Abstract
Motivation
Receptor mediated entry is the first step for viral infection. However, the question of how viruses select receptors remains unanswered.
Results
Here, by manually curating a high-quality database of 268 pairs of mammalian virus–host receptor interaction, which included 128 unique viral species or sub-species and 119 virus receptors, we found the viral receptors are structurally and functionally diverse, yet they had several common features when compared to other cell membrane proteins: more protein domains, higher level of N-glycosylation, higher ratio of self-interaction and more interaction partners, and higher expression in most tissues of the host. This study could deepen our understanding of virus–receptor interaction.
Availability and implementation
The database of mammalian virus–host receptor interaction is available at http://www.computationalbiology.cn: 5000/viralReceptor.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Over the past decades, great achievements have been made in prevention and control of infectious diseases, but the recent serial outbreaks of Zika virus (Mlakar et al., 2016), Ebola virus (EBOV) (Maganga et al., 2014) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) (Breban et al., 2013) indicate that the viral infectious diseases still pose a serious threat to human health and global security. The virus is the most abundant biological entity on Earth and exists in all habitats of the world (Paez-Espino et al., 2016). Nearly all cellular organisms are prey to viral attacks. Humans were reported to be infected by hundreds of viruses (Geoghegan et al., 2016; Mihara et al., 2016). Most of the human emerging infectious diseases are zoonotic, with viruses that originate in mammals of particular concern (Olival et al., 2017), such as the Human Immunodeficiency Virus (HIV) (Sharp and Hahn, 2011) and Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) (Ge et al., 2013). Mammals are the most closely related animals to human in phylogeny, and contact with human most frequently (Olival et al., 2017), especially the livestock and pet. For effective control of human viral diseases, great attentions should be paid to mammalian viruses.
Receptor-binding is the first step for viral infection of host cells (Baranowski et al., 2001; Dimitrov, 2004; Grove and Marsh, 2011; Li, 2015a,b). Multiple types of molecules including protein (Li, 2016; Wang, 2002; Yan et al., 2012), carbohydrate (Isa et al., 2006; Peng et al., 2017) and lipid (Mazzon and Mercer, 2014), could be used as viral receptors (Baranowski et al., 2001; Casasnovas, 2013). The important question of why viruses show strong preferential binding to particular host receptors remains unanswered (Casasnovas, 2013; Grove and Marsh, 2011; Li, 2016; Backovic and Rey, 2012). Specificity and affinity are the most important factors for receptor-virus interactions (Casasnovas, 2013). Widely distributed on host cell surfaces, carbohydrates and lipids are easy targets for viruses (Dimitrov, 2004; Li, 2015a,b). Compared to these molecules, proteins were reported to be more suitable receptors because of stronger affinity and higher specificity for viral attachment, which could increase the efficiency of viral entry and facilitate viruses to expand their host ranges and alter their tropisms (Baranowski et al., 2001; Casasnovas, 2013; Dimitrov, 2004; Li, 2015a,b; Wang, 2002). Previous studies have shown that proteins that were abundant in the host cell surface or had relatively low affinity for their natural ligands, such as proteins involved in cell adhesion and recognition (Dimitrov, 2004; Wang, 2002), were preferred by viruses as receptors. This suggests that the selection of proteins by viruses as receptors should not be a random process. A systematic analysis of the characteristics of the viral receptor could help understand the mechanisms behind receptor selection by viruses.
In this study, structural, functional, evolutionary and tissue-specific expression characteristics of mammalian virus receptors were investigated using computational methods by manually curating a high-quality database of 268 pairs of mammalian virus–receptor interaction, which included 128 unique viral species or sub-species and 119 virus receptors. This study helps to understand the mechanism behind virus–receptor interactions, as well as predict and identify viral receptors.
2 Results
2.1 Database of mammalian virus–host receptor interaction
To understand how viruses select receptors, we manually curated a high-quality database of 268 pairs of mammalian virus–host receptor interactions (see Methods in Supplementary Information), which included 128 unique viral species or sub-species from 21 viral families and 119 virus receptors from 13 mammal species (Supplementary Fig. S1 and Table S1). The viral receptor collected belonged to 13 mammal species (Supplementary Fig. S2A), among which the human accounted for the most (74/119). The viruses included in the database covered all groups of viruses in the Baltimore classification (Supplementary Fig. S1). Among them, the single-stranded RNA (ssRNA) virus accounted for over half of all viruses (76/128), while the double-stranded RNA (dsRNA) virus accounted for the least (3/128). On the level of family, the family of Picornaviridae of ssRNA virus, Retroviridae of Retro-transcribing viruses (RT) and Herpesviridae of double-stranded DNA (dsDNA) viruses were the most abundant ones in the database (Supplementary Fig. S1 and Table S1).
Analysis of the association between the virus and their receptors showed that 60% of the viruses (77/128) have one specific receptor only (Fig. 1A), while certain viruses [e.g. the Human alphaherpesvirus 1 (HSV-1) and Hepatitis C virus (HCV)] had over five receptors. Then, the receptor usage was analyzed on the level of viral family. Indeed, all fifteen viral families containing two or more viruses in the database used two or more sets of receptors, suggesting that different viruses in the same family prefer different receptors. For instance, in the family of Togaviridae, the Chikungunya virus (CHIKV), the Rubella virus (RBV) and the Sindbis virus (SBV) preferred prohibitin (PHB), myelin oligodendrocyte glycoprotein (MOG) and ribosomal protein SA (RPSA) as the receptor, respectively. Meanwhile, viruses in different families or groups preferred one specific receptor (Supplementary Fig. S1). For instance, HIV-2 and EBOV, which are from the family of Retroviridae (RT group) and Filoviridae (ssRNA group) respectively, used the CD209 molecule (CD209) (marked with an asterisk in Supplementary Fig. S1) as receptor. On average, each receptor was used by more than two viruses. Among the 119 virus receptors, 45 were used by more than one virus (Supplementary Figs S1 and S2B), 21 were used by viruses in different families, and 15 were used by viruses in different groups (Supplementary Fig. S1).
Fig. 1.
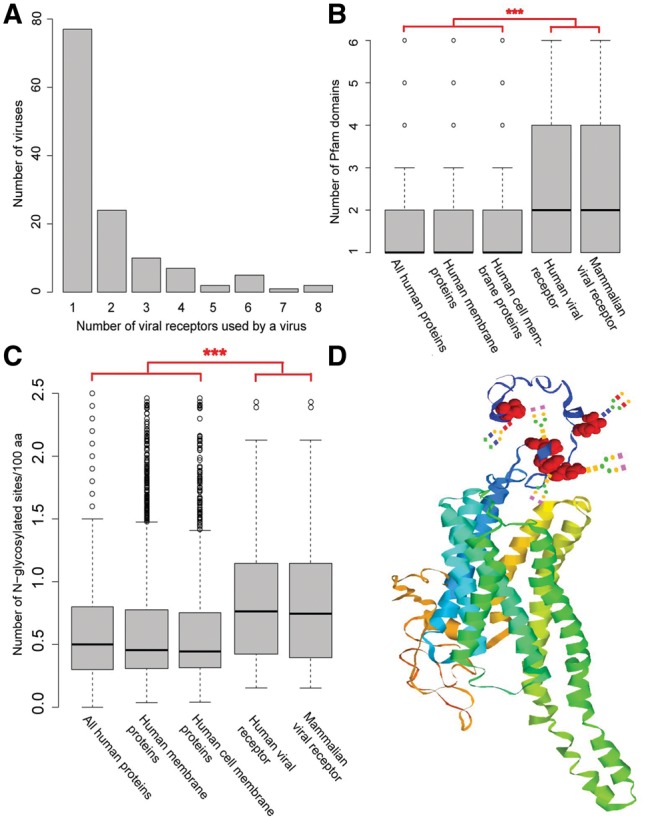
Analysis of structure features of mammalian virus receptors. (A) Distribution of the number of receptors used by a virus. (B) Comparing the number of Pfam domains within a protein for the set of human proteins, human membrane proteins, human cell membrane proteins, human viral receptors and mammalian viral receptors. For clarity, all the outliers greater than six were removed from the figure. “***”, P-value < 0.001. (C) Comparison of the N-glycosylation level between mammalian viral receptors, human viral receptors, human cell membrane proteins, human membrane proteins and all human proteins. For clarity, the outliers greater than 2.5 were removed. “***”, P-value < 0.001. (D) The modeled 3D-structure of HTR2A. Five N-glycosylation sites are highlighted in black. Artificial glycans were manually added onto the site
2.2 Mammalian virus receptors were structurally diverse
Firstly, the structural characteristics of mammalian virus receptor proteins were investigated (Supplementary Methods). As observed, all mammalian virus receptor proteins were transmembrane proteins with at least one transmembrane alpha helix (Supplementary Fig. S3A). Among these proteins, 24 had more than five helixes, such as 5-hydroxytryptamine receptor 2A (HTR2A) and NPC intracellular cholesterol transporter 1 (NPC1). Basically, virus receptor proteins are distributed on cell membrane. Specifically, 43 virus receptor proteins were distributed in the cytoplasm and 13 were distributed in the nucleus.
Then, the protein domain composition of the mammalian virus receptor protein was analyzed. According to the Pfam database, the mammalian virus receptor proteins consist of 336 domains, and each viral receptor protein contains more than two domains on average (Fig. 1B), which was significantly higher than that of human proteins or human membrane proteins (P-values < 0.001 in the Wilcoxon rank-sum test). Additionally, certain viral receptor proteins, including complement C3d receptor 2 (CR2) and low density lipoprotein receptor (LDLR), contain over 10 domains. The 336 protein domains in mammalian virus receptor proteins could be categorized into 77 families, indicating great structure diversity of the mammalian virus receptor protein. The most commonly observed Pfam families include Immunoglobulin V-set domain, Immunoglobulin C2-set domain, Integrin beta chain VWA domain, Integrin plexin domain and so on (Supplementary Fig. S3B).
2.3 Mammalian virus receptors had high level of N-glycosylation
Protein glycosylation is commonly observed in eukaryote cell. Here, the glycosylation level of the mammalian virus receptor was checked (Supplementary Methods). The results indicated that 93 out of 119 mammalian virus receptors were N-glycosylated with an average of 0.94 glycosylation sites per 100 amino acids (Fig. 1C), which increased to 0.97 glycosylation sites per 100 amino acids for the human viral receptor (Fig. 1C), among which 62 were N-glycosylated. Twelve human viral receptors had ten or more N-glycosylation sites, such as complement C3b/C4b receptor 1 (CR1) and lysosomal associated membrane protein 1 (LAMP1). Figure 1D illustrates the modeled 3D-structure of HTR2A, which is the receptor for JC polyomavirus (JCPyV). The black highlighted areas refer to N-glycosylation sites, which were reported to be important for viral infection (Maginnis et al., 2010). As a comparison, N-glycosylation level of human cell membrane proteins, human membrane proteins and all human proteins were also checked (Fig. 1C). The results indicated that the N-glycosylation level of these proteins were significantly lower than that of human and mammalian virus receptors (P-values < 0.001 in the Wilcoxon rank-sum test), demonstrating the significant role of N-glycosylation for the viral receptor. O-glycosylation is another common glycosylation. However, it is only observed in a small protion of mammalian virus receptors (14/119). Besides, no significant difference was observed between the quantity of O-glycosylation sites in mammalian virus receptor proteins and that in human proteins (Supplementary Fig. S3C).
2.4 Functional characterization of human virus receptors
We next characterized the function of viral receptors. As mentioned above, 74 of 119 mammalian virus receptors belonged to human and 36 of the remaining non-human mammalian virus receptors were homologs of human virus receptors (Supplementary Table S2). Therefore, functional characterization was applied to human virus receptors only in this study. Firstly, function enrichment analysis was conducted based on the database of Gene Ontology (GO) and KEGG and the results indicated that human viral receptors were mainly enriched in the GO terms or KEGG pathways related to virus entry into the host (see Supplementary Information for details). Also, it has been observed that some pathways associated to heart diseases, including ‘Dilated cardiomyopathy’, ‘Hypertrophic cardiomyopathy’, ‘Arrhythmogenic right ventricular cardiomyopathy’ and ‘Viral myocarditis’, were enriched.
Then, the role of human viral receptors in the human protein–protein interaction (PPI) network was investigated. Herein, a human PPI network (PPIN) was constructed based on previous reports (Menche et al., 2015). The PPIN consists of 13 460 human proteins, which are interconnected by 141 296 interactions. To evaluate the role of proteins in the human PPIN, the degree of each protein, which was defined as the number of connections the protein has to other proteins in the human PPIN, was calculated. The results showed that the degrees for human membrane proteins and cell membrane proteins were significantly smaller than those of other human proteins (Fig. 2A and Supplementary Fig. S4A, P-value < 0.001 in Wilcox rank-sum test) in the PPIN. Notably, the human virus receptor protein, which is a subset of the human cell membrane protein, had significantly larger degrees than other human proteins did in the PPIN (Fig. 2A and Supplementary Fig. S4, P-value < 0.001 in Wilcox rank-sum test). Additionally, the human virus receptors had a median of 13 interaction partners in the PPIN (Fig. 2A), which was nearly twice as much as that of all human proteins. Specifically, six viral receptors, including the epidermal growth factor receptor (EGFR), the heat shock protein family A member 8 (HSPA8), the PHB, the RPSA, the CD4 molecule (CD4) and the integrin subunit beta 1 (ITGB1), were observed to have more than 100 interaction partners (Fig. 2B). For robustness of these results, the node betweenness (another measure of node centrality in network) of each protein was also calculated based on the human PPIN. The results showed that the betweenness of human viral receptors was significantly higher than that of other sets of proteins (Supplementary Fig. S4B and C), further demonstrating the importance of human viral receptors in human PPIN.
Fig. 2.
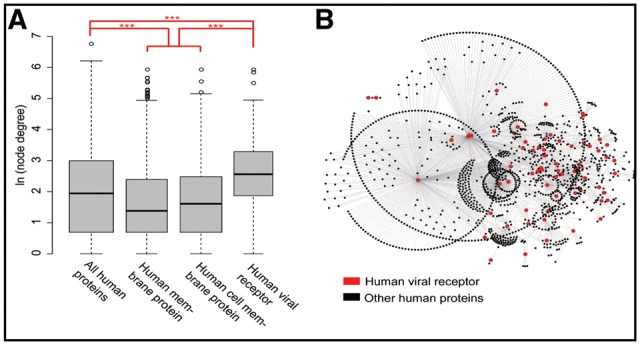
Analysis of protein-protein interactions of human virus receptors. (A) Comparison of the degree of proteins between human viral receptors, human cell membrane proteins, human membrane proteins and all human proteins in the human PPIN which were derived from Menche's work and included 13460 human proteins and 141296 interactions. For clarity, the node degree was transformed in natural logarithm. “***”, p-value < 0.001. (B) A partial human PPI network derived from Menche's work. It was only composed of the PPIs which involved at least one viral receptor protein (in triangle), and it included 1475 human proteins (67 virus receptors (in triangle) and 1408 other proteins (in circle)) and 2280 PPIs (edges in gray)
As human viral receptors (highlighted in triangle in Fig. 2B) interact with various human proteins (colored in circles in Fig. 2B) in the PPIN, the functional enrichment of these proteins was further investigated using GO enrichment analysis, which may also reflect the influence of human viral receptors on cellular activities. Interestingly, six of the top ten enriched terms in the domain of Biological Process were related to protein targeting or localization (Supplementary Table S3).
When looking at the interactions between human virus receptor proteins (Supplementary Fig. S4D), we found that only 50 virus receptor proteins interacted with itself or other virus receptors. Surprisingly, 38 of these virus receptor proteins interacted with itself. This ratio (38/50 = 76%) was much higher than that of human proteins (22%), membrane proteins (11%) and human cell membrane proteins (14%). Other than self-interactions, only 50 PPIs were observed between 25 human virus receptor proteins, with each human virus receptor protein interacting with two other virus receptor proteins on average. This suggested that human virus receptors tended to avoid interactions with other virus receptors.
2.5 Conservation of mammalian virus receptors in mammals
In this section, we conducted sequence and structure conservation analysis of mammalian virus receptors in 108 mammal species which were richly annotated in the NCBI Reference Sequences (RefSeq) database (Supplementary Methods and Table S4). The results showed that all mammalian virus receptors (119 receptors) involved in this study had homologs in 25–108 mammal species and a viral receptor had homologs in 103 mammal species on average (Supplementary Fig. S5A). For instance, Claudin-1, which is a major component of the tight junction complexes that regulate the permeability of epithelia, has homologs in all 108 mammal species. Then, we calculated the average pairwise sequence identities between mammalian virus receptors and their homologs in mammal species. They ranged from 0.42 to 0.98, with a median of 0.80 (Supplementary Fig. S5B). Further analysis of the conservation of amino acid sites on mammalian virus receptors showed that over 60% of sites had conservation scores (defined as the ratio of the dominant amino acid in the site) larger than 0.9 in viral receptors, although these amino acid sites had conservation scores ranging from 0.15 to 1 (Supplementary Fig. S5C). Finally, the conservation of protein structural domains of mammalian virus receptors in mammals was analyzed. As shown in Supplementary Table S5, each viral receptor has at least one structural domain that is completely conserved in the receptor and its homologs (Supplementary Table S5). Besides, 47 categories of structural domains were observed in certain viral receptors and their homologs. For instance, all homologs of Integrin beta-8 have both Integrin_beta and EGF_2 domains, while 68% of homologs of Integrin beta-8 have a PSI_integrin domain.
As a comparison, we also analyzed the sequence conservation of human proteins by randomly selecting 1000 human proteins from the NCBI RefSeq database. These human proteins were observed to have comparable homolog quantity and sequence conservation level with human viral receptors (74 receptors) (Supplementary Fig. S5D and E and Table S6).
2.6 High expression of human viral receptors in 32 major human tissues
As virus competes with other proteins to bind to receptors, proteins with relatively high expression are supposed to be preferred by viruses as receptors. Therefore, the average expression level of human viral receptors, human membrane genes, human cell membrane genes and all human genes in 32 major human tissues were calculated and compared with each other (Supplementary Methods and Supplementary Table S7). As shown in Figure 3, the expression level of human cell membrane genes (in hollow circle) was comparable to that of all human genes (in solid circle) in these tissues, while both were lower than that of human membrane genes (in hollow triangle). However, the expression levels of human viral receptor genes (highlighted in solid triangle), which were part of the human cell membrane gene set, were two times higher than other set of genes in most examined tissues (Fig. 3). The median expression value in all 32 tissues was 24 transcripts per million (TPM) for human virus receptors, and 8 TPM, 4 TPM and 4 TPM for the human membrane gene, the human cell membrane gene and all human genes, respectively (see the black arrow in Fig. 3).
Fig. 3.
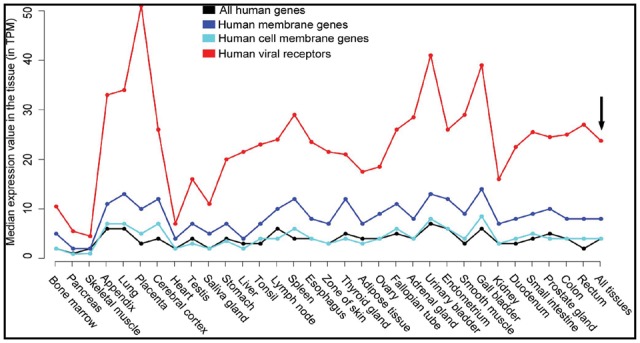
The average expression level of human viral receptors (in solid triangle), human cell membrane genes (in hollow circle), human membrane genes (in hollow triangle) and all human genes (in solid circle) in 32 major human tissues. The expression level was measured with transcripts per million (TPM). The black arrow refers to the average expression level of genes in all 32 tissues
3 Discussion
This study proposes a systematic investigation of the structural, functional, evolutionary and tissue-specific expression characteristics of mammalian virus receptors based on the largest dataset ever reported about the mammalian virus–host receptor interaction. It was found that the viral receptors were a subset of structurally and functionally diverse cell membrane proteins. They were enriched in GO terms and KEGG pathways related to junctions, adhesion and binding, which were typical features of viral receptors reported elsewhere (Backovic and Rey, 2012; Baranowski et al., 2001; Casasnovas, 2013; Dimitrov, 2004; Wang, 2002). This study identified several features that have not been reported for viral receptors. Firstly, the viral receptor had a higher level of N-glycosylation than other proteins. As we know, glycosylation of proteins is widely observed in eukaryote cells (Corfield, 2017) and plays a key role in multiple cellular activities such as folding and stability of glycoprotein, immune response, cell–cell adhesion. Glycans are abundant on host cell surfaces and are probably the primordial and fallback receptors for the virus (Li, 2015a,b). To take glycans as their receptors, various viruses employed a host galectin as its viral lectin (Krupovic and Koonin, 2017; Li, 2015a,b). For instance, the single jelly roll (SJR) fold, which was responsible for glycan recognition and binding in cellular proteins, was observed in viral capsid proteins of over 25% of all viruses (Krupovic and Koonin, 2017). Hence, the protein with high level of glycosylation could provide a basal attachment ability for the virus and might be the preferred receptor for viruses during the protein receptor searching process.
Secondly, this study demonstrated that the viral receptor protein had a tendency for self-interaction and possessed more interaction partners than other membrane proteins. Besides functioning as viral receptor, the receptor protein also functions in the host cell by interacting with other proteins of the host, including signal molecules and ligands. Therefore, viruses have to compete with these proteins for binding to the receptor (Wang, 2002) and the proteins with less interaction partners are supposed to be preferred by viruses. One possible reason for viruses selecting proteins with multiple interaction partners as receptors is that the receptor proteins are closely related to the ‘door’ of the cell, so that proteins have to interact with them for in-and-out of the cell. This is partially validated by the observation that six of the top ten enriched terms in the domain of GO Biological Process were related to protein targeting or localization for the interaction partners of human viral receptors (Supplementary Table S3). For entry into the cell, the virus also selects these proteins as receptors. Another possible reason is that viral entry into the cell needs cooperation of multiple proteins which were not identified as viral receptors. Additionally, previous studies suggested that viruses could structurally mimic native host ligands (Drayman et al., 2013), which help them bind to the host receptor. Therefore, membrane proteins with multiple interaction partners are more likely to be selected by viruses as receptors.
Thirdly, the viral receptor had a significant higher level of expression than other genes in all 32 human tissues. This is consistent with the finding that the viral receptor has multiple interaction partners. On one hand, the viral receptor needs multiple copies to interact with multiple proteins; on the other hand, membrane proteins with high expression are preferred by viruses as receptors as viruses have to compete with other proteins.
To date, various antiviral drugs have been developed for targeting viruses (Wikipedia, 2018). A full understanding of the virus–receptor interaction would facilitate therapeutic intervention of viruses. Several therapeutic approaches to target the virus–receptor interactions have been reported, including agents blocking virus–receptor interaction (Florea et al., 2003; Idemyor, 2005; Pugach et al., 2008), mimicking virus-associated protein and cellular receptors (Smith et al., 2002), and inhibiting membrane-fusion process (Lalezari et al., 2003; Leneva et al., 2009). Also, this study revealed that one specific receptor could be selected by multiple viruses (e.g. CD209 and CD55). In this case, similar strategies or antiviral drugs could be employed to combat these viruses.
Nevertheless, this study has several limitations. Firstly, the viral receptor was biased towards the human owing to the bias of studies towards human viruses. Fortunately, the viral receptor was conserved in mammal species to a large extent, resulting in reduced influence of the bias on the diversity of viral receptors. Secondly, the database of mammalian virus–host receptor interaction was still limited in its size due to the difficulties of identifying viral receptors (Li, 2015a,b; Pillay et al., 2016; Yan et al., 2012). Therefore, effective methods, either experimental or computational (Drayman et al., 2013), shall be developed for identifying viral receptors, while this study may facilitate this process. Thirdly, the interfaces of receptors and viruses need further investigations in most cases. The residues on the interfaces have a direct effect on the affinity and specificity of virus–receptor interactions. Previous studies demonstrated that mutations of these residues could alter virus–receptor interactions and hinder, if not abolish, virus infections (Bosch et al., 2013; Colon-Moran et al., 2017; Ng et al., 2015). For example, a single amino acid change in the NPC1 could greatly reduce the affinity of EBOV-NPC1 interaction in African straw-colored fruit bats (Ng et al., 2015). Hence, great efforts must be invested to identify the interaction interface between receptors and viruses.
In summary, the structural, functional, evolutionary and tissue-specific expression characteristics identified here contributes the understanding of viral receptor selection and facilitates the development of effective methods for viral receptor identification.
Supplementary Material
Acknowledgements
The authors would like to thank Pro. Xiangjun Du in Sun Yat-sen University for helpful suggestions.
Funding
This work was supported by the National Key Plan for Scientific Research and Development of China (2016YFD0500300 and 2016YFC1200200), the National Natural Science Foundation of China (31500126, 31671371, 81730064, 81571985 and U1603126), National Science and Technology Major Project (2017ZX10202201), the Chinese Academy of Medical Sciences (2016-I2M-1-005) and Fundamental Research Funds for the Central Universities of China.
Conflict of Interest: none declared.
References
- Backovic M., Rey F.A. (2012) Virus entry: old viruses, new receptors. Curr. Opin. Virol., 2, 4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowski E. et al. (2001) Evolution of cell recognition by viruses. Science, 292, 1102–1105. [DOI] [PubMed] [Google Scholar]
- Bosch B.J. et al. (2013) Spiking the MERS-coronavirus receptor. Cell Res., 23, 1069–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breban R. et al. (2013) Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet, 382, 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasnovas J.M. (2013) Virus–receptor interactions and receptor-mediated virus entry into host cells In: Mateu M.G. (ed.). Structure and Physics of Viruses: An Integrated Textbook, Springer, Dordrecht. [Google Scholar]
- Colon-Moran W. et al. (2017) Three cysteine residues of SLC52A1, a receptor for the porcine endogenous retrovirus-A (PERV-A), play a critical role in cell surface expression and infectivity. Virology, 507, 140–150. [DOI] [PubMed] [Google Scholar]
- Corfield A. (2017) Eukaryotic protein glycosylation: a primer for histochemists and cell biologists. Histochem. Cell Biol., 147, 119. 147, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D.S. (2004) Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol., 2, 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayman N. et al. (2013) Pathogens use structural mimicry of native host ligands as a mechanism for host receptor engagement. Cell Host Microbe, 14, 63–73. [DOI] [PubMed] [Google Scholar]
- Florea N.R. et al. (2003) Pleconaril, a novel antipicornaviral agent. Pharmacotherapy, 23, 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Y. et al. (2013) Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature, 503, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J.L. et al. (2016) Virological factors that increase the transmissibility of emerging human viruses. Proc. Natl. Acad. Sci. USA, 113, 4170–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J., Marsh M. (2011) The cell biology of receptor-mediated virus entry. J. Cell Biol., 195, 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idemyor V. (2005) Human immunodeficiency virus (HIV) entry inhibitors (CCR5 specific blockers) in development: are they the next novel therapies? HIV Clin. Trials, 6, 272–277. [DOI] [PubMed] [Google Scholar]
- Isa P. et al. (2006) Role of sialic acids in rotavirus infection. Glycoconjugate J., 23, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Koonin E.V. (2017) Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. USA, 114, E2401–E2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalezari J.P. et al. (2003) A phase II clinical study of the long-term safety and antiviral activity of enfuvirtide-based antiretroviral therapy. Aids, 17, 691–698. [DOI] [PubMed] [Google Scholar]
- Leneva I.A. et al. (2009) Characteristics of arbidol-resistant mutants of influenza virus: implications for the mechanism of anti-influenza action of arbidol. Antiviral Res., 81, 132–140. [DOI] [PubMed] [Google Scholar]
- Li F. (2015a) Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol., 89, 1954–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. (2015b) The hepatitis B virus receptor. Annu. Rev. Cell Dev. Biol., 31, 125–147. [DOI] [PubMed] [Google Scholar]
- Li F. (2016) Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol., 3, 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maganga G.D. et al. (2014) Ebola Virus Disease in the Democratic Republic of Congo. New. Engl. J. Med., 371, 2083–2091. [DOI] [PubMed] [Google Scholar]
- Maginnis M.S. et al. (2010) Role of N-linked glycosylation of the 5-HT2A receptor in JC virus infection. J. Virol., 84, 9677–9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzon M., Mercer J. (2014) Lipid interactions during virus entry and infection. Cell. Microbiol., 16, 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menche J. et al. (2015) Uncovering disease-disease relationships through the incomplete interactome. Science, 347, 1257601.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara T. et al. (2016) Linking virus genomes with host taxonomy. Viruses-Basel, 8, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J. et al. (2016) Zika virus associated with microcephaly. N. Engl. J. Med., 374, 951–958. [DOI] [PubMed] [Google Scholar]
- Ng M. et al. (2015) Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. eLife, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival K.J. et al. (2017) Host and viral traits predict zoonotic spillover from mammals. Nature, 546, 646.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D. et al. (2016) Uncovering Earth's virome. Nature, 536, 425. [DOI] [PubMed] [Google Scholar]
- Peng W.J. et al. (2017) Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe, 21, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay S. et al. (2016) An essential receptor for adeno-associated virus infection. Nature, 530, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach P. et al. (2008) Neutralizing antibody and anti-retroviral drug sensitivities of HIV-1 isolates resistant to small molecule CCR5 inhibitors. Virology, 377, 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Hahn B.H. (2011) Origins of HIV and the AIDS Pandemic. Csh. Perspect. Med., 1, a006841.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B.J. et al. (2002) Structural studies of the resistance of influenza virus neuramindase to inhibitors. J. Med. Chem., 45, 2207–2212. [DOI] [PubMed] [Google Scholar]
- Wang J.H. (2002) Protein recognition by cell surface receptors: physiological receptors versus virus interactions. Trends Biochem. Sci., 27, 122–126. [DOI] [PubMed] [Google Scholar]
- Wikipedia. (2018) List of antiviral drugs.
- Yan H. et al. (2012) Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife, 1, e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


