase Sda1 allows invasive M1T1 group A Streptococcus to prevent TLR9-dependent recognition. PLoS Pathog. 2012;8:e1002736. doi: 10.1371/journal.ppat.1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Parker D, Prince A. Staphylococcus aureus induces type I IFN signaling in dendritic cells via TLR9. J Immunol. 2012;189:4040–4046. doi: 10.4049/jimmunol.1201055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bafica A, Santiago HC, Goldszmid R, Ropert C, Gazzinelli RT, Sher A. Cutting edge: TLR9 and TLR2 signaling together account for MyD88-dependent control of parasitemia in Trypanosoma cruzi infection. J Immunol. 2006;177:3515–3519. doi: 10.4049/jimmunol.177.6.3515. [DOI] [PubMed] [Google Scholar]
- 128.Drennan MB, Stijlemans B, Van den Abbeele J, Quesniaux VJ, Barkhuizen M, Brombacher F, De Baetselier P, et al. The induction of a type I immune response following a Trypanosoma brucei infection is MyD88 dependent. J Immunol. 2005;175:2501–2509. doi: 10.4049/jimmunol.175.4.2501. [DOI] [PubMed] [Google Scholar]
- 129.Abou Fakher FH, Rachinel N, Klimczak M, Louis J, Doyen N. TLR9-dependent activation of dendritic cells by DNA from Leishmania major favors Th1 cell development and the resolution of lesions. J Immunol. 2009;182:1386–1396. doi: 10.4049/jimmunol.182.3.1386. [DOI] [PubMed] [Google Scholar]
- 130.Coban C, Igari Y, Yagi M, Reimer T, Koyama S, Aoshi T, Ohata K, et al. Immunogenicity of whole parasite vaccines against plasmodium falciparum involves malarial hemozoin and host TLR9. Cell Host Microbe. 2010;7:50–61. doi: 10.1016/j.chom.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 131.Parroche P, Lauw FN, Goutagny N, Latz E, Monks BG, Visintin A, Halmen KA, et al. Malaria hemozoin is immunologically inert but radically enhances innate responses by presenting malaria DNA to Toll-like receptor 9. Proc Natl Acad Sci USA. 2007;104:1919–1924. doi: 10.1073/pnas.0608745104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ramirez ZG, Specht OC, Wang JP, Lee CK, Bartholomeu DC, Gazzinelli RT, Levitz SM. Toll-like receptor-9 dependent immune activation by unmethylated CpG motifs in Aspergillus fumigatus DNA. Infect Immunol. 2008;76:2123–2129. doi: 10.1128/IAI.00047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kasperkovitz PV, Khan NS, Tam JM, Mansour MK, Davids PJ, Vyas JM. Toll-like receptor 9 modulates macrophage antifungal effector function during innate recognition of candida albicans and saccharomyces cerevisiae. Infect Immun. 2011;79:4858–4867. doi: 10.1128/IAI.05626-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bordon Y. Innate immunity: TLR13, unlucky, but just for some. Nat Rev Immunol. 2012;12:618–619. doi: 10.1038/nri3284. [DOI] [PubMed] [Google Scholar]
- 135.Hidmark A, von Saint Paul A, Dalpke AH. Cutting Edge: TLR13 is a receptor for bacterial RNA. J Immunol. 2012;189:2717–2721. doi: 10.4049/jimmunol.1200898. [DOI] [PubMed] [Google Scholar]
- 136.Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, et al. Sequence-and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lai Y, Yi G, Chen A, Bhardwaj K, Tragesser BJ, Rodrigo A Valverde, Zlotnick A, et al. Viral double-strand RNA-binding proteins can enhance innate immune signaling by Toll-like receptor 3. PLoS One. 2011;6:e25837. doi: 10.1371/journal.pone.0025837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lai Y, Adhikarakunnathu S, Bhardwaj K, Ranjith-Kumar CT, Wen Y, Jordan JL, Wu LH, et al. LL37 and cationic peptides enhance TLR3 signaling by viral double-stranded RNAs. PLoS One. 2011;6:e26632. doi: 10.1371/journal.pone.0026632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu J, Xu C, Hsu LC, Luo Y, Xiang R, Chuang TH. A five-amino-acid motif in the undefined region of the TLR8 ectodomain is required for species-specific ligand recognition. Mol Immunol. 2010;47:1083–1090. doi: 10.1016/j.molimm.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Govindaraj RG, Manavalan B, Basith S, Choi S. Comparative analysis of species-specific ligand recognition in Toll-like receptor 8 signaling: a hypothesis. PLoS One. 2011;6:e25118. doi: 10.1371/journal.pone.0025118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gorden KK, Qiu XX, Binsfeld CC, Vasilakos JP, Alkan SS. Cutting edge: activation of murine TLR8 by a combination of imidazoquinoline immune response modifiers and polyoligodeoxynucleotides. J Immunol. 2006;177:6584–6587. doi: 10.4049/jimmunol.177.10.6584. [DOI] [PubMed] [Google Scholar]
- 142.Demaria O, Pagni PP, Traub S, de Gassart A, Branzk N, Murphy AJ, Valenzuela DM, et al. TLR8 deficiency leads to autoimmunity in mice. J Clin Invest. 2010;120:3651–3662. doi: 10.1172/JCI42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Forsbach A, Nemorin JG, Montino C, Müller C, Samulowitz U, Vicari AP, Jurk M, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 144.Ruscanu S, Pascale F, Bourge M, Hemati B, Elhmouzi-Younes J, Urien C, Bonneau M, et al. The double-stranded RNA bluetongue virus induces type I interferon in plasmacytoid dendritic cells via a MYD88-dependent TLR7/8-independent signaling pathway. J Virol. 2012;86:5817–5828. doi: 10.1128/JVI.06716-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 146.Barrat FJ, Meeker T, Gregorio J, Barrat FJ, Meeker T, Gregorio J, Chan JH, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Baumann CL, Aspalter IM, Sharif O, Pichlmair A, Blüml S, Grebien F, Bruckner M, et al. CD14 is a coreceptor of Toll-like receptors 7 and 9. J Exp Med. 2010;207:2689–2701. doi: 10.1084/jem.20101111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cros J, Cagnard N, Woollard K, Patey N, Zhang SY, Senechal B, Puel A, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–386. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 150.Hanagata N. Structure-dependent immunostimulatory effect of CpG oligodeoxynucleotides and their delivery system. Int J Nanomed. 2012;7:2181–2195. doi: 10.2147/IJN.S30197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pauls E, Shpiro N, Peggie M, Young ER, Sorcek RJ, Tan L, Choi HG, et al. Essential role for IKKβ in production of type I interferons by plasmacytoid dendritic cells. J Biol Chem. 2012;287:19216–1928. doi: 10.1074/jbc.M112.345405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Siggs OM, Berger M, Krebs P, Arnold CN, Eidenschenk C, Huber C, Pirie E, et al. A mutation of Ikbkg causes immune deficiency without impairing degradation of IkappaB alpha. Proc Natl Acad Sci USA. 2010;107:3046–3051. doi: 10.1073/pnas.0915098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 154.Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, Murthy N, Pulendran B. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI3K-mTOR-p70S6K pathway. Nat Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tsujimura H, Tamura T, Kong HJ, Nishiyama A, Ishii KJ, Klinman DM, Ozato K. Toll-like receptor 9 signaling activates NF-κB through IFN regulatory factor-8/IFN consensus sequence binding protein in dendritic cells. J Immunol. 2004;172:6820–7682. doi: 10.4049/jimmunol.172.11.6820. [DOI] [PubMed] [Google Scholar]
- 156.Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 2012;33:449–458. doi: 10.1016/j.it.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 157.Zeng W, Xu M, Liu S, Sun L, Chen ZL. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol Cell. 2009;36:315–25. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shi HX, Liu X, Wang Q, Tang PP, Liu XY, Shan YF, Wang C. Mitochondrial ubiquitin ligase MARCH5 promotes TLR7 signaling by attenuating TANK action. PLoS Pathog. 2011;7:e1002057. doi: 10.1371/journal.ppat.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Saitoh T, Satoh T, Yamamoto N, Uematsu S, Takeuchi O, Kawai T, Akira S. Antiviral protein viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity. 2011;34:352–363. doi: 10.1016/j.immuni.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 160.Renner F, Saul VV, Pagenstecher A, Wittwer T, Schmitz ML. Inducible SUMO modification of TANK alleviates its repression of TLR7 signaling. EMBO Rep. 2011;12:129–135. doi: 10.1038/embor.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao XT. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat Immunol. 2009;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 162.Nahid MA, Satoh M, Chan EK. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol Immunol. 2011;8:388–403. doi: 10.1038/cmi.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yang G, Yang L, Zhao Z, Wang JJ, Zhang XB. Signature miRNAs involved in the innate immunity of invertebrates. PLoS One. 2012;7:e39015. doi: 10.1371/journal.pone.0039015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-κB signaling. J Mol Cell Biol. 2011;3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol. 2004;5:495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- 166.Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappaB activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 167.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 168.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Saitoh T, Yamamoto M, Miyagishi M, Taira K, Nakanishi M, Fujita T, Akira S, et al. A20 is a negative regulator of IFN regulatory factor 3 signaling. J Immunol. 2005;174:1507–1512. doi: 10.4049/jimmunol.174.3.1507. [DOI] [PubMed] [Google Scholar]
- 170.Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O'Rourke KM, Eby M, et al. A deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 171.Friedman CS, O'Donnell MA, Legarda-Addison D, Ng A, Cárdenas WB, Yount JS, Moran TM, et al. The tumor suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li S, Zheng H, Mao AP, Zhong B, Li Y, Liu Y, Gao Y, et al. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010;285:4291–4297. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kawagoe T, Takeuchi O, Takabatake Y, Kato H, Isaka Y, Tsujimura T, Akira S. TANK is a negative regulator of Toll-like receptor signaling and is critical for the prevention of autoimmune nephritis. Nat Immunol. 2009;10:965–972. doi: 10.1038/ni.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.An H, Zhao W, Hou J, Zhang Y, Xie Y, Zheng YJ, Xu HM, et al. SHP-2 phosphatase negatively regulates the TRIF adaptor protein-dependent type I interferon and proinflammatory cytokine production. Immunity. 2006;25:919–928. doi: 10.1016/j.immuni.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 176.Croker BA, Lawson BR, Rutschmann S, Berger M, Eidenschenk C, Blasius AL, Moresco EM, et al. Inflammation and autoimmunity caused by a SHP1 mutation depend on IL-1, MyD88, and a microbial trigger. Proc Natl Acad Sci USA. 2008;105:15028–15033. doi: 10.1073/pnas.0806619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bao M, Hanabuchi S, Facchinetti V, Du Q, Bover L, Plumas J, Chaperot L, et al. CD2AP/SHIP1 complex positively regulates plasmacytoid dendritic cell receptor signaling by inhibiting the E3 ubiquitin ligase Cbl. J Immunol. 2012;189:786–792. doi: 10.4049/jimmunol.1200887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chockalingam A, Cameron JL, Brooks JC, Leifer CA. Negative regulation of signaling by a soluble form of toll-like receptor 9. Eur J Immunol. 2011;41:2176–2184. doi: 10.1002/eji.201041034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Carty M, Goodbody R, Schröder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- 180.Janssens S, Burns K, Tschopp J, Beyaert R. Regulation of interleukin-1- and lipopolysaccharide-induced NF-kappaB activation by alternative splicing of MyD88. Curr Biol. 2002;12:467–471. doi: 10.1016/s0960-9822(02)00712-1. [DOI] [PubMed] [Google Scholar]
- 181.Kobayashi K, Hernandez LD, Galán JE, Janeway CA, Jr, Medzhitov R, Flavell RA. IRAK-M is a negative regulator of toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 182.Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J. Inhibition of interleukin 1 receptor/toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med. 2003;197:263–268. doi: 10.1084/jem.20021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.O'Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signaling. Nat Revs Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 184.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA. 2009;106:2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pifer R, Benson A, Sturge CR, Yarovinsky F. UNC93B1 is essential for TLR11 activation and IL-12- dependent host resistance to Toxoplasma gondii. J Biol Chem. 2011;286:3307–3314. doi: 10.1074/jbc.M110.171025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yokota SI, Okabayashi T, Fujii N. The battle between virus and host: modulation of Toll-like receptor signaling pathways by virus infection. Mediators Inflamm. 2010;2010:184328. doi: 10.1155/2010/184328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Celhar T, Magalhães R, Fairhurst AM. TLR7 and TLR9 in SLE: when sensing self goes wrong. Immunol Res. 2012;53:58–77. doi: 10.1007/s12026-012-8270-1. [DOI] [PubMed] [Google Scholar]
- 189.Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 190.Krieg A, Vollmer J. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol Rev. 2007;220:251–269. doi: 10.1111/j.1600-065X.2007.00572.x. [DOI] [PubMed] [Google Scholar]