Abstract
Severe acute respiratory syndrome (SARS) has emerged as a highly contagious, sometimes fatal disease. To find disease-specific B cell epitopes, phage-displayed random peptide libraries were panned on serum immunoglobulin (Ig) G antibodies from patients with SARS. Forty-nine immunopositive phage clones that bound specifically to serum from patients with SARS were selected. These phageborne peptides had 4 consensus motifs, of which 2 corresponded to amino acid sequences reported for SARS-associated coronavirus (SARSCoV). Synthetic peptide binding and competitive-inhibition assays further confirmed that patients with SARS generated antibodies against SARS-CoV. Immunopositive phage clones and epitope-based peptide antigens demonstrated clinical diagnostic potential by reacting with serum from patients with SARS. Antibody-response kinetics were evaluated in 4 patients with SARS, and production of IgM, IgG, and IgA were documented as part of the immune response. In conclusion, B cell epitopes of SARS corresponded to novel coronavirus. Our epitope-based serologic test may be useful in laboratory detection of the virus and in further study of the pathogenesis of SARS.
Since November 2002, case reports of a life-threatening, highly contagious, febrile respiratory illness of unknown etiology from Guangdong Province in southern China have been followed by case reports from Vietnam, Hong Kong, Singapore, Canada, and the United States [1–9]. In late February 2003, this illness was identified as a new clinical entity called “severe acute respiratory syndrome,” or “SARS” [10–13]. The ready transmissibility of SARS has led to outbreaks not only among household members and health care workers but also within the community at large [1, 3–8, 14, 15]. Reports have indicated that a SARS-associated coronavirus (SARSCoV) or a human metapneumovirus (hMPV) might be responsible for SARS [16–20]. One recent study has indicated that SARS-CoV fulfils all of Koch's postulates for the primary etiological agent for SARS [21].
By activating host humoral immunity, viral infections trigger production of antibodies directed against viral protein epitopes. Knowledge of viral protein epitopes is pivotal in understanding the pathogenesis of viral infections and in developing diagnostic reagents and effective vaccines. Phage display has been used to identify immunogenic targets or epitopes recognized by antibodies. Selection of peptide libraries on monoclonal antibodies [22, 23] and in complex serum from patients with disease [24, 25] has led to the isolation of immunoreactive peptide epitopes. Disease-specific epitopes that are useful for the development of diagnostic or preventive reagents have been identified by screening of phage-displayed peptide libraries in serum or cerebrospinal fluid samples from patients with viral infections [24, 25], rheumatoid arthritis [26], multiple sclerosis [27], autoimmune thrombocytopenic purpura [28, 29], and neurocysticercosis [30].
To identify peptides corresponding to or mimicking natural epitopes, we applied phage-display technology to the characterization of specific antibodies present in serum samples from patients with SARS. Results of this study will contribute to the development of a sensitive, simple test for diagnosis of SARS and will help in the investigation of the pathogenesis of SARS.
Materials and Methods
Patient serum samples. From 8 March to 3 May 2003, 7 patients with illnesses that met the Centers for Disease Control and Prevention (CDC) case definition (1 May 2003) of probable SARS [31 ] were identified and treated at the National Taiwan University Hospital (NTUH) in Taipei. Serum samples from the 7 patients were sent to the hospital's Department of Laboratory Medicine for routine serologic and biochemical analysis. The laboratory diagnostic test, clinical manifestations of illness, and treatment of these patients have been described in detail in our recently published report [31 ]. Serum samples were collected from patients with SARS and were tested by use of the following methods [31 ]: virus isolation in Vero E6 cells (American Type Culture Collection), serotype-specific reversetranscriptase (RT) polymerase chain reaction (PCR) [16 ], and a standard indirect fluorescent antibody (IFA) assay for the detection of antibody to SARS-CoV [19 ]. All 7 patients with SARS were positive for SARS RNA by RT-PCR, and 6 of them were positive for SARS antibody by IFA assay (patient SP7 was seronegative) [31 ]. Although the patients had received ribavirin, corticosteroid, and intravenous immunoglobulin treatment during the early stage of the disease, the antibody was detected by IFA assay as early as 10–12 days after onset of illness [31 ]. Written informed consent was obtained from all volunteers. The study was approved by the ethics committee of the College of Medicine, National Taiwan University (Taipei), and the human experimentation guidelines of the US Department of Health and Human Services were followed in the conduct of this research.
Affinity selection of phages, by biopanning. Protein G magnetic microbeads (Dynabeads protein G; Dynal) were used to purify IgG from serum samples from patients with SARS. To enhance selection of peptides binding to IgG specifically associated with patients with SARS, we designed a preclearing step to remove nonspecific clones by preabsorption of the phage-displayed 12-mer peptide library (4×1010 phage particles; New England BioLabs) onto purified IgG from normal serum pooled from 5 control subjects (SARS-CoV seronegative by IFA assay) who were blood donors. Next, the precleared phage library was selected onto IgG purified from the serum of patients with SARS. Affinity selection on immobilized IgG from the serum of patients with SARS, for 1 h at 4°C, was used for screening. Unbound phage particles were removed, and magnetic beads were washed extensively with PBST0.5 (PBS plus 0.5% [wt/vol] Tween-20). Bound phage particles were eluted with glycine buffer (pH 2.2), neutralized with Tris buffer (pH 9.1), and amplified for subsequent rounds of selection. Three rounds of selection were performed for each patient. The biopanning protocol for the second and third rounds was identical to that of the first round, with the addition of 2×1011 pfu of phage particles for biopanning. Titration of unamplified third-round phage eluate was done on Luria broth/isopropyl β-D-thiogalactoside/5- bromo-4-chloro-3-indolyl-β-D-galactoside plates (Falcon; Becton Dickinson). Remaining eluate was stored at 4°C.
Identification of immunopositive phage clones, by ELISA. The ELISA plate was coated with 10 µg/mL purified anti- human IgG capture antibodies (Jackson ImmunoResearch Labs) in 0.1 mol/L NaHCO3 (pH 8.6), at room temperature for 2 h, and was blocked with PBSB (1% bovine serum albumin in PBS), at 4°C overnight. Serially diluted phage particles were added to plates coated with serum antibodies from patient SP1 and were incubated at room temperature for 1 h. The plate was washed 6 times with PBST0.5 , and a 5000-fold dilution of horseradish peroxidase (HRP)-conjugated anti-M13 antibody (Pharmacia product no. 27-9411-01) was added. The plate was incubated at room temperature for 1 h, with agitation, and was washed 6 times with PBST0.5 . Color was subsequently developed in the dark, with o-phenylenediamine dihydrochloride (Sigma) and hydrogen peroxide. The reaction was stopped with 3 N HCl, and absorbance was measured, at 490 nm, by use of an ELISA reader (Versa Max Tunable Microplate Reader; Molecular Devices). Immunopositive phage clones were further characterized by DNA sequencing, according to our method described elsewhere [22 ].
DNA sequencing and computer analysis. DNA sequences of purified phage clones were determined according to the dideoxynucleotide chain-termination method, by using an automated DNA sequencer (ABI PRISM 377; Perkin-Elmer). The primer used for phage DNA sequencing was 5'-CCCTCATAG TTAGCGTAA-3'. This primer is located in the antisense strand of gene III of the M13 phage and is separated from the inserted DNA by 96 nt. Inserted oligonucleotide sequences of phage DNA were selected and translated to peptide sequences. Peptide sequences were aligned by using MacDNASIS software (Hitachi Software Engineering). We aligned phage-displayed peptide sequences and the published protein sequences of several viruses, including 3 SARS-CoVs, dengue virus (DEN-1-4), Japanese encephalitis virus, hMPV, human rhinovirus type 14, human enterovirus type 71, respiratory syncytial virus, and influenza A virus, which were retrieved from GenBank (accession nos. NC004718, AY278554, AY278741, M87512, M20558, M93130, M14931, U14163, NC_004148, K02121, Q66478, NC_001803, and NC_004518∼004525, respectively).
Antibody competitive-inhibition assay. For the competitive- inhibition assay, 109 immunopositive phage particles were incubated with individual peptide antigens or with control peptide before being transferred to the antibody-coated plate and incubated for 1 h. The plates were incubated with HRP-conjugated anti-M13 antibody, and the procedures described above were followed.
Detection of serum samples from patients with SARS, by use of immunopositive phage clones. The ELISA plates were coated with 10 µg/mL purified anti-human IgM or anti-human IgG capture antibodies (Jackson ImmunoResearch Labs), were blocked with PBSB, and then were incubated with the tested serum samples, diluted 1:100, at room temperature for 1 h. We added 109 immunopositive phage particles to the antibodycoated plates; the plates were washed 6 times with PBST0.5 , and the procedures described above were followed.
Detection of serum samples from patients with SARS, by use of epitope-based synthetic peptides. The ELISA plates were coated with 10 µg/mL individual peptide antigens, at 50 µL/well, and were incubated at 4°C for 6 h. After being washed with PBST0.1 (PBS plus 0.1% [wt/vol] Tween-20), the plates were blocked with PBSB, at 4°C overnight, and then were incubated with the tested serum samples, diluted 1:100, at room temperature for 1 h. HRP-conjugated, goat anti-human IgG (Jackson ImmunoResearch Labs), diluted 1:20,000, was added to the microtiter plates, following the procedures described above.
Results
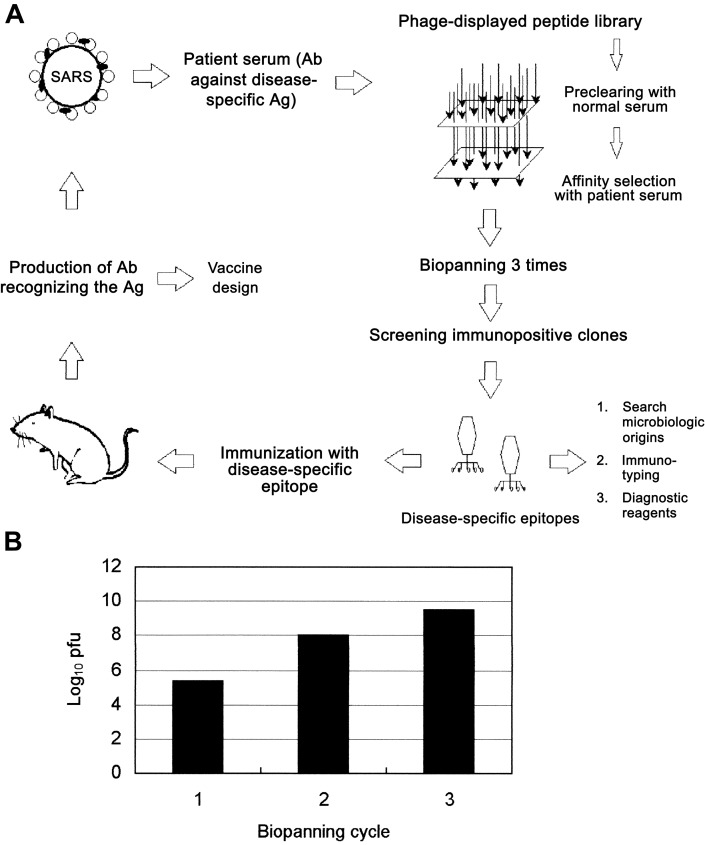
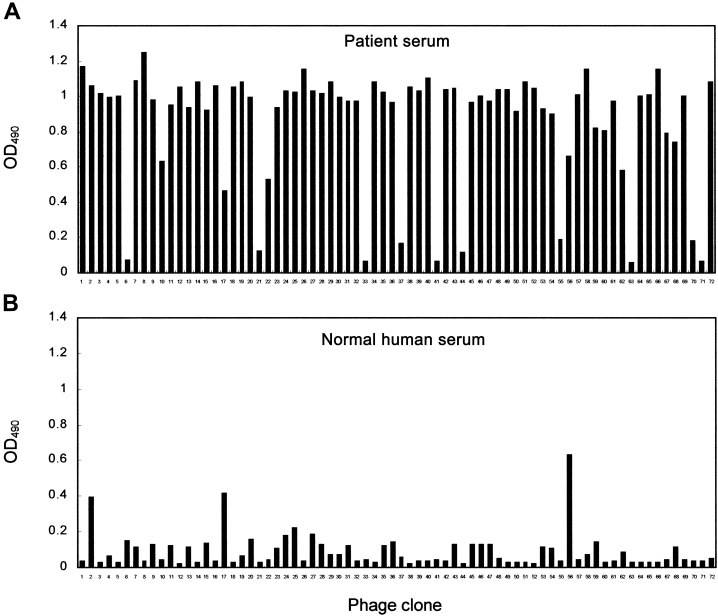
Screening of serum samples, with the phage-displayed peptide library. Patient SP1, the first documented case patient with SARS in Taiwan, developed a febrile illness and was admitted to NTUH on 8 March 2003. During the patient's convalescence, serum samples were collected on 8 April 2003, to study diseasespecific epitopes of SARS. Selection of immunopositive phage clones by SARS-specific serum antibodies was done by immobilization on protein G magnetic beads; the bound phage clones were selected after biopanning. To enhance selection of peptides binding to IgG specifically associated with patients with SARS, we designed a preclearing and selection procedure (see Materials and Methods). The strategy for identification of SARS-specific epitopes is shown in figure 1A. A marked enrichment (log10 scale) of as much as 14,000-fold followed the third round of selection, compared with the first round of selection (figure 1B). Individual phage clones from the third round of biopanning were randomly selected. ELISA was performed to determine whether antibodies present in serum samples from patient SP1 were specifically recognized by selected phage clones derived from biopanning. Of 72 selected phage clones, 49 had significant enhancement of binding activity to SARS-specific serum antibodies (figure 2A). These clones did not bind to normal human serum (figure 2B).
Figure 1.
Phage-displayed peptide-library screening of serum samples from patients with severe acute respiratory syndrome (SARS). A, Illustration of principle of selection of the SARS-specific epitopes used in this study. The phage-displayed peptide library was precleared by use of normal human serum and by affinity selection with serum antibodies (Abs) from patients with SARS. After biopanning 3 times, immunopositive phage clones were selected by ELISA. Disease-specific epitopes were further identified and characterized by synthetic peptide binding and competitive-inhibition assay. Phage-displayed disease-specific epitopes can be used to determine microbiologic origins, to study immunotyping, and to provide information for the development of diagnostic reagents and vaccines. B, Selection of a phage-displayed peptide library on immunoglobulins purified from serum samples from patient SP1. After the third round of biopanning, marked enrichment (up to 14,000-fold) was evident, compared with the first round of selection. Ag, antigen; pfu, plaque-forming units.
Figure 2.
Identification of serum antibody-selected phage clones, by ELISA, in serum samples from a patient with severe acute respiratory syndrome (SP1). A phage-displayed random peptide library was screened with serum antibodies from patient SP1. After 3 screening rounds, 49 phage clones from 72 selected phage clones were significantly reactive to antibodies in serum samples from patient SP1 (A) but not to samples of normal human serum (B). OD490 , optical density at 490 nm.
Identification of SARS-specific B cell epitopes. Forty-nine immunopositive phage clones (SP1-1, −3–5, −7–9, −12–14, −16, −18–20, −24–26, −28–32, −34–36, −38–40, −42, −43, −45–54, −57, −58, −60, −61, −64–66, −69, and −72) that were highly reactive with SARS-specific serum antibodies and less reactive with antibodies in normal serum were amplified, and phage DNA was isolated for sequencing. Inserted nucleotides of selected phage clones were sequenced, and all contained 36 nt (translated into 12 aa residues) (table 1). Peptide sequences were aligned by use of MacDNASIS software, to analyze epitopes and binding motifs of SARS antibodies. From 49 immunopositive clones, we obtained 4 binding motifs (table 1).
Table 1.
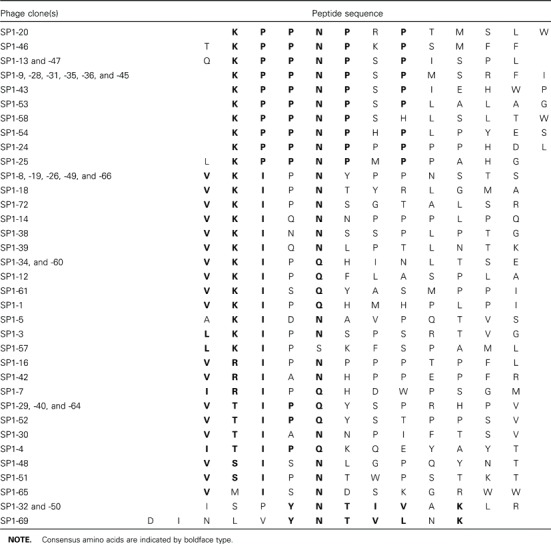
Alignment of phage-displayed peptide sequences selected by serum antibodies from a patient with severe acute respiratory syndrome (patient SP1).
Alignment of phage-displayed peptide sequences with the complete genome of several viruses, described in Materials and Methods, showed that 2 binding motifs were highly conserved in many immunopositive phage clones and that they corresponded exactly to amino acid residues of SARS-CoV (table 2). One binding motif was a residue of 3 aa, proline (Pro [P])- Pro-asparagine (Asn [N]), that was exhibited in all 16 immunopositive phage clones and only corresponded to amino acid residues 1184-1186 (PPN) of the CDS2 gene of SARS-CoV (table 2). The other binding motif was a residue of 5 aa, valine (Val [V])-lysine (Lys [K])-isoleucine (Ile [I])-X-Asn (N), that was highly conserved in 16 immunopositive phage clones and only corresponded to amino acid residues 18-22 (VKIDN) of the CDS4 gene of SARS-CoV (table 2). The peptide sequences of 2 immunopositive phage clones, SP1-32 and -50 (ISPYNTIVAKLR; table 1), were similar to amino acid residues 375– 386 of CDS4 in hMPV (LSPLGALVACYK; consensus sequences are indicated by boldface type).
Table 2.
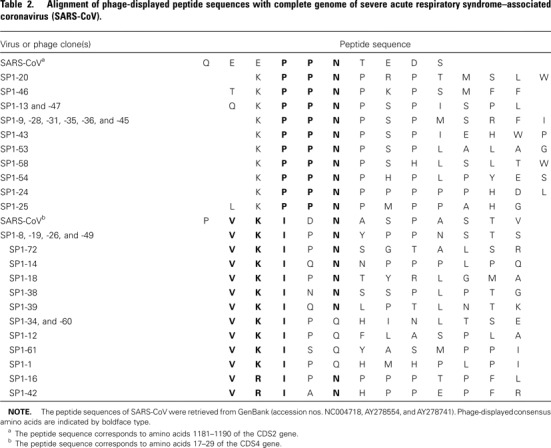
Alignment of phage-displayed peptide sequences with complete genome of severe acute respiratory syndrome-associated coronavirus (SARS-CoV).
To prove that selected phage clones bound specifically to serum from patients with SARS, antibodies from 1 patient with SARS (SP1) and from 2 healthy control subjects were incubated in a 3-fold serial dilution of immunopositive phage clone SP1- 1. SP1-1 bound serum from patients with SARS specifically and dose dependently. Two control serum samples did not react with SP1-1 (figure 3A). For further confirmation that selected phage clones specifically bound serum from patients with SARS, 8 immunopositive phage clones (SP1–1, −8, −29, −30, −32, −39, −54, and −72) were incubated with serum samples from 4 patients with SARS and with serum samples from 4 healthy control subjects. ELISA indicated that all immunopositive phage clones had high antibody specificity to the SP1 phage clones (figure 3B). Three phage clones (SP1-1, −8, and −29) were highly reactive with serum samples from the 4 patients with SARS. None of these phage clones reacted with serum samples from the 4 healthy control subjects (figure 3B).
Figure 3.
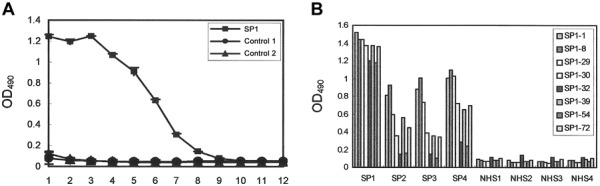
Specific reactivity of selected phage clones with serum antibodies from patients with severe acute respiratory syndrome (SARS). A, Serum antibodies from patient SP1 and from 2 healthy control subjects were incubated with a 3-fold serial dilution, from 1010 to 1.69×105 pfu, of selected phage clone SP1-1 (1, 1010 pfu; 2, 3.33×109 pfu; 3, 1.11×109 pfu; 4, 3.70×108 pfu; 5, 1.23×108 pfu; 6, 4.11×107 pfu; 7, 1.37×107 pfu; 8, 4.57×106 pfu; 9, 1.52×106 pfu; 10, 5.08×105 pfu; 11, 1.69×105 pfu; and 12, 0 pfu). SP1-1 reacted specifically with serum antibodies from patient SP1 but not with serum antibodies from the 2 control subjects. B, Analysis of serum from 4 patients with SARS (SP1–SP4) and of normal serum from 4 healthy subjects (NHS1-NHS4). Serum samples were incubated with 1×109 pfu of immunopositive phage clones (SP1-1, −8, −29, −30, −32, −39, −54, and −72). Immunopositive phage clones SP1-1, −8, and −29 were highly reactive to serum from patients with SARS but not to serum from the healthy subjects. OD490 , optical density at 490 nm.
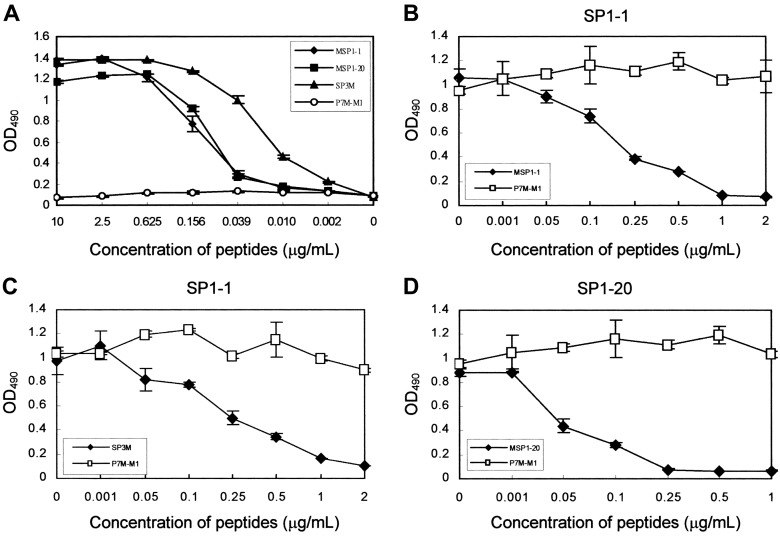
Characterization of SARS-specific B cell epitopes. To verify that peptide sequences identified by phage display were recognized by SARS-specific serum antibodies, we performed synthetic peptide-binding assays. Phage-displayed peptides were synthesized in multiple-antigen peptide (MAP) form, because ELISA sensitivity for an 8-chain MAP is greater than that for a singlechain peptide [22, 32]. As shown in figure 4A, synthetic peptides MSP1-1 (VKIPQHMHPLPI), MSP1-20 (VKPPNPRPTMSLW), and SP3M (VKIDNASPAS), which corresponded to immunopositive phage clones SP1-1 and SP1-20 and amino acid residues 18-–27 of CDS4 of SARS-CoV, bound antibodies in a concentration-dependent manner. The unrelated control MAP P7M-M1 (SLHNTMPSES) [33] was not reactive.
To further confirm that the phage-displayed peptide was the epitope of SARS-specific serum antibodies, a peptide competitive- inhibition assay was performed to determine whether the synthetic peptide MSP1-1 and the selected phage clone SP1-1 competed for the same antibody-binding site. Binding activity of SARS-specific serum antibodies with phage clone SP1-1 was inhibited by the synthetic peptide MSP1-1 in a dosedependent manner. The arbitrary control peptide P7M-M1 (SLHNTMPSES) had no effect on the ability of the phage particles to bind SARS-specific serum antibodies (figure 4B). One microgram per milliliter of peptide MSP1-1 inhibited 92.4% of the phage-clone binding to SARS-specific serum antibodies. Binding of the other phage clone, SP1-20, to SARS-specific serum antibodies also was inhibited by the synthetic peptide MSP1-20 in a dose-dependent manner. One microgram per milliliter of peptide MSP1-20 inhibited 92.7% of phage-clone binding to SARS-specific serum antibodies (figure 4D).
Figure 4.
A, Binding of synthetic peptides with serum antibodies from a patient with severe acute respiratory syndrome (SP1). Epitope-based synthetic peptides MSP1-1, MSP1-20, and SP3M reacted specifically and dose dependently with serum antibodies from patient SP1. Control peptide P7M-M1 was not reactive with serum antibodies from patient SP1. B-D, Competitive inhibition of phage clones binding to serum antibodies from patient SP1, by use of synthetic peptides. B, Binding of phage clone SP1-1 inhibited by peptide MSP1-1. C, Binding of phage clone SP1-1 inhibited by peptide SP3M. D, Binding of phage clone SP1-20 inhibited by peptide MSP1-20. Control peptide P7M-M1 had no effect in any of the 3 assays. Error bars indicate SDs. OD490 , optical density at 490 nm.
The synthetic peptide SP3M (VKIDNASPAS), which corresponded to amino acid residues 18–27 of CDS4 of SARS-CoV, bound antibody in a concentration-dependent manner (figure 4A). For further confirmation that phage-displayed peptides were epitopes of SARS-specific serum antibodies, a peptide competitive- inhibition assay was conducted to determine whether the SP3M peptide and phage clone SP1-1 competed for the same antibody-binding site. Our results indicated that the binding activity of SARS-specific serum antibodies with phage clone SP1- 1 was inhibited by SP3M in a dose-dependent manner. One microgram per milliliter of peptide SP3M inhibited 83.8% of phage-clone binding to SARS-specific serum antibodies, whereas the arbitrary control peptide P7M-M1 (SLHNTMPSES) had no effect (figure 4C).
To confirm that epitope-based synthetic peptides generated antibodies against SARS-CoV, we immunized BALB/c mice with peptides MSP1-1 and SP3M. Serum samples from both MSP1-1- and SP3M-immunized mice were found, by ELISA, to generate antibodies against the corresponding peptide antigens (figure 5A). Four of 5 MSP1-1-immunized mice (MSP1-1 MS-1–3 and −5) and 2 SP3M-immunized mice (SP3M MS- 1 and −2) were found, by IFA assay, to generate antibodies that recognized SARS-CoV-infected Vero E6 cells (antibody titers from ⩾1:20 to Δ1:40) (figure 5). Peptides MSP1-1 and SP3M competitively inhibited antibodies binding to SARS-CoV (figure 5B, panelsB and F, respectively), but unrelated control peptide P7M-M1 (SLHNTMPSES) had no such effect (figure 5B, panels C and G). Normal mouse serum was not reactive (figure 5B, panel D).
Figure 5.
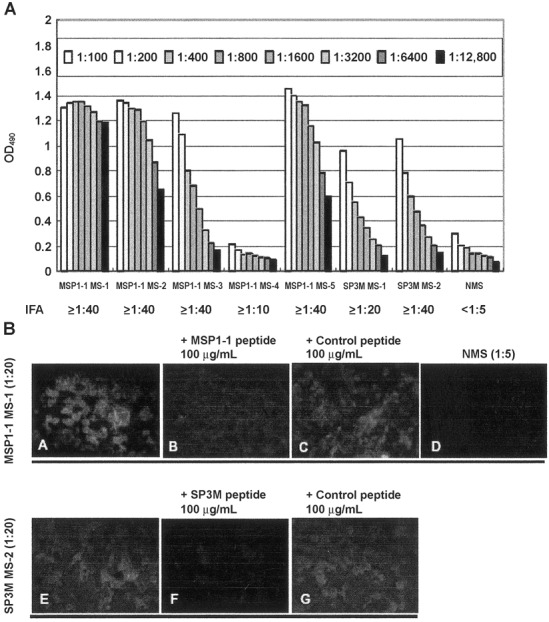
Phage-displayed epitopes immunogenic to severe acute respiratory syndrome-associated coronavirus (SARS-CoV). Vero E6 cells were infected with SARS-CoV and immunostained with antibodies against epitope-based synthetic peptides. Reactivity between peptide antigens and SARSCoV in serum samples from 5 mice immunized with peptide MSP1-1 (MSP1-1 MS-1-5) and 2 mice immunized with peptide SP3M (SP3M MS-1 and -2) was assayed by ELISA (A) and indirect fluorescent antibody (IFA) assay (B). SARS-CoV-infected cells were detected in serum samples from MSP1- 1-immunized mouse MS-1 (B, top row, panel A) and SP3M-immunized mouse MS-2 (B, bottom row, panel E) (20-fold dilution). The reactivity of antibodies with SARS-CoV was inhibited by 100 mg/mL of peptides MSP1-1 (B, panel B) and SP3M (B, panel F); 100 mg/mL of control peptide P7MM1 had no effect (B, panels C and G). Normal mouse serum (NMS; 5-fold dilution; B, panel D) did not react with SARS-CoV-infected cells. OD490 , optical density at 490 nm.
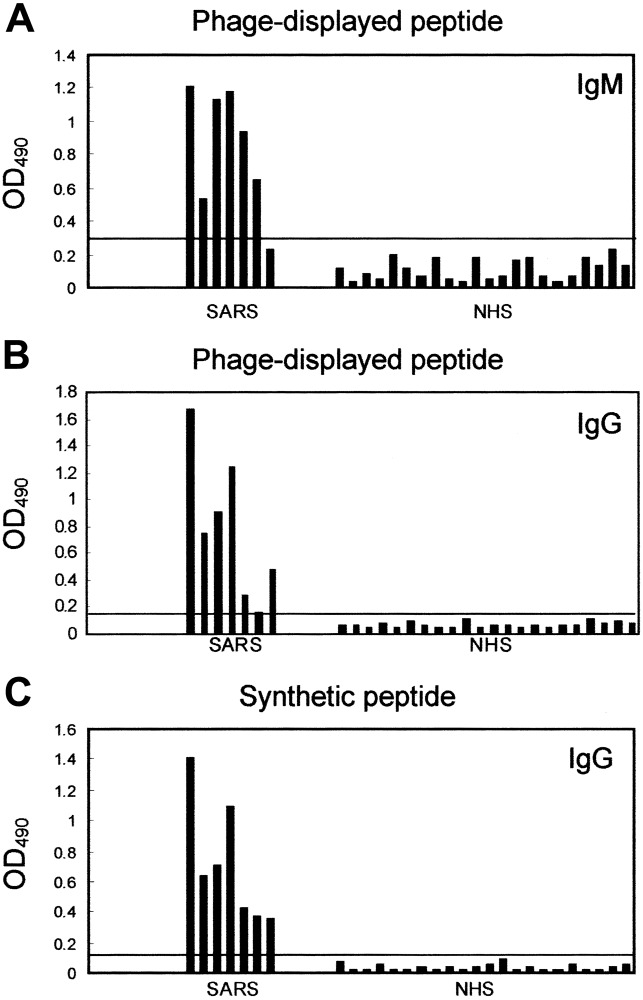
Detection of serum samples from patients with SARS, using epitope-based peptide antigens. We evaluated whether epitope- based peptide antigens in immunopositive phage clones or synthetic peptides could be used as diagnostic reagents for the detection of serum samples from patients with SARS. Between 8 March and 8 May 2003, we collected serum samples from 7 patients with illnesses that met the CDC case definition of probable SARS, to test the detection efficacy of the peptide antigens. During the patients' convalescence, serum samples were obtained <18 days after disease onset. For patients SP1- SP7, serum samples were obtained 42, 33, 30, 30, 25, 24, and 19 days, respectively, after onset of illness. To diagnose SARS, we tested these serum samples with phage clone SP1-1, using IgM- or IgG-capture ELISA.
Results indicated that, for samples from patients SP1-SP6, detection was positive by IgM-capture ELISA (figure 6A) and that, for samples from patients SP1-SP5 and SP7, detection was positive by IgG-capture ELISA (figure 6B). None of 22 healthy control subjects was detected as having SARS (figure 6A and 6B). Mean optical density plus 3× SD (0.108 + 0.186, measured at 490 nm) was used to determine the cutoff value (0.294) for IgM-capture ELISA. The sensitivity of this serologic test was 85.7%. Mean optical density plus 3× SD (0.069+0.079, measured at 490 nm) was used to determine the cutoff value (0.148) for IgG-capture ELISA. The sensitivity of this serologic test was 85.7%. If we combined the results of IgM- and IgGcapture ELISAs, samples from all 7 patients would be detected, and sensitivity would increase to 100% (figure 6A and 6B). In contrast, all serum samples obtained from the 22 healthy control subjects were seronegative (figure 6A and 6B). The specificity of this test was 100% for healthy control subjects.
Figure 6.
ELISA reactivity of phage clone SP1-1 and synthetic peptide SP3M with serum samples from patients with severe acute respiratory syndrome (SARS) vs. samples of normal human serum (NHS) from healthy control subjects. A, Serum IgM was captured by anti-human IgM antibodies and was incubated with 1×109 pfu of phage clone SP1-1. In patient samples, IgM against SP1-1 was further detected as described in Materials and Methods. The cutoff value was calculated as 0.294 (horizontal line). B, Serum IgG was captured by anti-human IgG antibodies and was incubated with 1×109 pfu of phage clone SP1-1. In patient samples, IgG against SP1-1 was further detected as described in Materials and Methods. The cutoff value was calculated as 0.148 (horizontal line). C, Detection of SARS-specific serum antibodies by synthetic peptide SP3M. In patient samples, IgG was further detected as described in Materials and Methods. The cutoff value was calculated as 0.139 (horizontal line). OD490 , optical density at 490 nm.
To test the reactivity of serum antibodies from patients with SARS, we used epitope-based synthetic peptide SP3M, which contains 10 aa residues in CDS4 of SARS-CoV. SP3M detected samples from all 7 patients with SARS, by ELISA (figure 6C). In contrast, all serum samples obtained from the 22 healthy control subjects were seronegative. Mean optical density + 3 (0.035 + 0.104, measured at 490 nm) was used to determine the cutoff value (0.139). Sensitivity and specificity were both 100% when we used epitope-based synthetic peptide SP3M for this serologic test (figure 6C).
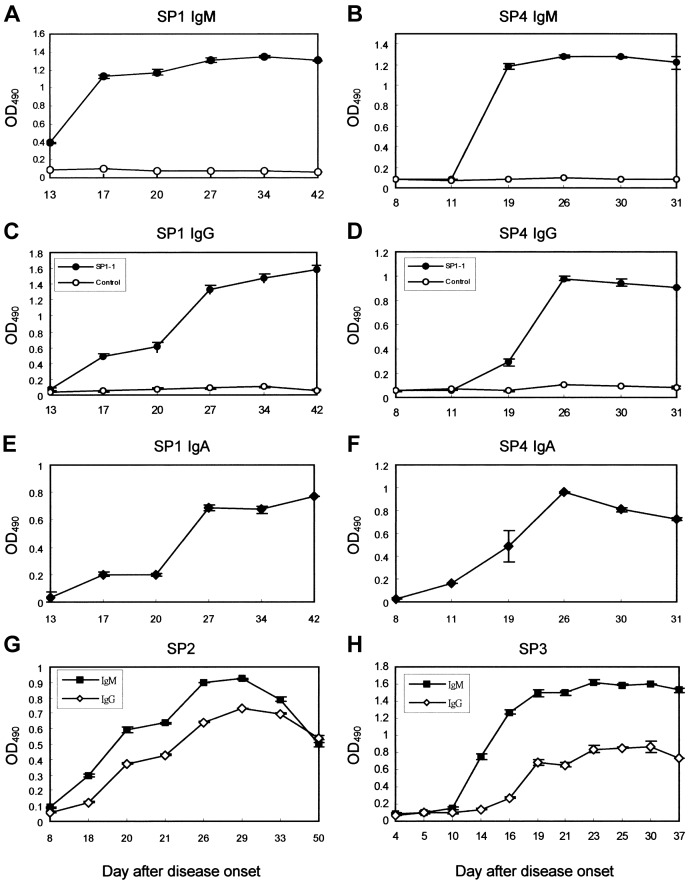
Kinetics of antibody response to SARS viral infection. We further analyzed antibody-response kinetics, by using ELISA, in 2 patients (SP1 and SP4) who had higher SARS-CoV-specific antibody titers. In patient SP1, IgM antibodies appeared on day 13 after disease onset, and levels peaked on day 17 (figure 7A). IgG antibody titers were not detected until day 17 after disease onset, and levels increased exponentially until day 27 (figure 7C). The kinetics of the IgA antibody response was similar to that for IgG: IgA antibody titers were not detected until day 17 after disease onset, and levels increased exponentially until day 27 (figure 7E). In patient SP4, IgM antibodies were not detected until day 11 after disease onset, and levels peaked on day 19 (figure 7B). IgG antibody titers were not detected until day 19 after disease onset, and levels increased exponentially until day 26 (figure 7D). In patient SP4, levels of IgA antibody titers increased exponentially from onset of the disease until day 26 (figure 7E). The kinetics of antibody response also were studied in 2 patients (SP2 and SP3) who had lower SARS-CoV-specific antibody titers (figure 7G and 7H). Patient SP2 tested seronegative for IgM at day 8 after the onset of symptoms (figure 7G), and patient SP3 tested seronegative for IgM and IgG at days 10 and 14, respectively, after the onset of disease (figure 7H).
Figure 7.
Kinetics of antibody response to a severe acute respiratory syndrome (SARS) viral infection. Kinetics of IgM, IgG, and IgA responses in patients with SARS were analyzed with phage clone SP1-1. IgM (A and B), IgG (C and D), and IgA (E and F) responses in patients SP1 and SP4 were measured by using phage clone SP1-1, as described in Materials and Methods. IgM and IgG responses of 2 additional patients with SARS, SP2 (G) and SP3 (H), were also measured by using phage clone SP1-1, as described in Materials and Methods. Control phage clones did not react with serum samples from the patients with SARS. To study the kinetics of antibody response, we used phage clone SP1-1 but not synthetic peptide SP3M, because the background of ELISA increased when we used synthetic peptides as antigens for the detection of human IgM. OD490 , optical density at 490 nm.
Discussion
Identification of viral B cell epitopes is important in understanding virus-antibody interactions at a molecular level and provides information for the development of virus-specific serologic diagnostic reagents and subunit vaccines. This is the first study to characterize serotype-specific B cell epitopes of SARS-CoV, using the phage-display method. Selected phage-displayed peptide sequences had 2 consensus motifs, Pro-Pro-Asn and Val-Lys-Ile- X-Asn, which corresponded to amino acid residues 1184–1186 of CDS2 (PPN) and 18–22 of CDS4 (VKIDN) in SARS-CoV, respectively. Results of synthetic peptide-binding and competitive- inhibition assays further confirmed that patients with SARS generated antibodies against SARS-CoV. We also detected antibodies in serum samples from patients with SARS by using phage-displayed and epitope-based peptide antigens.
Tissue culture, electron microscopy, IFA tests, and PCR amplification of specific genomic sequences and of the complete genome of SARS-CoV have provided evidence of novel coronavirus infection in patients with SARS [8, 16, 34]. Using phagedisplay methods, we found B cell epitopes in the antibody response of patients with SARS that corresponded to SARS-CoV, further confirming the microbiologic origin of SARS. While studying the alignment of phage-displayed peptide sequences and the published protein sequences of several viruses (described in Materials and Methods), 16 immunopositive phage clones displayed a consensus motif of PPN, which only corresponded to amino acid residues 1184–1186 of CDS2 of SARSCoV (table 2). The gene product of CDS2 (GenBank accession no. AAP30029) of SARS-CoV is an orfla polyprotein containing 4382 aa residues. Another 16 immunopositive phage clones displayed the consensus motif VKIXN, which only corresponded to amino acid residues 18–22 of CDS4 of SARS-CoV (table 2). The gene product of CDS4 (GenBank accession no. P59632) of SARS-CoV is a hypothetical transmembrane protein containing 274 aa residues.
We also found the peptide sequences of 2 immunopositive phage clones, SP1-32 and -50 (ISPYNTIVAKLR; table 8), to be similar to amino acid residues 375–386 of CDS4 in hMPV (LSPLGALVACYK; consensus sequences are indicated by boldface type). The serum samples from all 7 patients with SARS were detected by use of ELISA with phage clone SP1-1, but only the serum samples from patient SP1 were detected by phage clone SP1-32 (figures 3 and 6; data not shown). These findings suggest that SARS is caused by SARS-CoV. We also cannot exclude the possibility that hMPV infection may have exacerbated the disease in some of the patients with SARS. Correlations between SARS and hMPV infection need further investigation. We also compared phage-displayed peptide sequences with those from reference viruses representing each species in the 3 groups of coronaviruses [34], but no similarity was found.
Serum samples from patients with SARS were accurately detected only by use of ELISA with phage clone SP1-1 and synthetic peptide SP3M (figure 6). Sensitivity for IgM- or IgG-capture ELISA alone was 85.7%; however, when IgM- and IgGcapture ELISAs were combined, serum samples from all 7 patients with SARS were detected, and sensitivity increased to 100%. Using the same serologic test, we found that serum samples from all 22 healthy adults were seronegative, yielding a specificity of 100% for healthy control subjects (figure 6A and 6B. Epitopebased synthetic peptide SP3M, corresponding to amino acid residues 18–27 of CDS4 in SARS-CoV, detected serum samples from all 7 patients with SARS, by ELISA, and was nonreactive with serum samples from all 22 healthy adults (figure 6C). Sensitivity and specificity were both 100% when we used the 2 methods for this serologic test. For all 7 patients with SARS and the 22 healthy adults, a blinded analysis also was done by ELISA. Serum samples from all 7 patients with SARS were detected, and the 22 healthy adults were found to be seronegative, further confirming the efficacy of this means of detecting the disease (data not shown).
In our study of antibody-response kinetics in patients with SARS, antibodies were detectable at 13–18 days after onset of disease (figure 7). IgM was not found earlier than days 8, 10, and 11 after onset of symptoms in patients SP2, SP3, and SP4, respectively (figure 7B, 7G, and 7H). During acute SARS-CoV infection, IgM antibodies did not appear until days 8–11, and levels peaked on days 17–19 after onset of disease. IgG antibody titers were detectable only by days 17–19 and continued to increase exponentially until days 26 and 27 (figure 7). Delayed induction of IgM and IgG responses to SARS needs further investigation. To some extent, this delay may explain the severity of SARS-CoV infections.
The experimental approach described in this study identifies peptides that react with disease-specific antibodies. We isolated the B cell epitope of SARS-CoV and confirmed that the Pro- Pro-Asn and Val-Lys-Ile-X-Asn motifs were crucial for peptide antigen-antibody binding. From the study of antibody kinetics (figure 7), we found that the peptides were able to detect patient serum samples during the convalescent phase of SARS but not during the acute phase. Peptide-binding and competitive-inhibition assay showed that synthetic peptide SP3M corresponded to amino acid residues 18–27 of CDS4 of SARS-CoV, further confirming that patients with SARS generated antibodies against SARS-CoV (figure 4). To further confirm that epitope-based synthetic peptides generated antibodies against SARS-CoV, we generated antibodies against peptides MSP1-1 and SP3M. Both MSP1-1- and SP3M-immunized serum samples were found, by IFA, to generate antibodies that recognized SARS-CoV-infected Vero E6 cells (figure 5), further proving that the phage-displayed epitope was immunogenic to SARS-CoV. We also studied the neutralizing activity of these antibodies (using 100 TCID50 of SARS-CoV). The neutralizing activity of patient SP1 antiserum was ⩾1:60. No notable neutralization of SARS-CoV was observed in MSP1-1- and SP3M-immunized mouse serum samples. Identification of neutralizing epitopes by using spike protein- purified antibodies is currently being investigated.
Our findings also indicate that B cell epitopes of these antibodies are linear. Typically, B cell epitopes identified by this method have an easily recognizable consensus sequence, often corresponding to the peptide sequence found in the natural antigen [22]. These epitopes can be used to study microbiologic origins, to develop epitope-based diagnostic reagents and immunogens against viral infection, and to dissect the antibody response in SARS. The phage itself is an excellent immunogen. Immunization of mice with phage particles elicits a T cell-dependent response against the phage-displayed epitope [35, 36] and antibodies against the antigen [23, 24]. Development of a SARS vaccine consisting of a cocktail of phage-displayed neutralizing epitopes may be possible.
To date, we have analyzed results for 7 patients with illnesses that met the CDC case definition of probable SARS, and we have studied serum samples obtained from convalescing patients >18 days after onset of disease. Further investigation regarding the applicability of this serologic test to human serum samples is in progress with more cases of SARS. Specifically, we are evaluating the percentage of subclinical or mild infections and the relationship between antibody response and disease severity.
Serologic diagnostic reagents for SARS that are highly sensitive, specific, and convenient need to be developed.We found that using synthetic peptides for SARS diagnosis was relatively simple. Because selected phage clones and epitope-based peptide antigens in our study were highly specific for serum from patients with SARS, we feel that they may be useful not only in developing diagnostic and prognostic reagents but also in understanding the pathogenesis of SARS.
Acknowledgments
We thank C. J. Chen, C. C. King, and the SARS Study Group of National Taiwan University, for their help, and C. D. Wu, for preparation of the manuscript.
References
- 1.Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–94. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 2.Global surveillance for severe acute respiratory syndrome (SARS) Wkly Epidemiol Rec. 2003;78:100–19. [PubMed] [Google Scholar]
- 3.Update: outbreak of severe acute respiratory syndrome worldwide, 2003. MMWR Morb Mortal Wkly Rep. 2003;52:241–6. [PubMed] [Google Scholar]
- 4.Update: outbreak of severe acute respiratory syndrome worldwide,2003. MMWR Morb Mortal Wkly Rep. 2003;52:269–72. [PubMed] [Google Scholar]
- 5.Tsang KW, Ho PL, Ooi GC, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1977–85. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 6.Poutanen SM, Low DE, Henry B, et al. Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 7.Drazen JM. Case clusters of the severe acute respiratory syndrome. N Engl J Med. 2003;348:6–7. doi: 10.1056/NEJMe030062. [DOI] [PubMed] [Google Scholar]
- 8.Gerberding JL. Faster...but fast enough? Responding to the epidemic of severe acute respiratory syndrome. N Engl J Med. 2003;348:2030–1. doi: 10.1056/NEJMe030067. [DOI] [PubMed] [Google Scholar]
- 9.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med. 2000;342:1334–49. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Severe acute respiratory syndrome (SARS) updated interim case definition, 2003. Available at: http://www.cdc.gov/ncidod/sars/casedefinition.htm. Accessed April 2003.
- 11.World Health Organization Cumulative number of reported cases of severe acute respiratory syndrome (SARS) Available at: http://www.who.int/csr/sarscountry/2003_04_02/en. Accessed April 2003.
- 12.World Health Organization Case definitions for surveillance of severe acute respiratory syndrome (SARS), 2003. Available at: http://www.who.int/csr/sars/casedefinition/en. Accessed April 2003.
- 13.Update: severe acute respiratory syndrome—United States, 2003. MMWR Morb Mortal Wkly Rep. 2003;52:357–60. [PubMed] [Google Scholar]
- 14.World Health Organization Severe acute respiratory syndrome(SARS) Available at: http://www.who.int/csr/sars/en. Accessed April 2003.
- 15.Benitez MA. Hong Kong health chief falls ill with suspected SARS virus. Lancet. 2003;361:1106. doi: 10.1016/S0140-6736(03)12912-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 17.Peiris JSM, Lai ST, Poon LLM, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1312–3. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falsey AR, Walsh EE. Novel coronavirus and severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13084-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ksiazek TG, Erdman D, Goldsmith G, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 20.Severe acute respiratory syndrome (SARS) and coronavirus testing—United States, 2003. MMWRMorb Mortal Wkly Rep. 2003;52:297–302. [PubMed] [Google Scholar]
- 21.Fouchier RAM, Kuiken T, Schutten M, et al. Aetiology: Koch's postulates fulfilled for SARS virus [brief communication] Nature. 2003;423:240–1. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu HC, Huang YL, Chao TT, et al. Identification of B-cell epitope of dengue virus type 1 and its application in diagnosis of patients. J Clin Microbiol. 2001;39:977–82. doi: 10.1128/JCM.39.3.977-982.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu HC, Yeh CT, Huang YL, Tarn LJ, Lung CC. Characterization of neutralizing antibodies and identification of neutralizing epitope mimics on the Clostridium botulinum neurotoxin type A. Appl Environ Microbiol. 2001;67:3201–7. doi: 10.1128/AEM.67.7.3201-3207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folgori A, Tafi R, Meola A, et al. A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J. 1994;13:2236–43. doi: 10.1002/j.1460-2075.1994.tb06501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prezzi C, Nuzzo M, Meola A, et al. Selection of antigenic and immunogenic mimics of hepatitis C virus using sera from patients. J Immunol. 1996;156:4504–13. [PubMed] [Google Scholar]
- 26.Dybwad A, Forre O, Kjeldsen-Kragh J, Natvig JB, Sioud M. Identification of new B cell epitopes in the sera of rheumatoid arthritis patients using a random nanopeptide phage library. Eur J Immunol. 1993;23:3189–93. doi: 10.1002/eji.1830231222. [DOI] [PubMed] [Google Scholar]
- 27.Cortese I, Tafi R, Grimaldi LME, et al. Identification of peptides specific for cerebrospinal fluid antibodies in multiple sclerosis by using phage libraries. Proc Natl Acad Sci USA. 1996;93:11063–7. doi: 10.1073/pnas.93.20.11063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowditch RD, Tani P, McMillan R. Characterization of autoantigenic epitopes on platelet glycoprotein IIb/IIIa using random peptide libraries. Blood. 1996;88:4579–84. [PubMed] [Google Scholar]
- 29.Gevorkian G, Manoutcharian K, Almagro JC, Govezensky T, Dominguez V. Identification of autoimmune thrombocytopenic purpura-related epitopes using phage-display peptide library. Clin Immunol Immunopathol. 1998;86:305–9. doi: 10.1006/clin.1997.4502. [DOI] [PubMed] [Google Scholar]
- 30.Manoutcharian K, Sotelo J, Garcia E, Cano A, Gevorkian G. Characterization of cerebrospinal fluid antibody specificities in neurocysticercosis using phage display peptide library. Clin Immunol. 1999;91:117–21. doi: 10.1006/clim.1998.4669. [DOI] [PubMed] [Google Scholar]
- 31.Hsueh PR, Hsiao CH, Wang WK, et al. Microbiologic characteristics, serologic responses, and clinical manifestations in severe acute respiratory syndrome, Taiwan. Emerg Infect Dis. 2003;9:1163–7. doi: 10.3201/eid0909.030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tam JP, Zavala F. Multiple antigen peptide: a novel approach to increase detection sensitivity of synthetic peptides in solid-phase immunoassays. J Immunol Methods. 1989;124:53–61. doi: 10.1016/0022-1759(89)90185-3. [DOI] [PubMed] [Google Scholar]
- 33.Wu HC, Jung MY, Chiu CY, Lai SC, Jan JT, Shaio MF. Identification of a dengue virus type 2 (DEN-2) serotype-specific B-cell epitope and detection of DEN-2-immunized animal serum samples using an epitope-based peptide antigen. J Gen Virol. 2003;84:2271–9. doi: 10.1099/vir.0.19228-0. [DOI] [PubMed] [Google Scholar]
- 34.Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–9. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 35.Greenwood J, Willis AE, Perham RN. Multiple display of foreign peptides on a filamentous bacteriophage. J Mol Biol. 1991;220:821–7. doi: 10.1016/0022-2836(91)90354-9. [DOI] [PubMed] [Google Scholar]
- 36.Willis AE, Perham RN, Wraith D. Immunological properties of foreign peptides in multiple display on a filamentous bacteriophage. Gene. 1993;128:79–83. doi: 10.1016/0378-1119(93)90156-w. [DOI] [PubMed] [Google Scholar]