Abstract
Identification of potential virus–host interactions is useful and vital to control the highly infectious virus-caused diseases. This may contribute toward development of new drugs to treat the viral infections. Recently, database records of clinically and experimentally validated interactions between a small set of human proteins and Ebola virus (EBOV) have been published. Using the information of the known human interaction partners of EBOV, our main objective is to identify a set of proteins that may interact with EBOV proteins. Here, we first review the state-of-the-art, computational methods used for prediction of novel virus–host interactions for infectious diseases followed by a case study on EBOV–human interactions. The assessment result shows that the predicted human host proteins are highly similar with known human interaction partners of EBOV in the context of structure and semantics and are responsible for similar biochemical activities, pathways and host–pathogen relationships.
Keywords: virus–host interaction prediction, infectious diseases, Ebola glycoprotein, Ebola–human interactions, computational methods
Introduction
Infectious diseases are still among the major and prevalent death causes in human mostly because of the unknown pathogenic mechanisms of viruses [1]. The eukaryotic hosts are directly affected by the viral pathogens through complex interaction mechanisms [2]. The molecular-level interactions between the virus and their host play a key role here. Thus, the virus–host protein–protein interactions (PPIs) are crucial for better understanding of the infection mechanism and pathogenesis of infectious diseases [3, 4].
In proteomic studies, PPI prediction remains a hot topic for decades. Owing to the limitation of proteomic data, most of the previous studies were focused on predicting intra-species PPIs, i.e. interactions within a single organism. In recent analysis, several improvements have been reported in PPI predictions between different organisms, i.e. inter-species. These types of PPI prediction methods offer important information for further analysis of infection mechanisms between different species. PPIs between virus and host proteins allow pathogenic microorganisms to manipulate host mechanisms to use host capabilities and to escape from host immune responses [5–8]. Therefore, a complete understanding of infection mechanisms through PPIs is crucial for the development of new and more effective therapeutics.
The computational PPI prediction methods primarily use sequence information [9–14], domain-based [5, 15–17], protein structure [18–21], physiochemical properties [22], semantic analysis [23, 24] and known interactions between virus and host proteins [25]. Classical machine learning techniques are well-accepted tools for PPI predictions when there are sufficient numbers of known interactions. In contrast, prediction of inter-species (virus–host) PPI is relatively young field of study, which requires new model-based approaches. To tackle the problem of data scarcity, eliciting and transferring data from related domains to a desired formulation can be a promising solution [26, 27]. Multitask learning [28–30] uses relationship among different domains and learns the problem simultaneously, which leads to a better performance rather conducting learning task on individual domain.
In this article, we concentrate on the computational approaches regarding the prediction of virus–host interactions, followed by a case study on prediction of novel interactions between Ebola virus (EBOV) and human host proteins. We also present a brief cluster analysis on the predicted host proteins. This analysis includes the infection pathways and functional annotations that can be valuable in prevention, diagnosis and treatment of EBOV-infected diseases.
Computational approaches
Several computational approaches have been developed to predict novel host–virus protein interactions. Depending on the availability of the interaction information, different predictive models have been proposed in novel host–virus interaction predictions. Becerra et al. [31] have proposed short linear motifs-based predicting method for PPIs between HIV-1 and human proteins. They have implemented three filtering methods to obtain linear motif sets: (i) conserved in viral proteins (C), (ii) located in disordered regions (D) and (iii) rare or scarce in a set of randomized viral sequences (R). Finally, these three sets have used to find the disordered protein regions among the HIV-1 sequences and host sequences. Their study shows that the majority of conserved linear motifs in the virus are located in disordered regions. In [32], Segura et al. proposed a method to model viral–human interaction network using motif–domain interactions.
Kharrat et al. [33] used structure sequence to cluster human viral proteomes. They analyzed the charged residue of amino acid composition (AC) between the viral and target host proteins and provided a better understanding of several pathological and biological processes (BPs). The charged residues in protein sequences mediate the interactions and play an important role in protein transport, localization and regulatory functions.
Mukhopadhyay et al. [34, 35] proposed a bi-clustering approach to predict HIV-1-infected human proteins applying interaction-based analysis. A set of association rules was mined by bi-clustering technique from the interaction between HIV-1 proteins. Finally, a set of high-confidence rule was extracted to predict novel human protein interactions. As a further improvement to their work, Mukhopadhyay et al. [36] introduced type and direction (host-to-virus and virus-to-host)-based bi-clustering to existing interactions to predict novel host proteins. Mondal et al. [37] also proposed a HIV–human interaction prediction method using hierarchical bi-clustering and minimal covers of association rule mining.
An approach for predicting future dominant hemagglutinin gene of influenza A virus was proposed by Plotkin et al. [38] and antigenic evolution over the host genes were analyzed. The spatiotemporal distribution of viral swarm and the evolution of hemagglutinin structure were analyzed for clustering. Finally, a critical length scaling in amino acid space was applied to cluster the viral sequences.
A sequence-based hierarchical clustering approach over the EBOV and influenza virus was introduced in [39]. In a study by Spencer et al. [40], phosphorylation clustering was applied over the infection of bronchitis virus protein. Sequence similarity-based domain–domain interaction detection was proposed by Schleker et al. [41]. Salmonella–human interaction was predicted by this method. Several approaches proposed on Salmonella–human interaction predictions [42, 43].
Support Vector Machine (SVM) based approaches were successfully applied in virus–host protein interaction prediction studies [44, 45]. Cui et al. [44] proposed a SVM-based approach, where they used fixed-length feature vector indicating relative frequency of consecutive amino acids in the protein sequence.
Doolittle et al. [46] proposed a method to predict the interactions between HIV-1 and human proteins based on protein structural similarity, where two crystal structures are compared with compute the structural similarity between host and pathogen proteins. The assumption is that, human proteins that have high structural similarity to a HIV protein are identified and their known interacting partners are considered as targets. They applied similar approach for developing interaction network between Dengue virus and the host [47].
Cao et al. [48] proposed a network-based approach to predict EBOV–human interaction. They introduced a principle called ‘guilt by association’ (GBA) for their prediction method. The GBA principle was stated as proteins interacting with each other are likely to function similarly or the same. Based on this assumption, they predicted EBOV infection-related human genes from a PPI network using Dijkstra algorithm.
In several works [49–52], it has been reported that, molecular mimicry plays a key role in viral–host pathogen interactions, where a viral protein mimics similar structural binding surface similar to that of a host protein. As a result, viral protein competitively binds to another host protein and spread over the host. From available experimental data [53–56], it has been suggested that pathogenic agents extensively use the molecular mimicry to their advantages.
Mei et al. [57] proposed transfer learning-based technique with three different classifier, where each individual classifier was executed on three gene ontology (GO)-based features. Finally, the classifier ensembling was applied to produce final result using weighting probability outputs of individual classifiers.
In addition, to analyze the biological activity of proteins, GO-based semantic similarity creates an evolutionary orientation in PPI [58–63]. GO annotation-based semantic similarity has been regarded as one of the most powerful indicators for interaction [23, 64]. Thus, structural and semantic similarity becomes valuable features for interaction prediction involving in novel host protein. Table 1 summarizes the list of methods that have been successfully applied in virus–host (human) interaction prediction in the literature.
Table 1.
Computational approaches on virus host (human) interaction prediction
| Interactors | Approaches/methods | References |
|---|---|---|
| Ebola–human | Graph-based multitask learning-based approach | [26] |
| Network similarity-based approach | [48] | |
| Dengue virus–human | Sequence- and structure-based method | [42] |
| Structural motif–domain interaction-based approach | [32] | |
| Structural similarity of DENV proteins to human proteins having known interactions | [47] | |
| Human papillomaviruse–human | Relative frequency of amino acid triplets (RFATs), frequency difference of amino acid triplets (FDATs) and AC | [45] |
| Fixed-length feature vector of protein sequence | [44] | |
| Hepatitis C virus–human | Graph-based multitask learning-based approach | [26] |
| RFATs, FDATs and AC | [45] | |
| Sequence, network topology, domain, GO and pathway-based kernel method | [12] | |
| Topological and functional properties of interaction network and domain interaction-based method | [25] | |
| Fixed-length feature vector of protein sequence | [44] | |
| Salmonella–human | Sequence- and structure-based method | [42] |
| Multi-instance homolog transfer-based approach | [43] | |
| Virus–host domain interaction, gene expression, pathway sharing and sequence-based | [26, 28] | |
| Obtain host–pathogen interactome using sequence and interacting domain similarity to known PPIs | [41] | |
| Homology detection method using template PPI databases | [17] | |
| Plasmodium falciparum–human | K-mer-based sequence homology and pathway-based approach | [14] |
| GO annotation and sequence filtering-based approach | [10] | |
| Homology detection method using template PPI databases | [11] | |
| Domain–domain interaction probability-based approach | [6] | |
| Influenza A–human | Graph-based multitask learning-based approach | [26] |
| Structural homology-based approach | [21 | |
| HIV-1–human | Short linear motifs-based approach | [31] |
| Bi-clustering with association rule mining | [34–36] | |
| Sequence-based classifier ensembling | [13] | |
| Differential gene expression between virus and host | [24] | |
| Hierarchical bi-clusters and minimal covers of association rule-based approach | [37] | |
| Supervised learning and prediction of physical interactions | [5] | |
| Escherichia coli–human | Homology detection method using template PPI databases | [17] |
| Mycobacterium tuberculosis (H37Rv)–human | Stringent homology which uses inter-species template PPI | [4] |
In the following sections, a case study on prediction of novel EBOV–human interaction is discussed. The structural and GO-based semantic assessment scores are considered as effective features of these predictive models with four classifiers. Finally a cluster of predicted EBOV host is extracted from the prediction result. The known host proteins are considered as seed for the predictions, where unknown human proteins are considered as target; thus, seed and target creates the interaction pair. The predictive analysis on these EBOV–human protein pairs is discussed in the following sections. This assessment will facilitate the diagnosis and treatment of EBOV infections.
The role of Ebola glycoprotein in virus–host interaction
The Ebola hemorrhagic fever by EBOV infection causes pervasive human fatality and mortality. EBOV, a member of the Filoviridae family, is a negative-sense RNA virus [65]. The EBOV genome consists of seven genes, namely, nucleoprotein (NP), viral polymerase complex protein 35 (VP35), matrix protein (VP40), glycoprotein (GP), minor NP (VP30), membrane-associated protein (VP24) and polymerase (L-protein) [66]. Among the seven genes of EBOV, GP is the only viral protein that is present on the EBOV surface and is responsible for mediating attachment to host cell surface receptors and entry of the virus into host cells. Mature GP is a trimer of three disulfide-linked GP1-GP2 heterodimers [67–69]. GP1 mediates adhesion of the virus to host cells and regulates GP2, which carries out membrane fusion [70–72]. The EBOV virus initially targets specific cell types, including liver cells, immune system cells and endothelial cells. EBOV GP can damage cell adhesion, so that cells do not remain attached to each other and to the extracellular matrix. By targeting liver cells, EBOV disrupts the mechanism of removal toxins from the bloodstream. The infection leads to organ failure, fever and severe internal bleeding, which ultimately leads to death [73].
The first step in EBOV infection is attachment to host cell surface receptors. Specifically, EBOV GP allows the virus to introduce its contents into monocytes, macrophages, dendritic cells and/or endothelial cells [74–77], which causes the release of cytokines associated with inflammation and fever. EBOV GP has been found to be the most important factor required for EBOV entry into a host cell by binding to surface receptors on the host cell [78]. EBOV binds to human cells through various receptors expressed on the cell membrane surface—C-type lectins, which interact with glycans on EBOV GP [79–84], phosphatidylserine receptors, which interact with phosphatidylserine on EBOV GP [85–92], and integrins [93–95]. The cholesterol transporter, Niemann–Pick C1 (NPC1), facilitates EBOV entry into host cells and the release of the virus from the vacuole into the cytoplasm of the host [96, 97]. EBOV entry into endosomal compartments is primarily achieved through macropinocytosis, as well as other entry mechanisms, such as clathrin-dependent endocytosis [91, 98–100].
As EBOV GP mediates the entry of the virus into host cells, its role is important and essential to understand the interactions between EBOV GP and human cell surface receptors. The list of known interactions between EBOV GP and human with domain annotations is given in Table 2.
Table 2.
Known human target proteins of EBOV GP
| Functional group | Gene name | Protein name |
|---|---|---|
| C-type lectin domain family | CLEC4M [82] | Liver/lymph node-specific ICAM-3 grabbing non-integrin (L-SIGN) |
| CLEC4G [83] | Liver and lymph node sinusoidal endothelial cell C-type lectin (LSECTIN) | |
| CLEC10A [81] | Human macrophage galactose-and N-acetyl-galactosamine-specific C-type lectin (hMGL) | |
| Dendritic cell-specific intercellular adhesion molecule (ICAM) | CD209 [82] | Dendritic cell-specific intercellular adhesion molecule-3-grabbingnon-integrin (DC-SIGN) |
| Tyrosine-protein kinase receptor | AXL [90] | Tyrosine-protein kinase receptor UFO |
| TYRO3 [89] | Tyrosine-protein kinase receptor TYRO3 | |
| MER [89] | Tyrosine-protein kinase Mer | |
| T-cell immunoglobulin and mucin domain | TIM1 [85] | T-cell immunoglobulin and mucin domain-containing protein 1 |
| TIM4 [79] | T-cell immunoglobulin and mucin domain-containing protein 4 | |
| Integrin domain | ITGB1 [94] | Integrin beta-1 |
| ITGA5 [93] | Integrin alpha-5 | |
| Lactadherin | MFGE8 [92] | Lactadherin |
| Growth arrest-specific protein | GAS6 [79] | Growth arrest-specific protein 6 |
Structure-based similarity assessment
In structure-based analysis, each residue on the surface is compared with the target residue to extract the structural neighbors. A variety of features derived in different analysis [101–110] from the structural component of virus–host protein pairs. In this analytic approach, three-dimensional structure-based protein features are incorporated to find the structural neighbors. Five scoring metrics, Template modeling-score (TM-score) [111], RMSD [112], MaxSub-score [113], GDT_score [114] and Tm-Rm score [Equation (1)], are used to quantify the structural similarity of two proteins.
TM-score [111] gives a value in the range (0, 1), where 1 indicates a perfect match in topological similarity of two protein structures. Scores <0.17 indicates no structural similarity, whereas a score >0.5 suggests that the two structures have similar fold. We use the TM align algorithm [115] for comparing the structures of two proteins. This algorithm identifies the best structural alignment between two proteins.
After the optimal superposition, RMSD [112] represents the root-mean-square deviation of all the equivalent atom pairs of two protein structures. In general, lower RMSD indicates better superposition. For similar structural domain identification, data sets like SCOP and CATH set a RMSD threshold of 5 Å. An RMSD value <3 Å indicates a high degree of structural similarity. However, a lower RMSD and higher TM-score indicate a better structural similarity; thus, they are inversely related. In addition, RMSD value 0 and TM-score 1 represent optimal structural similarity.
MaxSub-score method [113] identifies the largest subset of atom of a protein structure that superimposes well over another structure and provides a single normalized score. MaxSub score ranges from 0 to 1, where 0 indicates a wrong superimposing and 1 indicates perfect superimposition [113].
The global distance test (GDT) score [114] is calculated as the largest set of residue-based atoms in a structure falling within a defined distance cutoff of their position with respect to other structure. An increase in GDT may indicate an extreme divergence between a structure pair, such that no additional atoms are included in any cutoff of a reasonable distance [116].
Finally, a new structural similarity-based property is defined using both TM-score and RMSD.
| (1) |
Here, the RMSD score is restrict up to 3 Å for higher structural similarity. In Equation (1), any RMSD value <3 Å will contribute positively with TM-score.
GO-based semantic assessment
The semantic similarity between human proteins is estimated by combining the similarities of their annotating GO term pairs belonging to a particular ontology [e.g. molecular function (MF), BP, cellular component (CC)]. Similarity of a GO term pair is determined by considering certain topological properties (shortest path length) of the GO graph and the average information content (IC) of the disjunctive common ancestors (DCAs) of the GO terms as proposed in [23].
In this measure, to estimate the semantic similarity between two GO terms t1 and t2, first certain GO terms are selected as cluster centers based on a value called propTerms(t) assigned to each GO term t in the GO graph, which gives the proportion of GO terms connected directly and indirectly to t in the ontology. The GO terms for which this propTerms value is above a given threshold are selected as cluster centers. The threshold values for selecting cluster centers with respect to MF, CC and BP ontologies are given in Table 3. Depending on the interaction prediction result, the threshold values are chosen. Initially, threshold value is started from 0.1 and gradually increases. With the increasing threshold value, the area under receiver operating characteristic (ROC) curve (AUC) is determined with the varying k (width of Gaussian function) values (from 1 to 10). Finally, the threshold and k are chosen for which the AUC score is highest. After selecting the cluster centers, the degree of membership of a GO term to each of the selected cluster centers is calculated using its respective shortest path lengths to the corresponding cluster centers. The membership of the GO term to a cluster decreases with increase in its shortest path length to the cluster center. Next, using the difference in membership values of the GO terms t1 and t2 with respect to each cluster center, a weight parameter is defined as one minus the maximum membership difference value. This weight value determines how dissimilar two GO terms can be with respect to the cluster centers. Next, the average IC [117], of the DCAs of GO terms t1 and t2, is determined. Finally, the semantic similarity between the two GO terms t1 and t2 is defined as the product of the weight parameter and the average IC of the DCAs of the two GO terms.
Table 3.
GO-based cluster center threshold with k-value
| GO | Cluster center threshold | Width k |
|---|---|---|
| MF | 0.18 | 2 |
| CC | 0.55 | 1 |
| BP | 0.31 | 3 |
To determine the semantic similarity scores for protein pairs, the semantic similarity scores of their respective GO terms are combined using the best-match average approach [118, 119]. Here, the semantic similarity is estimated with respect to BP, CC and MF ontologies of the GO database [120] separately.
Data sources
Ontology data
Ontology data are downloaded from the GO database [121, 120] (dated July 2015) containing 43 368 ontology terms subdivided into 28 539 BP terms, 10 868 MF terms and 3961 CC terms.
GO annotation data
GO annotations for human proteins are downloaded from the Uniprot database [122, 123].
Seed proteins
The seed human proteins (referred as set SD) for clustering are those proteins that are known to interact with EBOV GP. These known human seeds are assimilated from literature survey. The list of known target human proteins (seeds in our approach) of EBOV GP is given in Table 2.
Target proteins
The target human proteins are collected from Uniprot database [122, 123].These target proteins are selected such that they are the first-level interaction partner of the human seed proteins and have structural information in Protein Data Bank (PDB) [124]. In addition, human protein interaction information is collected from DIP [125], MINT [126], BioGrid [127], STRING [128] and iRefWeb [129] databases.
Clustering analysis
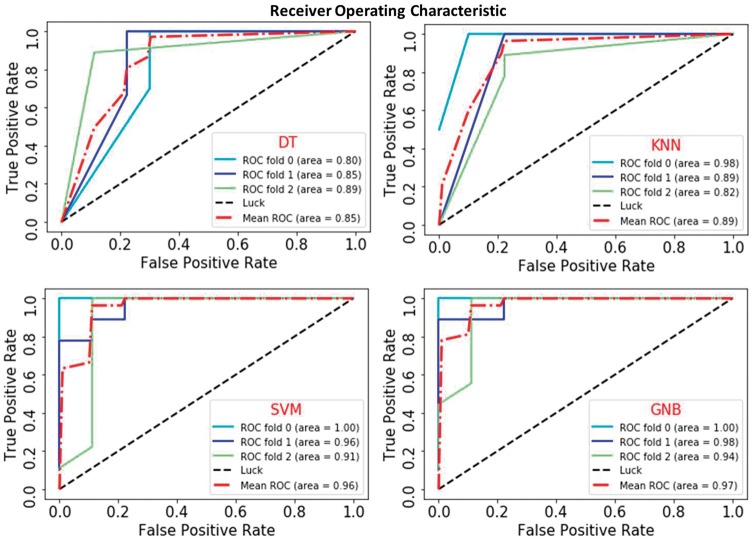
The cluster analysis strategy is designed by integrating four classifiers, namely, Decision Tree (DT) Classifier [130], KNeighbors (KNN) Classifier [131], SVM [132] and Gaussian Naive Bayes (GNB) [133, 134]. A 3-fold cross-validation is done in case of all four classifiers and their respective ROC curves [135] are given in Figure 1. A pairwise relation with respect to seed proteins is generated with the above-defined structural and semantic features. All pairwise combinations, (Seedi, Seedj) within the set SD, are considered as the positive samples for the classifiers. The negative data have created as a pair of proteins (Seedk, Nseedp) where and Nseedp are the proteins that have no interaction evidence with seed proteins and EBOV proteins. Finally, all classifier results are aggregated for final cluster. In this proposed work, we consider only those novel interactions where classification results are in agreement with all the classifiers to obtain more accurate cluster. The basic workflow of clustering is shown in Figure 2.
Figure 1.
ROC curves of DT, KNN, SVM and GNB. (A colour version of this figure is available online at: https://academic.oup.com/bfg)
Figure 2.
The workflow of cluster analysis based on known human target proteins of EBOV GP using DT, KNN, SVM and GNB. (A colour version of this figure is available online at: https://academic.oup.com/bfg)
Discussion
The predictive model is able to retrieve the potential human proteins, which may interact with EBOV GP and facilitate entry of the virus into host cells. Structural and GO-based functional annotation is considered as the key point of this analysis. In this analysis, total 10 EBOV GP hosts are selected as seed (Table 2). Among them, only 28 pairs of structural comparison are possible and considered as positive data pair for training. In addition, 28 pairs of negative data are manually created for the classifiers. A new test data are created from human proteins with respect to each seed proteins and those have structural information. These proteins are selected as the first-level interactor of EBOV GP host and . Finally, total 116 proteins are resulted from the cluster as the potential EBOV host. To establish the involvement of these proteins in viral infection, we have found some common pathway from KEGG database (http://www.genome.jp/kegg/). A set of pathways (hsa05162:Measles, hsa04145:Phagosome, hsa05203:Viral carcinogenesis, hsa05152:Tuberculosis, hsa05133:Pertussis and hsa05205:Proteoglycans in cancer) is found as common between the EBOV GP host and these cluster proteins (shown in Table 4). A GO-based functional analysis over these proteins is shown in Tables 5–7. These proteins share many biological activities related to virus receptor, immune response and viral genome replication with the known EBOV GP interactor.
Table 6.
Significant common GO terms (molecular function) found in known and new human proteins
| Serial number | KEGG | Term | Host |
New host |
||
|---|---|---|---|---|---|---|
| % of proteins | P-value | % of proteins | P-value | |||
| 1 | GO:0001618 | Virus receptor activity | 61.54 | 1.20E-14 | 19.43 | 3.20E-06 |
| 2 | GO:0001786 | Phosphatidylserine binding | 33.17 | 2.10E-06 | 17.3 | 4.20E-07 |
| 3 | GO:0004714 | Transmembrane receptor protein tyrosine kinase activity | 23.08 | 3.20E-04 | 9.8 | 1.60E-03 |
Table 4.
Significant common KEGG pathways found in known and new human proteins
| Serial number | KEGG | Term | Host |
New host |
||
|---|---|---|---|---|---|---|
| % of proteins | P-value | % of proteins | P-value | |||
| 1 | hsa05162 | Measles | 15.38 | 5.7E-2 | 22.52 | 7.8E-4 |
| 2 | hsa04145 | Phagosome | 30.77 | 1.1E-5 | 27.27 | 3.10E-06 |
| 3 | hsa05203 | Viral carcinogenesis | 18.22 | 1.30E-11 | 33.21 | 5.30E-09 |
| 4 | hsa05152 | Tuberculosis | 15.31 | 7.5E-2 | 17.07 | 4.80E-02 |
| 5 | hsa05133 | Pertussis | 25.81 | 3.2E-2 | 14.34 | 5.0E-3 |
| 6 | hsa05205 | Proteoglycans in cancer | 12.23 | 8.40E-03 | 18.79 | 2.90E-02 |
Table 5.
Significant common GO terms (biological process) found in known and new human proteins
| Term | Host |
New host |
||
|---|---|---|---|---|
| % of proteins | P-value | % of proteins | P-value | |
| Antigen processing and presentation | 35.38 | 3.50E-02 | 21.73 | 7.40E-06 |
| Modulation by virus of host morphology or physiology | 15.32 | 4.60E-03 | 24.22 | 5.60E-04 |
| Innate immune response | 30.7 | 2.40E-03 | 17.39 | 2.50E-02 |
| Viral genome replication | 23.08 | 3.50E-05 | 13.62 | 1.80E-02 |
| Integrin-mediated signaling pathway | 14.83 | 6.30E-02 | 28.19 | 4.70E-04 |
| Platelet activation | 16.61 | 5.00E-05 | 18.4 | 6.80E-02 |
Table 7.
Significant common GO terms (cellular component) found in known and new human proteins
| Serial number | KEGG | Term | Host |
New host |
||
|---|---|---|---|---|---|---|
| % of proteins | P-value | % of proteins | P-value | |||
| 1 | GO:0005615 | Extracellular space | 31.87 | 5.40E-02 | 19.56 | 5.50E-06 |
| 2 | GO:0009986 | Cell surface | 26.14 | 4.70E-03 | 17.97 | 7.10E-03 |
| 3 | GO:0005886 | Plasma membrane | 53.84 | 3.50E-02 | 28.2 | 7.70E-02 |
| 4 | GO:0001726 | Ruffle | 22.15 | 5.80E-02 | 9.6 | 4.50E-03 |
Conclusion
In this article, we have reviewed the diverse level of host–virus interaction predictions across the variety of pathogenic species and their human host. These computational methods may have important roles in paving the way of experimental verification of virus–host interactions by highlighting high potential interactions. Depending on the availability of the required data, some virus–host interaction mechanisms are well studied and targeted in more research. The main challenge for computational virus-interaction predictions is the lack of available verified interactions and the relevant feature information in most of the prediction methods. Finally, a case study-based analysis is proposed on EBOV–human interaction prediction. Here, four different classifiers, DT Classifier [130], KNN Classifier [131], SVM [132] and GNB [133, 134], are used for predictions. In this approach, a cluster of potential human proteins is retrieved from the predicted novel interactions. These sets of proteins have close structural and semantic similarities with known EBOV GP human target proteins, and this may facilitate EBOV entry into host cells through interaction with EBOV GP. For defining the structural similarity feature, we use five scores, namely, TM-score [111], RMSD [112], MaxSub-score [113], GDT_score [114] and Tm-Rm score [Equation (1)]. The semantic similarity feature is determined by GO graph-based properties. The proteins predicted by this method are highly likely to interact with EBOV GP and facilitate EBOV entry into human cells. This method would enlighten the promising future direction for novel host–virus interactions.
Key Points
The review presents computational approaches toward the host–virus interaction-based predictive models.
EBOV, a member of Filoviridae family, is a negative-sense RNA virus that causes high fatality rate in humans. Among the seven genes of Ebola, GP is responsible for mediating attachment to host cell surface receptors and entry of the virus into host cells.
A case study-based analysis on novel EBOV–human protein interactions prediction using structural and semantic similarity features.
Structural similarity feature is defined using five structural alignment scores, namely, TM-score, RMSD, MaxSub-score and Tm-Rm score. The semantic similarity feature is determined by using properties of the GO graph and IC of GO terms.
Finally, the pathway and GO-based functional annotation is provided for novel EBOV–human interactions.
Funding
This work is partially supported by the CMATER research laboratory of the Computer Science and Engineering Department, Jadavpur University, India, PURSE-II and UPE-II project and Research Award [F.30-31/2016(SA-II)] from University Grants Commission, Government of India and Visvesvaraya PhD scheme from DeitY, Government of India.
Anup Kumar Halder is a PhD scholar in the Department of Computer Science and Engineering at Jadavpur University, India. He received his ME degree in Computer Science and Engineering from Jadavpur University, India in 2014.
Pritha Dutta received her BTech degree in Computer Science and Engineering from B. P. Poddar Institute of Management and Technology, India in 2013 and ME degree in Computer Science and Engineering from Jadavpur University, India in 2015.
Mahantapas Kundu is a Professor in the Department of Computer Science and Engineering, Jadavpur University, India. He received his B.E.E, M.E.Tel.E and PhD (Engg.) degrees from Jadavpur University, in 1983, 1985 and 1995, respectively.
Subhadip Basu is an Associate Professor in the Department of Computer Science and Engineering, Jadavpur University, India. He has published around 150 research articles in various International Journals, Conference proceedings and book chapters in the areas of Bioinformatics, Biomedical Image Analysis and Character Recognition.
Mita Nasipuri is a Professor in the Department of Computer Science and Engineering, Jadavpur University, India. She has published around 400 research articles in various International Journals, Conference proceedings and book chapters in the areas of Bioinformatics, Pattern Recognition, Computer Vision, Face Recognition, etc.
References
- 1. Bennett JE, Dolin R, Blaser MJ.. Principles and Practice of Infectious Diseases. Philadelphia: Elsevier Health Sciences, 2014. [Google Scholar]
- 2. Arnold R, Boonen K, Sun MGF, et al. Computational analysis of interactomes: current and future perspectives for bioinformatics approaches to model the host–pathogen interaction space. Methods 2012;57(4):508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou H, Jin J, Wong L.. Progress in computational studies of host–pathogen interactions. J Bioinform Comput Biol 2013;11(02):1230001.. [DOI] [PubMed] [Google Scholar]
- 4. Zhou H, Gao S, Nguyen NN, et al. Stringent homology-based prediction of H. Sapiens-M. tuberculosis h37rv protein-protein interactions. Biol Direct 2014;9(1):5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dyer MD, Murali TM, Sobral BW.. Supervised learning and prediction of physical interactions between human and HIV proteins. Infect Genet Evol 2011;11(5):917–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dyer MD, Murali TM, Sobral BW.. Computational prediction of host-pathogen protein–protein interactions. Bioinformatics 2007;2313:i159–66. [DOI] [PubMed] [Google Scholar]
- 7. Dyer MD, Murali TM, Sobral BW.. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog 2008;4(2):e32.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dyer MD, Neff C, Dufford M, et al. The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS One 2010;5(8):e12089.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shen J, Zhang J, Luo X, et al. Predicting protein–protein interactions based only on sequences information. Proc Natl Acad Sci USA 2007;104(11):4337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee SA, Chan Ch, Tsai CH, et al. Ortholog-based protein-protein interaction prediction and its application to inter-species interactions. BMC Bioinformatics 2008;9(Suppl 12):S11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krishnadev O, Srinivasan N.. A data integration approach to predict host-pathogen protein-protein interactions: application to recognize protein interactions between human and a malarial parasite. Silico Biol 2008;8(3, 4):235–50. [PubMed] [Google Scholar]
- 12. Nourani E, Khunjush F, Durmu S.. Computational approaches for prediction of pathogen-host protein-protein interactions. Front Microbiol 2015;6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mei S. Probability weighted ensemble transfer learning for predicting interactions between hiv-1 and human proteins. PLoS One 2013;8(11):e79606.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wuchty S. Computational prediction of host-parasite protein interactions between P. falciparum and H. sapiens. PLoS One 2011;6(11):e26960.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chatterjee P, Basu S, Kundu M, et al. PPI_SVM: Prediction of protein-protein interactions using machine learning, domain-domain affinities and frequency tables. Cell Mol Biol Lett 2011;16(2):264.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Itzhaki Z. Domain-domain interactions underlying herpes virus-human protein-protein interaction networks. PLoS One 2011;6(7):e21724.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krishnadev O, Srinivasan N.. Prediction of protein–protein interactions between human host and a pathogen and its application to three pathogenic bacteria. Int J Biol Macromol 2011;48(4):613–9. [DOI] [PubMed] [Google Scholar]
- 18. Song H, Qi J, Haywood J, et al. Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses. Nat Struct Mol Biol 2016;23(5):456–8. [DOI] [PubMed] [Google Scholar]
- 19. Tyagi M, Hashimoto K, Shoemaker BA, et al. Large-scale mapping of human protein interactome using structural complexes. EMBO Rep 2012;13(3):266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Franzosa EA, Xia Y.. Structural principles within the human-virus protein-protein interaction network. Proc Natl Acad Sci USA 2011;108(26):10538–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Chassey B, Meyniel-Schicklin L, Aublin-Gex A, et al. Structure homology and interaction redundancy for discovering virus–host protein interactions. EMBO Rep 2013;14(10):938–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nourani E, Khunjush F, Durmuş S.. Computational prediction of virus–human protein–protein interactions using embedding kernelized heterogeneous data. Mol Biosyst 2016;12(6):1976–86. [DOI] [PubMed] [Google Scholar]
- 23. Dutta P, Basu S, Kundu M.. Assessment of semantic similarity between proteins using information content and topological properties of the Gene Ontology graph. IEEE/ACM Trans Comput Biol Bioinforma 2017, doi: 10.1109/TCBB.2017.2689762. [DOI] [PubMed] [Google Scholar]
- 24. Nouretdinov I, Gammerman A, Qi Y, et al. Determining confidence of predicted interactions between HIV-1 and human proteins using conformal method In Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. Proceedings of the Pacific Symposium Kohala Coast, Hawaii, USA: NIH Public Access, 2012, 311. [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng LL, Li C, Ping J, et al. The domain landscape of virus-host interactomes. Biomed Res Int 2014;2014: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kshirsagar M, Carbonell J, Klein-Seetharaman J.. Multitask learning for host–pathogen protein interactions. Bioinformatics 2013;29(13):i217–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kshirsagar M, Murugesan K, Carbonell JG, et al. Multitask matrix completion for learning protein interactions across diseases. J Comput Biol 2017;24(6):501–14. [DOI] [PubMed] [Google Scholar]
- 28. Kshirsagar M, Carbonell J, Klein-Seetharaman J.. Techniques to cope with missing data in host–pathogen protein interaction prediction. Bioinformatics 2012;28(18):i466–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kshirsagar M, Carbonell J, Klein-Seetharaman J.. Multisource Transfer Learning for Host-Pathogen Protein Interaction Prediction in Unlabeled Tasks. In: Proceedings of NIPS Workshop on Machine Learning for Computational Biology, 2013. [Google Scholar]
- 30. Xu Q, Yang Q.. A survey of transfer and multitask learning in bioinformatics. J Comput Sci Eng 2011;5(3):257–68. [Google Scholar]
- 31. Becerra A, Bucheli VA, Moreno PA.. Prediction of virus-host protein-protein interactions mediated by short linear motifs. BMC Bioinformatics 2017;18(1):163.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Segura-Cabrera A, García-Pérez CA, Guo X, et al. A viral-human interactome based on structural motif-domain interactions captures the human infectome. PLoS One 2013;8(8):e71526.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kharrat N, Belmabrouk S, Abdelhedi R, et al. Screening for clusters of charge in human virus proteomes. BMC Genomics 2016;17(9):758.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mukhopadhyay A, Maulik U, Bandyopadhyay S, et al. Mining association rules from HIV-human protein interactions. In: 2010 International Conference on Systems in Medicine and Biology (ICSMB), pp. 344–8. IIT Kharagpur, India: IEEE, 2010.
- 35. Mukhopadhyay A, Maulik U, Bandyopadhyay S.. A novel biclustering approach to association rule mining for predicting HIV-1-human protein interactions. PLoS One 2012;7(4):e32289.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mukhopadhyay A, Ray S, Maulik U.. Incorporating the type and direction information in predicting novel regulatory interactions between HIV-1 and human proteins using a biclustering approach. BMC Bioinformatics 2014;15(1):26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mondal KC, Pasquier N, Mukhopadhyay A, et al. Prediction of protein interactions on HIV-1-human PPI data using a novel closure-based integrated approach. In: International Conference on Bioinformatics Models, Methods and Algorithms, pp. 164–173. Vilamoura, Algrave, Portugal: SciTePress, 2012.
- 38. Plotkin JB, Dushoff J, Levin SA.. Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus. Proc Natl Acad Sci USA 2002;99(9):6263–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abdel-Azim G. New hierarchical clustering algorithm for protein sequences based on Hellinger distance. Appl Math 2016;10(4):1541–9. [Google Scholar]
- 40. Spencer KA, Dee M, Britton P, et al. Role of phosphorylation clusters in the biology of the coronavirus infectious bronchitis virus nucleocapsid protein. Virology 2008;370(2):373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schleker S, Garcia-Garcia J, Klein-Seetharaman J, et al. Prediction and comparison of salmonella? Human and salmonella? Arabidopsis interactomes. Chem Biodivers 2012;9(5):991–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mariano R, Wuchty S.. Structure-based prediction of host–pathogen protein interactions. Curr Opin Struct Biol 2017;44:119–24. [DOI] [PubMed] [Google Scholar]
- 43. Mei S, Zhu H.. Adaboost based multi-instance transfer learning for predicting proteome-wide interactions between salmonella and human proteins. PLoS One 2014;9(10):e110488.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cui G, Fang C, Han K.. Prediction of protein-protein interactions between viruses and human by an SVM model. BMC Bioinformatics 2012;13(7):S5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim B, Alguwaizani S, Zhou X, et al. An improved method for predicting interactions between virus and human proteins. J Bioinform Comput Biol 2017;15(01):1650024.. [DOI] [PubMed] [Google Scholar]
- 46. Doolittle JM, Gomez SM.. Structural similarity-based predictions of protein interactions between HIV-1 and Homo sapiens. Virol J 2010;7(1):82.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Doolittle JM, Gomez SM.. Mapping protein interactions between dengue virus and its human and insect hosts. PLoS Negl Trop Dis 2011;5(2):e954.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cao H, Zhang Y, Zhao J, et al. Prediction of the Ebola virus infection related human genes using protein-protein interaction network. Comb Chem High Throughput Screen 2017, doi: 10.2174/1386207320666170310114816. [DOI] [PubMed] [Google Scholar]
- 49. Prehna G, Ivanov MI, Bliska JB, et al. Yersinia virulence depends on mimicry of host Rho-family nucleotide dissociation inhibitors. Cell 2006;126:869–80. [DOI] [PubMed] [Google Scholar]
- 50. Stebbins CE, Galán JE.. Structural mimicry in bacterial virulence. Nature 2001;412(6848):701.. [DOI] [PubMed] [Google Scholar]
- 51. Abbasi I, Githure J, Ochola JJ, et al. Diagnosis of Wuchereria bancrofti infection by the polymerase chain reaction employing patients’ sputum. Parasitol Res 1999;85(10):844–9. [DOI] [PubMed] [Google Scholar]
- 52. Zhao N, Pang B, Shyu CR, et al. Structural similarity and classification of protein interaction interfaces. PLoS One 2011;6(5):e19554.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ogmen U, Keskin O, Aytuna AS, et al. PRISM: protein interactions by structural matching. Nucleic Acids Res 2005;33(Suppl 2):W331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Winter C, Henschel A, Kim WK, et al. SCOPPI: a structural classification of protein–protein interfaces. Nucleic Acids Res 2006;34(Suppl 1):D310–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Teyra J, Paszkowski-Rogacz M, Anders G, et al. SCOWLP classification: structural comparison and analysis of protein binding regions. BMC Bioinformatics 2008;9(1):9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keskin O, Nussinov R, Gursoy A.. PRISM: protein-protein interaction prediction by structural matching. Methods Mol Biol 2008;484:505–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mei S, Flemington EK, Zhang K.. A computational framework for distinguishing direct versus indirect interactions in human functional protein–protein interaction networks. Integr Biol 2017;9:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bohn-Wippert K, Tevonian EN, Megaridis MR, et al. Similarity in viral and host promoters couples viral reactivation with host cell migration. Nat Commun 2017;8:15006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu G, He QY.. Functional similarity analysis of human virus-encoded miRNAs. J Clin Bioinform 2011;1(1):15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang SB, Tang QR.. Protein–protein interaction inference based on semantic similarity of gene ontology terms. J Theor Biol 2016;401:30–7. [DOI] [PubMed] [Google Scholar]
- 61. Jain S. and Others. An improved method for scoring protein-protein interactions using semantic similarity within the gene ontology. BMC Bioinformatics 2010;11(1):562.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cui G, Han K.. Scoring protein-protein interactions using the width of gene ontology terms and the information content of common ancestors In: International Conference on Intelligent Computing. Nanning, China: Springer, 2013, 31–6. [Google Scholar]
- 63. Cui G, Kim B, Alguwaizani S, et al. Assessing protein-protein interactions based on the semantic similarity of interacting proteins. Int J Data Min Bioinform 2015;13(1):75–83. [DOI] [PubMed] [Google Scholar]
- 64. Ikram N, Qadir M, Afzal M.. Investigating correlation between protein sequence similarity and semantic similarity using gene ontology annotations. IEEE/ACM Trans Comput Biol Bioinforma 2017, doi: 10.1109/TCBB.2017.2695542. [DOI] [PubMed] [Google Scholar]
- 65. Geisbert TW, Jahrling PB.. Exotic emerging viral diseases: progress and challenges. Nat Med 2004;10:S110–21. [DOI] [PubMed] [Google Scholar]
- 66. Sanchez AS, Khan AS, Zaki SR, et al. Filoviridae: Marburg and Ebola viruses: pathology and pathogenesis In: Knipe DM, Howley PM (eds), Field’s Virol, Vol. 1, Philadelphia: Williams and Wilkins, 2001, 1293. [Google Scholar]
- 67. Volchkov VE. Processing of the Ebola virus glycoprotein. Curr Top Microbiol Immunol 1999;235:35–47. [DOI] [PubMed] [Google Scholar]
- 68. Sanchez A, Yang ZY, Xu L, et al. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J Virol 1998;72(8):6442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jeffers SA, Sanders DA, Sanchez A.. Covalent modifications of the Ebola virus glycoprotein. J Virol 2002;76(24):12463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Weissenhorn W, Carfí A, Lee KH, et al. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell 1998;2(5):605–16. [DOI] [PubMed] [Google Scholar]
- 71. Ito H, Watanabe S, Sanchez A, et al. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J Virol 1999;73(10):8907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Feldmann H, Volchkov VE, Volchkova VA, et al. Biosynthesis and role of filoviral glycoproteins. J Gen Virol 2001;82(12):2839–48. [DOI] [PubMed] [Google Scholar]
- 73. Chertow DS, Kleine C, Edwards JK, et al. Ebola virus disease in West Africa-clinical manifestations and management. N Engl J Med 2014;371(22):2054–7. [DOI] [PubMed] [Google Scholar]
- 74. Yang Zy, Duckers HJ, Sullivan NJ, et al. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat Med 2000;6(8):886–9. [DOI] [PubMed] [Google Scholar]
- 75. Ströher U, West E, Bugany H, et al. Infection and activation of monocytes by Marburg and Ebola viruses. J Virol 2001;75(22):11025–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Geisbert TW, Hensley LE.. Ebola virus: new insights into disease aetiopathology and possible therapeutic interventions. Expert Rev Mol Med 2004;6(20):1–24. [DOI] [PubMed] [Google Scholar]
- 77. Bray M, Geisbert TW.. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol 2005;37(8):1560–6. [DOI] [PubMed] [Google Scholar]
- 78. Lee JE, Saphire EO.. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol 2009;4(6):621–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Moller-Tank S, Maury W.. Ebola virus entry: a curious and complex series of events. PLoS Pathog 2015;11(4):e1004731.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dakappagari N, Maruyama T, Renshaw M, et al. Internalizing antibodies to the C-type lectins, L-SIGN and DC-SIGN, inhibit viral glycoprotein binding and deliver antigen to human dendritic cells for the induction of T cell responses. J Immunol 2006;176(1):426–40. [DOI] [PubMed] [Google Scholar]
- 81. Takada A, Fujioka K, Tsuiji M, et al. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J Virol 2004;78(6):2943–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Alvarez CP, Lasala F, Carrillo J, et al. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol 2002;76(13):6841–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Powlesland AS, Fisch T, Taylor ME, et al. A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J Biol Chem 2008;283(1):593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dahlmann F, Biedenkopf N, Babler A, et al. Analysis of Ebola virus entry into macrophages. J Infect Dis 2015;212(Suppl 2):S247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yuan S, Cao L, Ling H, et al. TIM-1 acts a dual-attachment receptor for Ebolavirus by interacting directly with viral GP and the PS on the viral envelope. Protein Cell 2015;6(11):814–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kondratowicz AS, Lennemann NJ, Sinn PL, et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci USA 2011;108(20):8426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Moller-Tank S, Kondratowicz AS, Davey RA, et al. Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J Virol 2013;87(15):8327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jemielity S, Wang JJ, Chan YK, et al. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog 2013;9(3):e1003232.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bhattacharyya S, Zagórska A, Lew ED, et al. Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell Host Microbe 2013;14(2):136–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Brindley MA, Hunt CL, Kondratowicz AS, et al. Tyrosine kinase receptor Axl enhances entry of Zaire Ebola virus without direct interactions with the viral glycoprotein. Virology 2011;415(2):83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hunt CL, Kolokoltsov AA, Davey RA, et al. The Tyro3 receptor kinase Axl enhances macropinocytosis of Zaire Ebola virus. J Virol 2011;85(1):334–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Morizono K, Chen ISY.. Role of phosphatidylserine receptors in enveloped virus infection. J Virol 2014;88(8):4275–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schornberg KL, Shoemaker CJ, Dube D, et al. α5β1-Integrin controls ebolavirus entry by regulating endosomal cathepsins. Proc Natl Acad Sci USA 2009;106(19):8003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Takada A, Watanabe S, Ito H, et al. Downregulation of β1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology 2000;278(1):20–6. [DOI] [PubMed] [Google Scholar]
- 95. Simmons G, Wool-Lewis RJ, Baribaud F, et al. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J Virol 2002;76(5):2518–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Carette JE, Raaben M, Wong AC, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 2011;477(7364):340–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Côté M, Misasi J, Ren T, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 2011;477(7364):344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bhattacharyya S, Hope TJ, Young JAT.. Differential requirements for clathrin endocytic pathway components in cellular entry by Ebola and Marburg glycoprotein pseudovirions. Virology 2011;419(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Saeed MF, Kolokoltsov AA, Albrecht T, et al. Cellular entry of Ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog 2010;6(9):e1001110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nanbo A, Imai M, Watanabe S, et al. Ebola virus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog 2010;6(9):e1001121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Nugent T, Jones DT.. Membrane protein structural bioinformatics. J Struct Biol 2012;179(3):327–37. [DOI] [PubMed] [Google Scholar]
- 102. Carl N, Konc J, Vehar B, et al. Protein-protein binding site prediction by local structural alignment. J Chem Inf Model 2010;50(10):1906–13. [DOI] [PubMed] [Google Scholar]
- 103. Chakrabarti P, Janin J.. Dissecting protein–protein recognition sites. Proteins Struct Funct Bioinforma 2002;47(3):334–43. [DOI] [PubMed] [Google Scholar]
- 104. Csaba G, Birzele F, Zimmer R.. Protein structure alignment considering phenotypic plasticity. Bioinformatics 2008;24(16):i98–104. [DOI] [PubMed] [Google Scholar]
- 105. Jones S, Thornton JM.. Analysis of protein-protein interaction sites using surface patches. J Mol Biol 1997;272(1):121–32. [DOI] [PubMed] [Google Scholar]
- 106. Konc J, Janežič D.. ProBiS algorithm for detection of structurally similar protein binding sites by local structural alignment. Bioinformatics 2010;26(9):1160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Li JJ, Huang DS, Wang B, et al. Identifying protein–protein interfacial residues in heterocomplexes using residue conservation scores. Int J Biol Macromol 2006;38(3):241–7. [DOI] [PubMed] [Google Scholar]
- 108. Jordan RA, Yasser EM, Dobbs D, et al. Predicting protein-protein interface residues using local surface structural similarity. BMC Bioinformatics 2012;13(1):41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mehio W, Kemp GJL, Taylor P, et al. Identification of protein binding surfaces using surface triplet propensities. Bioinformatics 2010;26(20):2549–55. [DOI] [PubMed] [Google Scholar]
- 110. Monji H, Koizumi S, Ozaki T, et al. Interaction site prediction by structural similarity to neighboring clusters in protein-protein interaction networks. BMC Bioinformatics 2011;12(1):S39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhang Y, Skolnick J.. Scoring function for automated assessment of protein structure template quality. Proteins Struct Funct Bioinforma 2004;57(4):702–10. [DOI] [PubMed] [Google Scholar]
- 112. Kabsch W. A discussion of the solution for the best rotation to relate two sets of vectors. Acta Crystallogr Sect A 1978;34(5):827–8. [Google Scholar]
- 113. Siew N, Elofsson A, Rychlewski L, et al. MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics 2000;16(9):776–85. [DOI] [PubMed] [Google Scholar]
- 114. Zemla A, Venclovas Č, Moult J, et al. Processing and analysis of CASP3 protein structure predictions. Proteins Struct Funct Bioinform 1999;37(S3):22–9. [DOI] [PubMed] [Google Scholar]
- 115. Zhang Y, Skolnick J.. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 2005;33(7):2302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Read RJ, Chavali G.. Assessment of CASP7 predictions in the high accuracy template-based modeling category. Proteins Struct Funct Bioinform 2007;69(S8):27–37. [DOI] [PubMed] [Google Scholar]
- 117. Couto FM, Silva MJ.. Disjunctive shared information between ontology concepts: application to Gene Ontology. J Biomed Semant 2011;2:5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Azuaje F, Al-Shahrour F, Dopazo J.. Ontology-driven approaches to analyzing data in functional genomics In: Bioinformatics and Drug Discovery. Springer, 2006, 67–86. [DOI] [PubMed] [Google Scholar]
- 119. Wang JZ, Du Z, Payattakool R, Yu PS, et al. A new method to measure the semantic similarity of GO terms. Bioinformatics 2007;23(10):1274–81. [DOI] [PubMed] [Google Scholar]
- 120. Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Harris MA, Clark J, Ireland A, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 2004;32(Suppl 1):D258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Boeckmann B, Bairoch A, Apweiler R, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 2003;31(1):365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Boutet E, Lieberherr D, Tognolli M, et al. UniProtKB/Swiss-Prot: the manually annotated section of the UniProt KnowledgeBase. Plant Bioinform Methods Protoc 2007;:89–112. [Google Scholar]
- 124. Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res 2000;28(1):235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Salwinski L, Miller CS, Smith AJ, et al. The database of interacting proteins: 2004 update. Nucleic Acids Res 2004;32(Suppl 1):D449–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Chatr-Aryamontri A, Ceol A, Palazzi LM, et al. MINT: the molecular interaction database. Nucleic Acids Res 2007;35(Suppl 1):D572–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res 2015;43(D1):D470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Turner B, Razick S, Turinsky AL, et al. iRefWeb: interactive analysis of consolidated protein interaction data and their supporting evidence. Database 2010;2010:baq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Swain PH, Hauska H.. The decision tree classifier: design and potential. IEEE Trans Geosci Electron 1977;15(3):142–7. [Google Scholar]
- 131. Kuncheva LI, Jain LC.. Nearest neighbor classifier: simultaneous editing and feature selection. Pattern Recognit Lett 1999;20(11):1149–56. [Google Scholar]
- 132. Scholkopf B, Smola AJ.. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond. Cambridge, MA, USA: MIT Press, 2001. [Google Scholar]
- 133. John GH, Langley P.. Estimating continuous distributions in Bayesian classifiers. In: Proceedings of the Eleventh conference on Uncertainty in artificial intelligence, p 338–45. Morgan Kaufmann Publishers Inc, 1995. [Google Scholar]
- 134. Friedman N, Geiger D, Goldszmidt M.. Bayesian network classifiers. Mach Learn 1997;29(2-3):131–63. [Google Scholar]
- 135. Hand DJ. Measuring classifier performance: a coherent alternative to the area under the ROC curve. Mach Learn 2009;77(1):103–23. [Google Scholar]