Abstract
The severe acute respiratory syndrome–associated coronavirus (SARS-CoV) is reported to have deletions of various sizes. Recently, the large 386-nucleotide deletion (L386del) comprising nucleotide positions 27719-28104 and spanning open reading frames 9–11 has been reported in the genomes of some human isolates from Hong Kong. In this study, archived specimens from 71 patients with SARS who were admitted to the New Territory East Cluster Hospitals in Hong Kong were analyzed to determine whether the L386del variant of SARS-CoV was present. There was no clear relationship between the presence of the L386del variant and SARS clinical severity as defined either by the need for intensive-care therapy and/or ventilation or by death. One patient had evidence of both the L386del variant and the wild-type variant in the same clinical specimen, supporting the idea that SARS-CoV exists as a quasispecies in some patients, although the clinical significance of these quasispecies remains unclear
Since the worldwide SARS epidemic in 2003, many studies have focused on the genomics of the SARS-associated coronavirus (SARS-CoV), to understand more about the pathogenic potential of this new pathogen. Several significant differences between the various SARS-CoV isolates from different patients and animal hosts have now been identified. Guan et al. [1] found that the majority of human SARS-CoV associated with the epidemic had a 29-nt deletion spanning open reading frames (ORFs) 10 and 11 (in Guan et al.’s original nomenclature), compared with the SARS-like coronavirus (SL-CoV) identified in Himalayan palm civet cats (Paguma larvata) from a Guangdong live-animal market; this 29-nt deletion in the human SARS-CoV genome comprised nucleotide positions (np) 27869–27897 when that genome was aligned with the TOR-2 SARS-CoV reference sequence (GenBank accession no. NC004718). The functional significance of this deletion is uncertain, although it may represent an adaptive mutation to a human host by SARS-CoV from a potential civet-cat animal reservoir [1, 2]. However, it has been reported that the human-adapted SARS-CoV with the 29-nt deletion remains just as able to infect the civet cat [3]. Other studies have suggested that bats may be an alternative animal reservoir for SARS-CoV [4, 5], which suggests that the civet cat is a secondary, amplifying host for SARS-CoV rather than the primary reservoir. The bat SL-CoV also has the additional 29 nt in ORFs 10 and 11 and, in this respect, is similar to the civet-cat SL-CoV [4, 5]. Most recently, experiments have shown that replacement of this 29-nt deletion does not alter the in vitro or in vivo replication, persistence, or duration of the viral load in the murine model, although this finding does not exclude the possibility that there could be more-dramatic effects if a civet-cat or raccoon-dog model were used [6]
Other deletions have been noted in other areas of the SARS-CoV genome. An 82-nt deletion in ORF 8 has been reported in a study [7] whose authors also suggested that the earliest phase of the SARS epidemic (i.e., when SARS was confined to China) were characterized by 2 major SARS-CoV genotypes, one of them characterized by the 29-nt deletion and the other harboring the 82-nt deletion. These 2 genotypes had also been isolated from animals in Guangdong and other provinces (e.g., Hubei) in China. The authors of that study also noted that these 2 genotypes were not restricted to the early part of the epidemic but were isolated during the middle phase as well; in addition, they reported the presence, during the late phase of the epidemic, of a third genotype, characterized by a 415-nt deletion resulting in the loss of ORF 8. They speculated that ORF 8 may have either no or an unimportant coding function, because the virus’s life-cycle can be completed even if ORF 8 is absent [7]
Even within the human population, it seems that SARS-CoV has continued to adapt during the course of the epidemic. A report from Hong Kong has documented the presence, in some human SARS-CoV genomes, of a large 386-nt deletion (L386del) comprising np 27719–28104 [8]. This larger deletion flanks the 29-nt deletion segment described above, disrupting ORF 9—and, simultaneously, eliminating ORFs 10 and 11—of the SARS-CoV genome. So far, there have been no other reports of this large genomic deletion
The present study reports additional data on the presence of the L386del variant of SARS-CoV in other Hong Kong Chinese patients during the SARS epidemic in 2003. In addition, it reports 1 patient from whom multiple clinical specimens were taken that had both the L386del variant and the wild-type variant, indicating that the 2 variants can coexist not only within a single host but also within the same clinical specimen. The present study also assesses whether this deletion is associated with more-severe SARS, which is clinically defined either as requiring admission to intensive care (IC) and/or ventilation or as resulting in death
Patients, Materials, and Methods
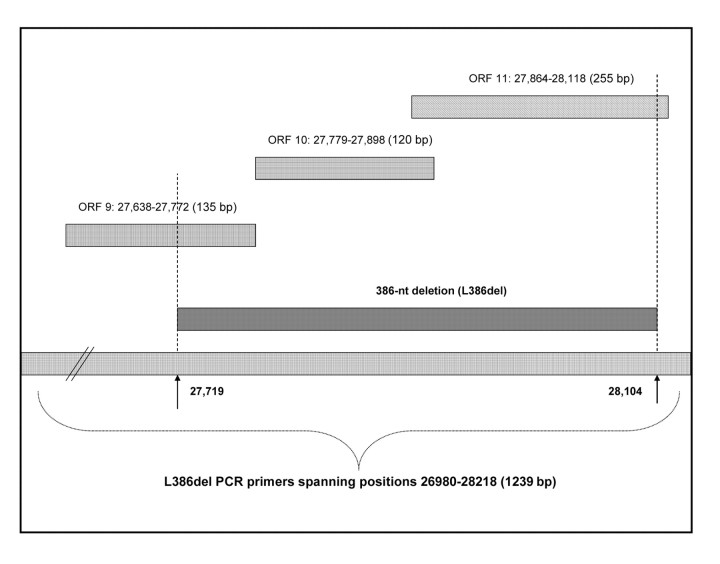
The clinical specimens used in the present study were obtained from patients with laboratory-confirmed SARS-CoV infection [9] who had been admitted to the New Territory East Cluster Hospitals during the Hong Kong SARS epidemic, which occurred in March–June 2003. All clinical specimens that had tested positive for SARS-CoV RNA during the routine investigation were retrieved for the present study, provided that sufficient remaining volumes were available; if the original clinical specimens were unavailable, their primary isolates, obtained from Vero cell culture, were used. A pair of specific primers, spanning np 26980–28218 (1239 bp), were designed for polymerase chain reaction (PCR) used to amplify the L386del region (figure 1); this region spans the entire length of ORFs 9–11, which are known to have—and to be disrupted by—this deletion [8]. All PCR products that gel electrophoresis revealed as having a size smaller than the 1239 bp expected for the wild type were then sequenced, to map the deletion site
Figure 1.
Location, in the SARS-associated coronavirus genome, of the large 386-nt deletion (L386del), in relation to open reading frames (ORFs) 9–11. PCR, polymerase chain reaction. (N.B.: the sizes of the bars are for illustrative purposes only and are not to scale.)
Results
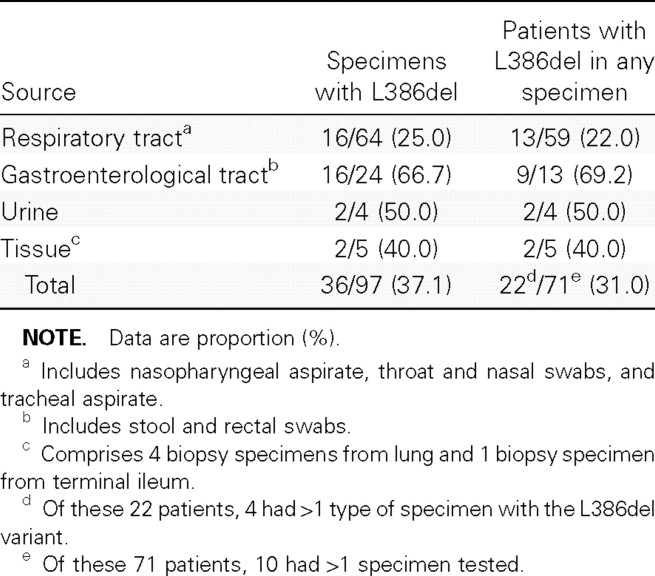
The present study included 71 patients (age range, 15–98 years; mean ± SD, 54.6 ± 21.2 years), 25 of whom were male. A total of 97 original specimens/primary isolates were available from these 71 patients: only 1 specimen was available from each of 61 (85.9%) of these patients, 2 specimens were available from each of 4 (5.6%) of these patients, and ⩾3 specimens were available from each of 6 (8.5%) of these patients. The distribution of specimen types is shown in table 1; the majority of the specimens studied were from the respiratory tract (66.0%) or stool/rectal swabs (24.7%)
Table 1.
Proportions of specimens and patients with the large 386-nt deletion (L386del) variant of SARS-associated coronavirus
A total of 36 (37.1%) of the 97 specimens had a PCR product with a size less than that expected for the wild-type variant. The sequencing results for these PCR products showed that they all had a 386-bp deletion corresponding to np 27719–28104 of the TOR-2 SARS-CoV reference sequence. All these sequences were submitted to GenBank (accession nos. DQ523267–DQ523302). The proportion of stool/rectal swab specimens with the L386del variant was significantly higher than that of respiratory specimens (66.7% vs. 25.0%, respectively [P<.001, by χ2 test]); the number of urine specimens was too few to allow for similar statistical analysis. In the patients, the distribution of variants was as follows: 49 (69.0%) of the 71 patients had only the wild-type variant, 20 (28.2%) of them had only the L386del variant, and 2 (2.8%) of them had both variants
Of the 71 patients studied, 36 (50.7%) had “mild” SARS illness (clinically defined either as not requiring admission to IC and/or ventilation or as recovered), and 8 (22.2%) of these 36 patients had the L386del variant; the remaining 35 (49.3%) of these 71 patients had “severe” SARS illness (clinically defined either as requiring admission to IC and/or ventilation or as resulting in death), and in 14 (40.0%) of these 35 patients ⩾1 specimen contained the L386del variant. Although a higher proportion of patients with severe SARS illness were found to have the L386del variant, the difference was not statistically significant (P=.1, by χ2 test)
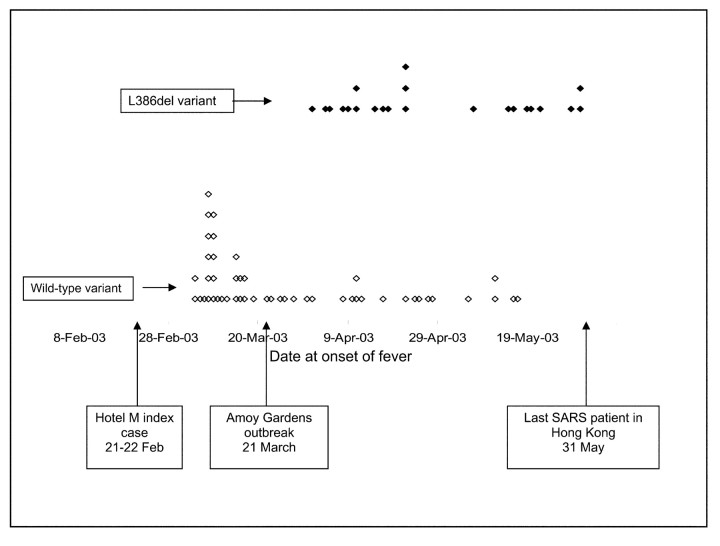
The correlation between the L386del variant and the time at onset of SARS is shown in figure 2. The L386del variant appeared at the end of March 2003, immediately after the major community (i.e., Amoy Gardens) outbreak, and then cocirculated with the wild-type variant and became predominant toward the end of the outbreak in Hong Kong
Figure 2.
Presence of the large 386-nt deletion (L386del) variant of SARS-associated coronavirus, in relation to date at onset of fever. Each diamond represents 1 patient examined in the present study; the diamonds are positioned according to the date at onset of fever, as indicated on the x-axis. Patients with the L386del variant (black diamonds) are clustered toward the upper part of the figure; patients without it (white diamonds) are clustered toward the lower portion of the figure
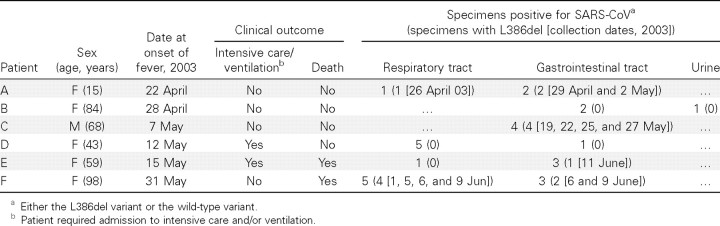
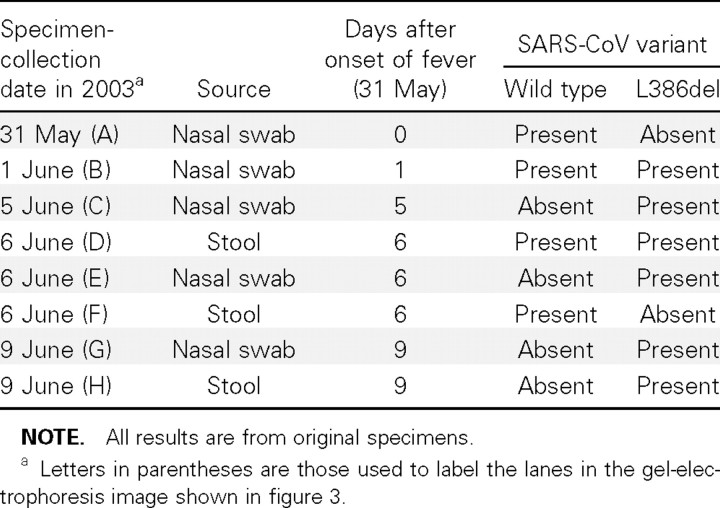
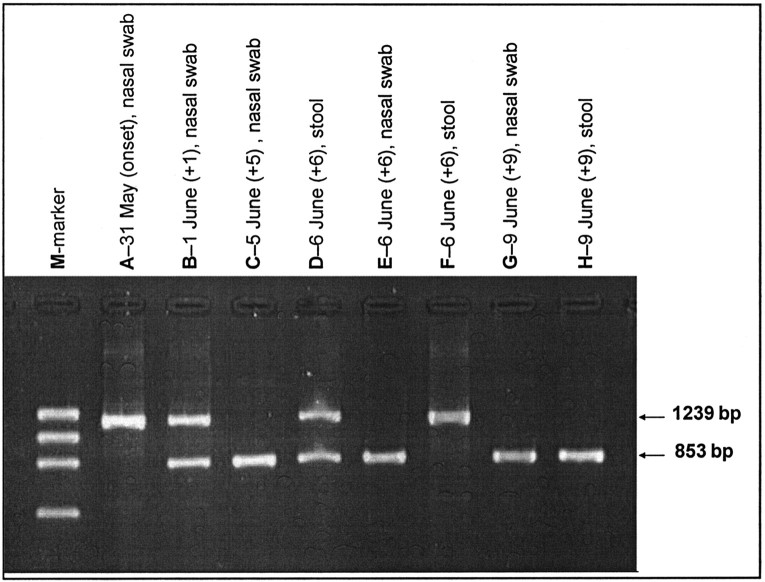
All 4 (100%) of the patients from whom 2 specimens each were available for study had the L386del variant, and it was present in all 8 specimens. The characteristics, specimen distribution, and results of the investigation of the 6 patients (patients A–F) from whom ⩾3 specimens were available for study are shown in table 2. In patient F, both the L386del variant and the wild-type variant were detected in the same specimen and also in specimens taken from different sites during the same day (table 3 and figure 3)
Table 2.
Distribution of the large 386-nt deletion (L386del) variant of SARS-associated coronavirus (SARS-CoV), in patients from whom ⩾3 specimens were available for study
Table 3.
Distribution of the large 386-nt deletion (L386del) variant and the wild-type variant of SARS-associated coronavirus (SARS-CoV), in serial specimens collected from patient F
Figure 3.
Gel-electrophoresis imaging of products of polymerase chain reaction (PCR) using primers specific for the large 386-nt deletion (L386del) in SARS-associated coronavirus. Lanes are labeled with specimen type and collection date (nos. in parentheses are no. of days after onset of fever). Lane M, marker; lanes A and F, wild-type variant only; lanes C, E, G, and H, L386del variant only; lanes B and D, mixture of the L386del variant and the wild-type variant. PCR-product sizes for the L386del variant and the wild-type variant are 853 bp and 1239 bp, respectively
Discussion
The present study used original clinical specimens or, if the latter were unavailable, primary isolates of SARS-CoV. Because Poon et al. [10] had documented some single-nucleotide polymorphisms in the SARS-CoV genome of cultured specimens, we also tested the effect that in vitro passage in Vero cells (which had been used in the culturing of the original clinical specimen) might have on the emergence of the L386del variant. No emergence of the L386del variant was observed; therefore, the present study’s results regarding these primary SARS-CoV isolates are likely to reflect the situation in the original clinical specimens
The results of the present study show that the L386del variant is more likely to be found in specimens from the gastrointestinal tract than in those from the respiratory tract. It has been shown that both the lungs and gastrointestinal tract are the predominant sites for SARS-CoV tropism [11], and high viral loads have been documented in both tissue types [12]. However, the duration of shedding from the 2 sites seems to be different: analysis based on the diagnostic yield of specimens taken at different times [9] has shown that virus shedding from the gastrointestinal tract persists longer than does that from the respiratory tract. This longer duration of infection might favor the generation of mutants, including the L386del variant investigated in the present study
At present, the clinical implications of the L386del variant remain uncertain. Although the present study has shown that a higher proportion (49.3% vs. 22.2%) of patients with severe SARS infection have the L386del variant, the difference is not statistically significant (P=.1, by χ2 test). Furthermore, because of the retrospective nature of the present study, it was not able to control for all possible confounding factors, such as variation in the number and nature of examined specimens from the patients, the timing of the collection of the specimens, and the treatment regimen used
At a population level, the L386del variant appeared soon after the SARS outbreak in Hong Kong and became dominant toward the end of this outbreak. This observation is in keeping with the hypothesis that the L386del variant has adapted to human infection and is being transmitted as efficiently as—or even more efficiently than—the wild-type variant
The present study has shown that, at any given time, both the L386del variant and the wild-type variant can be present within the same clinical specimen of a patient, an example being patient F in the present study; and this finding provides evidence that SARS-CoV evolves rapidly within the same host and, therefore, may exist as a quasispecies. The results observed in patient F are also consistent with what has been observed at the population level, where the L386del variant was found to have appeared during the course of the SARS outbreak and to have become dominant toward the end of this outbreak
The L386del variant disrupts or completely eliminates ORFs 9 (np 27638–27772), 10 (np 27779–27898), and 11 (np 27864–28118). The functions of these ORFs remain unknown. An earlier report on the SARS-CoV genome [13] predicted that ORFs 9, 10, and 11 would code for proteins comprising 44, 39, and 84 aa, respectively. That report also noted that ORF 9 was comparable to a putative sterol-C5 desaturase and a Clostridium perfringens protein but that ORF 10 was not comparable to any known proteins listed in public databases. The structure predicted for ORFs 9 and 10 was a single transmembrane helix, and that for ORF 11 was a soluble protein; the latter showed a slight match to the human coronavirus S glycoprotein precursor [13]. More recently, Li et al. [14] have suggested that ORF 10 may play a role by impairing the oxidoreductase system in the mitochondria of SARS-CoV–infected host cells, and those authors therefore have hypothesized that the clinical effects of SARS-CoV infection in humans may be due to a direct cytopathic effect of the virus, rather than to virus-induced immunopathological damage. A study of 33 complete SARS-CoV genomes [15] found that ORF 10 had the highest point-mutation rate, a rate that was higher than that in parts of the replicase, S, and N genes; because this high mutation rate has also been reported for a similar region (ORF 10′) in the SL-CoVs recently isolated from 3 species of horseshoe bats (genus Rhinolophus) [4], ORF 10 seems to be present in human and other mammalian SARS-CoV and SL-CoV species, although its function remains unknown. In both the human and civet-cat host, it is unclear whether the reported deletions in the various ORFs of the SARS-CoV genome have any significant clinical consequence; however, at least 1 of them, the 29-nt deletion, was considered to be a marker of a human-host origin when it was found in SARS-CoV isolated from a pig [16]. Therefore, at present, the region covered by ORFs 10 and 11 would seem to be useful as a genetic marker of a possible animal origin of SARS-CoV, rather than as a therapeutic or vaccine target—in contrast to its structural counterparts, such as the S gene (ORF 2) [17]
Footnotes
Potential conflicts of interest: none reported
Financial support: Research Grants Council of the Hong Kong Special Administrative Region, China (project HKU7542/03M)
References
- 1.Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–8. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 2.Song HD, Tu CC, Zhang GW, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA. 2005;102:2430–5. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu D, Tu C, Xin C, et al. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J Virol. 2005;79:2620–5. doi: 10.1128/JVI.79.4.2620-2625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–9. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 5.Lau SK, Woo PC, Li KS, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–5. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yount B, Roberts RS, Sims AC, et al. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J Virol. 2005;79:14909–22. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–9. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 8.Chiu RW, Chim SS, Tong YK, et al. Tracing SARS-coronavirus variant with large genomic deletion. Emerg Infect Dis. 2005;11:168–70. doi: 10.3201/eid1101.040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan PK, To WK, Ng KC, et al. Laboratory diagnosis of SARS. Emerg Infect Dis. 2004;10:825–31. doi: 10.3201/eid1005.030682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poon LL, Leung CS, Chan KH, Yuen KY, Guan Y. Recurrent mutations associated with isolation and passage of SARS coronavirus in cells from non-human primates. J Med Virol. 2005;76:435–40. doi: 10.1002/jmv.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.To KF, Tong JH, Chan PK, et al. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J Pathol. 2004;202:157–63. doi: 10.1002/path.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farcas GA, Poutanen SM, Mazzulli T, et al. Fatal severe acute respiratory syndrome is associated with multiorgan involvement by coronavirus. J Infect Dis. 2005;191:193–7. doi: 10.1086/426870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marra MA, Jones SJ, Astell CR, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 14.Li Q, Wang L, Dong C, et al. The interaction of the SARS coronavirus non-structural protein 10 with the cellular oxido-reductase system causes an extensive cytopathic effect. J Clin Virol. 2005;34:133–9. doi: 10.1016/j.jcv.2004.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang ZG, Li LJ, Luo Y, et al. Molecular biological analysis of genotyping and phylogeny of severe acute respiratory syndrome associated coronavirus. Chin Med J. 2004;117:42–8. [PubMed] [Google Scholar]
- 16.Chen W, Yan M, Yang L, et al. SARS-associated coronavirus transmitted from human to pig. Emerg Infect Dis. 2005;11:446–8. doi: 10.3201/eid1103.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang S, He Y, Liu S. SARS vaccine development. Emerg Infect Dis. 2005;11:1016–20. doi: 10.3201/eid1107.050219. [DOI] [PMC free article] [PubMed] [Google Scholar]