Abstract
The Sudan species of Ebola virus (SEBOV) causes severe, often fatal infection in ∼50% of infected humans. We sought to determine whether the human leukocyte antigen—B (HLA-B) locus has a role in the outcome of SEBOV disease by typing 77 cases from an outbreak in northern Uganda in 2000–2001. Sequence-based HLA-B typing was performed using leukocytes isolated from 77 patients. Statistical analysis and a predictive discriminant analysis (PDA) were applied to typing data. Epitope prediction software was also applied to SEBOV sequences. Statistically significant associations were found between certain sets of alleles and either fatal or nonfatal disease outcomes. Alleles B*67 and B*15 were associated with fatal outcomes, whereas B*07 and B*14 were associated with nonfatal outcomes. The PDA-derived functions that were produced were 81.8% accurate in classifying patients into their correct outcome group. Several epitopes predicted to bind strongly to HLA-B*07 molecules were identified in the viral polymerase, nucleoprotein, and VP35 protein. HLA-B alleles associated with either fatal or nonfatal outcomes of SEBOV disease were identified and can be used in a predictive model. Studies of HLA-B—restricted epitopes could contribute to characterization of protective host responses and to vaccine development.
In human and/or nonhuman primates, Ebola virus (EBOV) infections can cause a severe form of hemorrhagic fever that can lead to death if the infection is not controlled. In humans, infections caused by Zaire EBOV (ZEBOV) and Sudan EBOV (SEBOV) have accounted for most documented cases and result in mortality rates of up to 90% and, roughly, 50%, respectively [1]. Surviving these acute infections depends on the ability to efficiently clear virus and infected cells from the body before virus replication becomes unrestricted. Studies of EBOV infections in humans and animal models indicate that an important component of the immune response is cell-mediated immunity [2–4]. Humoral immunity appears to play only a minor role, if any, in surviving EBOV disease, because (1) antibody responses are characteristically delayed (i.e., they are absent early and usually are undetected in fatal cases) [5], (2) passive transfer of antiserum has not been effective in protecting primates [6, 7], and (3) antibody may actually enhance entry of EBOV into host cells [8]. NK cells may contribute to clearance, but there is no evidence suggesting that they play a significant role in controlling EBOV infections [9]. A study of infected macaques indicated a progressive depletion of these cells during the course of the disease [9].
In early September 2000, a large outbreak of SEBOV disease was identified in the northern region of Uganda and centered around the city of Gulu [10]. A team from the Centers for Disease Control and Prevention (CDC; Atlanta), participating in an international effort to control this outbreak of SEBOV disease, provided on-site diagnostic testing of blood samples and observed the progression of disease in many patients. All persons infected with SEBOV showed signs and symptoms of disease (no asymptomatic cases were identified), yet some patients were observed to have disease of milder severity and shorter duration. Analysis of the acute-phase peripheral blood mononuclear cells (PBMCs) from SEBOV-infected patients showed no significant difference in expression of cytokines, Fas, or Fas ligand when patients with fatal and nonfatal cases were compared with each other or were compared with uninfected, hospitalized patients [3]. However, persons with nonfatal outcomes differed from persons with fatal outcomes in that they had lower viral loads, as well as greater numbers of CD8+ T cells and activated (HLA-DR+) CD8+ T cells [3, 11]. These findings suggested that the production of cytotoxic T lymphocytes (CTLs) is important in limiting SEBOV replication and reducing disease severity. Because cellmediated immunity is likely to be a critical component in surviving SEBOV infections, it was postulated that the HLA class I composition of infected patients could influence the course of disease. To investigate this possibility, blood specimens collected from a variety of SEBOV-infected patients were used for sequence-based typing of the HLA-B locus (exons 2 and 3), the most divergent of the major histocompatibility complex (MHC) class I genes. Fortunately, the large number of cases sampled and the ∼50% fatality rate associated with the outbreak allowed statistical analysis to be performed on the data generated. The results of this investigation are the subject of the present article and implicate certain alleles in either nonfatal or fatal outcomes of disease due to SEBOV.
Materials and Methods
Human specimens used in genetic analysis. In accordance with CDC institutional review board policies, all samples were anonymized and identified only as fatal or nonfatal cases. Sequence information for patients infected with EBOV was generated from reverse-transcribed mRNA (cDNA) produced in another study [3]. In brief, total RNA was isolated fromPBMCs and cDNA produced by priming with oligo-d(T)12–18 and random hexamers that were extended with SuperScript II reverse transcriptase and RNA removed with RNase H (Invitrogen; catalog no. 11904-018).
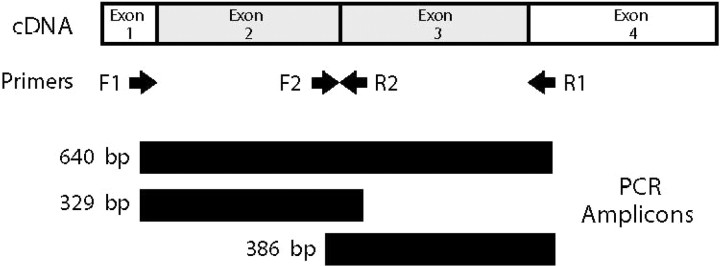
HLA-B locus sequence-based typing. Human HLA-B sequences were polymerase chain reaction (PCR) amplified from cDNA by use of Platinum PCR SuperMix (Invitrogen) or the Expand High Fidelity System (Roche Applied Science). To decrease background amplifications, 5% (v/v) dimethyl sulfoxide (D-2650; Sigma-Aldrich) was included in all reactions. Amplification of exons 2 through 3 was accomplished using the primers F1 (5′-CCGAACCGTCCTCCTGCTGCTC-3′; end of exon 1) and R1 (5′-GTGGTGGGTCACGTGTGTCTTTG-3′; beginning of exon 4). Separate amplification of exon 2 was accomplished using primers F1 and R2 (5′-CCCGCGGAGGAGGCGCCCGT-3′; beginning of exon 3), and amplification of exon 3 with primers F2 (5′-GCTACTACAACCAGAGCGAGGCC-3′; end of exon 2) and R1 (amplicon lengths of 640, 329, and 386 bp, respectively) (figure 1). Thermocycling conditions for amplification were as follows: 30–35 rounds of denaturing at 94°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 40 s. PCR amplification generally required only the use of F1 and R1 primers to produce enough template DNA for direct sequencing; however, at times, the use of F1 and R2 and/or F2 and R1 primer combinations was needed to increase amplicon synthesis. Amplicons were separated from reaction components by electrophoresis in 1.3% agarose and ethidium bromide—stained bands isolated using Qiagen Qiex II (with DNA eluted from binding matrix using nuclease-free water). Cycle sequencing of amplicons was performed using Prism BigDye chemistry (version 1; Applied Biosystems) in 20-µL reactions containing 100 ng of primer (4 separate reactions were performed using primers F1, R1, F2, and R2). Reactions were prepared in 0.2-mL thin-walled PCR tubes and were heated to 96°C for 1 min, followed by 25 cycles of denaturing at 96°C for 10 s, annealing at 50° for 5 s, and extension at 60°C for 4 min. Extension products were isolated by passing reactions through Centri-Sep columns (Princeton Separations) and thoroughly drying them in a speed vacuum. Products were analyzed on an automated sequencer (ABI 377 or ABI Prism 3100; Applied Biosystems) according to the manufacturer's protocols. Sequencher software (version 3.1.1; Gene Codes) was used to analyze chromatograph files in the generation of consensus HLA-B sequences and their exportation as ASCII files. HLA-B allele assignments were generated using the Assign software program (version 1.5.1) for sequence-based typing (developed by D. Sayer and J. Meier, Department of Clinical Immunology, Royal Perth Hospital, Perth, Australia). A description of Assign, version 2.0, has been published elsewhere [12], and the most recent version is available from Conexio Genomics. HLA-B assignment was limited to the allele type (i.e., B*07 and not B*0713), which provided sample sizes large enough to perform statistical analyses.
Figure 1.
Polymerase chain reaction (PCR) amplification of HLA-B sequences from cDNA. Shown is a diagram indicating the position of primers and amplicons generated from PCRs.
Epitope prediction. The encoded amino acid sequences for all genes of Sudan EBOV Gulu [13] were analyzed using Internet-based software/tools to identify epitopes potentially involved in cell-mediated immune responses. Analyses used CTLPred [14], a direct method for predicting CTL epitopes, as well as the following indirect methods, which were based on MHC antigen-binding properties: BIMAS [15], Rankpep [16], EpiJen [17], NetCTL [18], and ARB Matrix [19]. It is of note that some of these methods also factor in proteasomal C-terminal cleavage and TAP binding.
Statistical data analysis. Predictive discriminant analysis (PDA) techniques [20] were used to predict survival among patients with Ebola hemorrhagic fever (EHF) on the basis of the presence or absence of selected HLA-B alleles. The criterion variable was patient survival, with a patient either recovering from the EHF (n = 35) or dying as a result of the EHF (n = 42). Predictor variables were chosen froma set of 26HLAB alleles, alleles potentially associated with EHF survival. Fisher's exact test was used as the initial screening device to determine which of the 26 HLA-B alleles were associated with patient survival. An allele was considered for inclusion in the PDA model if its univariate association with patient survival resulted in a P value of ⩽.20. Once an initial set of predictor variables was selected from the univariate analyses, an all-possible subsets approach was used to select a set of HLA-B alleles that maximized classification accuracy.
Accuracy of the PDA model was assessed by examining the overall and individual group hit rates. The hit rates were estimated using the leave-one-out (L-O-O) method, a 1-sample cross-validation procedure that produces classification estimates that are “nearly” unbiased estimates of the true population hit rates. The overall hit-rate estimate was statistically compared with chance expectation by use of proportional chance criterion and maximum chance criterion methods. The group hit-rate estimates were statistically evaluated using the proportional chance criterion. If the overall and group hit rates were deemed to be statistically better than chance expectation, a descriptive measure (I) was calculated to denote percent improvement over chance expectation.
In this data set, a higher percentage of patients died (54.5%) than lived (45.5%). Because HLA-B allele information was not available for all patients, the data set does not include all patients who were infected with EHF in the outbreak in Uganda. Because the exact probability of surviving EHF is not exactly known, the prior probability of group membership was set at P = .5, which is a value close to the sample estimates in the current data set.
All PDA analyses were performed using the Discriminant procedure in SSPS software (version 12.0; SPSS), and all PDA analyses/interpretations were done according to established guidelines [20]. Because the HLA-B predictor variables were dichotomous variables, and because the discriminant analysis procedure assumes multivariate normality, the classification analyses were also done using Proc Logistic in SAS software (version 9.1; SAS Institute), as well as various kernel discriminant analysis options, by use of Proc Discrim in SAS software (version 9.1). Identical or near identical results were obtained using all statistical procedures.
Results
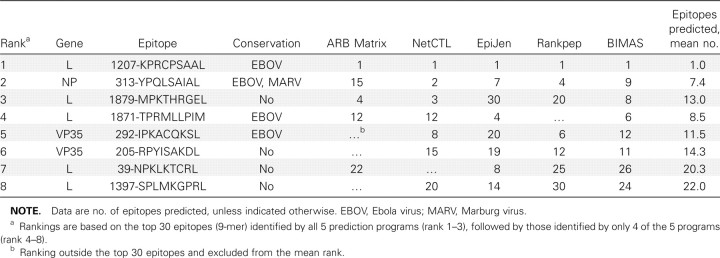
Results of HLA-B allele assignment for the 77 patients typed are presented in table 1 and show that all 11 patients with the B*07 allele survived SEBOV disease. Fortunately, this allele has been included in most MHCbinding/CTL prediction programs, which allowed the analysis of the predicted amino acid sequences among all the genes of Sudan EBOV Gulu for potential epitopes. Table 2 lists potential HLA-B*07 epitopes identified by prediction software, and, of the top 3 ranked epitopes, 2 are present in the L protein and 1 is present in the nucleoprotein. Lower-ranking epitopes are located in the L protein and VP35 protein. The highest-ranked epitope (1207- KPRCPSAAL) is highly conserved in all EBOVs and is only partially conserved in Marburg virus (MARV), a related filovirus. The nucleoprotein epitope 313-YPQLSAIAL, which is the second-ranked epitope, is highly conserved for both EBOV and MARV. The third-ranked epitope (1879-MPKTHRGEL) is not conserved. Of the other sequences, only the fourth- and fifthranked epitopes (1871-TPRMLLPIM and 292-IPKACQKSL, respectively) show any conservation (highly conserved for EBOV only).
Table 2.
HLA-B*07 epitopes predicted from Sudan Ebola virus Gulu-deduced protein sequences. Ranka
Univariate associations between patient survival and the presence of an HLA-B allele are presented in table 3. Alleles B*67, B*13, B*42, and B*15 were chosen for potential inclusion in the PDA model as predictors of fatal disease, because the P values determined by the Fisher's exact test were ⩽.20. As is evident in table 3, the patients with the aforementioned alleles were likely to die of EHF. Specifically, all 8 patients with the B*67 allele died of EHF, and >75% of the patients with alleles B67, B13, or B42 died of EHF. A second set of alleles (B*35, B*7, B*14, and B*40) were also chosen as potential predictors of patient survival. In contrast to patients with the first set of alleles, patients with the B*35, B*7, B*14, and B*40 alleles had an excellent chance of surviving EHF disease. All patients with the B*7, B*14, or B*40 alleles survived EHF infection, and 5 (83.3%) of 6 patients with the B35 allele survived EHF infection.
Table 3.
Univariate associations between patient survival and the presence of an HLA-B allele.
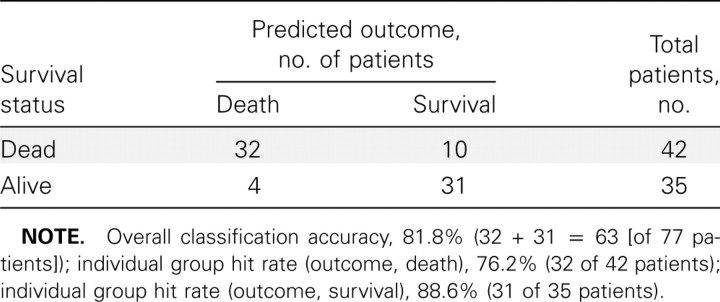
Initially, the PDA model contained 8 alleles (B*67, B*13, B*42, B*15, B*35, B*7, B*14, and B*40). On the basis of preliminary PDA analyses, alleles B35 and B14 were omitted from the set of predictors. Inclusion of either allele resulted in no gain or loss in classification accuracy. The L-O-O hit-rate estimates for the 6-allele system are presented in table 4. The overall rate of L-O-O classification accuracy was 81.8% (i.e., the outcomes for 63 of 77 patients were correctly classified), and the L-O-O group hit rates were 76.2% (32 of 42 patients) for the group of patients that died of EHF and 88.6% (31 of 35 patients) for the group of patients that survived EHF.
Table 4.
Leave-one-out (L-O-O) classification results for 77 patients.
By chance alone, it was expected that 38.5 (50%) of the patients would be classified into their correct outcome group. On the basis of results from the L-O-O method, the overall classification accuracy rate was 81.8% (63 of 77 patients), which is a marked improvement over chance expectation (I = 63.6%). By use of the proportional chance criterion (table 5), the overall classification accuracy of 81.8% was statistically better than chance, with z = 20.11 and P < .0001. On the basis of the maximum chance criterion (i.e., classification of all subjects into the largest group), 42 (54.5%) of 77 patients would be correctly classified by chance alone. On the basis of the maximum chance criterion, the overall hit rate (81.8%) was better than chance, with z = 16.63, P < .0001, and I = 60.0%. By use of the proportional chance criterion, the group hit rates were also statistically better than chance, with z = 7.76, P < .0001, and I = 52.4% for the group of patients who died of EHF and with z = 10.74, P < .0001, and I = 77.2% for the group of patients who survived EHF.
Table 5.
Proportional chance criterion results.
From the PDA model, 2 linear classification functions (LCFs) were created to predict disease outcome: these LCFs are as follows: LCFDead (fatal outcome) = −2.25 + (B*15 × 3.48) + (B*42 × 2.83) + (B*13 × 3.03) + (B*67 × 4.10) + (B*7 × .24) + (B*40 × −.86) and LCFAlive (nonfatal outcome) = −1.39 + (B*15 × .50) + (B*42 × .76) + (B*13 × .86) + (B*67 × .38) + (B*7 × 3.50) + (B*40 × 3.16). Insertion of a “0” value for an allele to denote genetic absence and a “1” value to denote genetic presence of an allele allows the calculation of LCFDead and LCFAlive values. The patient is assigned to the group with the higher LCF value.
Discussion
After the SEBOV outbreak in Uganda in 2000–2001, it was determined that persons with fatal outcomes of SEBOV disease had higher virus loads than did those who survived the disease [3, 11]. Survival is likely dependent on the rapid development of an effective CTL response that limits the level of virus replication before it can overwhelm the body, as is evidenced by increased numbers of activated (HLA-DR+) CD8+ lymphocytes in patients with nonfatal cases [3]. As our study suggests, the MHC class I composition appears to be an important factor in disease outcome, and the presence of certain HLA-B alleles can confer either a protective advantage or a disadvantage in patients infected with SEBOV. Our findings indicate that expression of the B*07 allele, which is highly associated with survival, confers a phenotype that efficiently drives T cell activation and CTL expansion early in the SEBOV infection, whereas alleles that are highly associated with fatalities (e.g., B*67 and B*15) contribute to an inefficient or unresponsive phenotype. One of the SEBOV-infected patients who survived had a B*07/B*15 profile (involving good and bad alleles), which suggests that, by itself, the B*07 allele may be sufficient to restrict SEBOV replication. With more data, one might be able to model the survival of patients with one good allele and one bad allele. However, in the present study, only 2 patients had such a profile, and, therefore, only univariate associations between allele type and patient survival were possible.
Correlations between HLA composition and disease susceptibility or severity have been identified for other viral infections, including those due to HIV [21–25], herpes viruses [26, 27], hepatitis B and C viruses [28–30], dengue virus [31], hantaviruses [32, 33], and severe acute respiratory syndrome coronavirus [34, 35]. It has also been reported that HLA-B-restricted epitopes exhibit a greater functional avidity (T cell receptor avidity for HLA-peptide complex) than do HLA-A-or HLA-C-restricted epitopes [36], which suggests a significant role for HLA-B alleles in acquired antiviral immunity. We believe that this is also the case for SEBOV infections.
Although we have shown that the HLA-B profile of a patient with SEBOV infection is associated with disease outcome, additional investigation is needed to further characterize this finding. The application of MHC class I tetramer technology [37] could be used to analyze PBMCs isolated from persons who have survived SEBOV disease or future cases of SEBOV disease, to determine whether any of the epitopes predicted in table 2 are immunodominant in HLA-B*07-restricted responses. There is also a need to examine the HLA-B profiles of patients infected with other filoviruses, which would be extremely helpful in gauging the influence of the HLA-B locus on filovirus clearance and could be useful in the development and refinement of vaccines.
The ability to predict the capacity of an individual to control acute and life-threatening infections, such as infections caused by filoviruses, could be useful in outbreaks. Current and future capabilities for rapid HLA typing could easily allow this type of testing to be performed in the field alongside other clinical assays. The classification functions (LCFdead and LCFAlive) generated from the PDA model could be used to gauge the risks to those providing medical assistance to patients with SEBOV infection (should they themselves become infected), and high-risk individuals might be assigned duties involving less contact with infected patients and infectious materials. Similarly, patients with SEBOV infection might be managed more appropriately if a fatal outcome is likely. Future typing of patients with SEBOV infection would be useful in assessing the reliability of such tools, and, if validated, these tools could help refine the classification functions and expand their scope.
In conclusion, the present study provides, to our knowledge, the first evidence of genetic predispositions to fatal and nonfatal disease outcomes in patients with EBOV infection, and it indicates that these traits are linked to alleles of the MHC HLAB locus. This association emphasizes the importance of the development of timely CTL responses to EBOV infections that lead to efficient virus clearance. It is hoped that this data set will be expanded to include predicted disease outcomes for ZEBOV and MARV, which would be an important resource for future investigations of human immune responses to filovirus infections.
Table 1.
HLA-B profiles of 77 patients with Sudan Ebola virus infection.
Acknowledgments
We thank the Ugandan Ministry of Health and the World Health Organization Response Team for their support in facilitating the collection and processing of patient samples. We acknowledge the clinical leadership and counsel of the late Matthew Lukwiya; the encouragement of the late Piero Corti; the isolation ward support of Daniel Bausch, Maria Di Santo, and Yoti Zebulon; the laboratory and logistical support of Franca Cian and Elio Croce; and the many efforts and sacrifices of the staffs of the St. Mary's Lacor and the Gulu Hospitals.
Supplement sponsorship. This article was published as part of a supplement entitled “Filoviruses: Recent Advances and Future Challenges,” sponsored by the Public Health Agency of Canada, the National Institutes of Health, the Canadian Institutes of Health Research, Cangene, CUH2A, Smith Carter, Hemisphere Engineering, Crucell, and the International Centre for Infectious Diseases.
References
- 1.Sanchez A, Geisbert TW, Feldmann H, et al. Filoviridae: Marburg and Ebola viruses. In: Knipe DM, Howley PM, Griffin DE, et al., editors. Fields virology. 5th ed. Vol 1. Philadelphia: Wolters Kluwer/Lippincott Williams and Wilkins; 2007. pp. 1409–48. [Google Scholar]
- 2.Baize S, Leroy EM, Georges-Courbot MC, et al. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5:423–6. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez A, Lukwiya M, Bausch D, et al. Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: cellular responses, virus load, and nitric oxide levels. J Virol. 2004;78:10370–7. doi: 10.1128/JVI.78.19.10370-10377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan N, Yang ZY, Nabel GJ. Ebola virus pathogenesis: implications for vaccines and therapies. J Virol. 2003;77:9733–7. doi: 10.1128/JVI.77.18.9733-9737.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ksiazek TG, Rollin PE, Williams AJ, et al. Clinical virology of Ebola hemorrhagic fever (EHF): virus, virus antigen, and IgG and IgM antibody findings among EHF patients in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis. 1999;179((Suppl 1)):S177–87. doi: 10.1086/514321. [DOI] [PubMed] [Google Scholar]
- 6.Jahrling PB, Geisbert J, Swearengen JR, et al. Passive immunization of Ebola virus-infected cynomolgus monkeys with immunoglobulin from hyperimmune horses. Arch Virol Suppl. 1996;11:135–40. doi: 10.1007/978-3-7091-7482-1_12. [DOI] [PubMed] [Google Scholar]
- 7.Jahrling PB, Geisbert TW, Beisbert JB, et al. Evaluation of immune globulin and recombinant interferon-α2b for treatment of experimental Ebola virus infections. J Infect Dis. 1999;179((Suppl 1)):S224–39. doi: 10.1086/514310. [DOI] [PubMed] [Google Scholar]
- 8.Takada A, Feldmann H, Ksiazek TG, Kawaoka Y. Antibody-dependent enhancement of Ebola virus infection. J Virol. 2003;77:7539–44. doi: 10.1128/JVI.77.13.7539-7544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed DS, Hensley LE, Geisbert JB, Jahrling PB, Geisbert TW. Depletion of peripheral blood T lymphocytes and NK cells during the course of Ebola hemorrhagic fever in cynomolgus macaques. Viral Immunol. 2004;17:390–400. doi: 10.1089/vim.2004.17.390. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Outbreak of Ebola viral hemorrhagic fever Uganda/ August 2000–January 2001. MMWR Morb Mortal Wkly Rep. 2001;50:73–7. [PubMed] [Google Scholar]
- 11.Towner JS, Rollin PE, Bausch DG, et al. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J Virol. 2004;78:4330–41. doi: 10.1128/JVI.78.8.4330-4341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayer DC, Goodridge DM, Christiansen FT. Assign 2.0: software for the analysis of Phred quality values for quality control of HLA sequencing- based typing. Tissue Antigens. 2004;64:556–65. doi: 10.1111/j.1399-0039.2004.00283.x. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez A, Rollin PE. Complete genome sequence of an Ebola virus (Sudan species) responsible for a 2000 outbreak of human disease in Uganda. Virus Res. 2005;113:16–25. doi: 10.1016/j.virusres.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 14.Bhasin M, Raghava GPS. Prediction of CTL epitopes using QM, SVM and ANN techniques. Vaccine. 2004;22:3195–201. doi: 10.1016/j.vaccine.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–75. [PubMed] [Google Scholar]
- 16.Reche PA, Glutting JP, Zhang H, Reinherz EL. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics. 2004;56:405–19. doi: 10.1007/s00251-004-0709-7. [DOI] [PubMed] [Google Scholar]
- 17.Doytchinova IA, Guan P, Flower DR. EpiJen: a server for multistep T cell epitope prediction. BMC Bioinformatics. 2006;7:131. doi: 10.1186/1471-2105-7-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen MV, Lundegaard C, Lamberth K, et al. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur J Immunol. 2005;35:2295–303. doi: 10.1002/eji.200425811. [DOI] [PubMed] [Google Scholar]
- 19.Bui HH, Sidney J, Peters B, et al. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–14. doi: 10.1007/s00251-005-0798-y. [DOI] [PubMed] [Google Scholar]
- 20.Huberty CJ. New York: John Wiley & Sons; 1994. Applied discriminant analysis. [Google Scholar]
- 21.Diouf K, Sarr AD, Eisen G, Popper S, Mboup S, Kanki P. Associations between MHC class I and susceptibility to HIV-2 disease progression. J Hum Virol. 2002;5:1–7. [PubMed] [Google Scholar]
- 22.Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 23.Klein MR, van der Burg SH, Hovenkamp E, et al. Characterization of HLA-B57-restricted human immunodeficiency virus type 1 Gagand RT-specific cytotoxic T lymphocyte responses. J Gen Virol. 1998;79:2191–201. doi: 10.1099/0022-1317-79-9-2191. [DOI] [PubMed] [Google Scholar]
- 24.Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97:2709–14. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNeil AJ, Yap PL, Gore SM, et al. Association of HLA types A1- B8-DR3 and B27 with rapid and slow progression of HIV disease. QJM. 1996;89:177–85. doi: 10.1093/qjmed/89.3.177. [DOI] [PubMed] [Google Scholar]
- 26.Khanna R, Burrows SR, Neisig A, Neefjes J, Moss DJ, Silins SL. Heirarchy of Epstein-Barr virus-specific cytotoxic T-cell responses in individuals carrying different subtypes of an HLA allele: implications for epitope-based antiviral vaccines. J Virol. 1997;71:7429–35. doi: 10.1128/jvi.71.10.7429-7435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lacey SF, Villacres MC, La Rosa C, et al. Relative dominance of HLAB*07 restricted CD8+ T-lymphocyte immune responses to human cytomegalovirus pp65 in persons sharing HLA-A*02 and HLA-B*07 alleles. Hum Immunol. 2003;64:440–52. doi: 10.1016/s0198-8859(03)00028-4. [DOI] [PubMed] [Google Scholar]
- 28.Hiroishi K, Kita H, Kojima M, et al. Cytotoxic T lymphocyte response and viral load in hepatitis C virus infection. Hepatology. 1997;25:705–12. doi: 10.1002/hep.510250336. [DOI] [PubMed] [Google Scholar]
- 29.Wang LY, Lin HH, Lee TD, et al. Human leukocyte antigen phenotypes and hepatitis C viral load. J Clin Virol. 2005;32:144–50. doi: 10.1016/j.jcv.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Wu YF, Wang LY, Lee TD, et al. HLA phenotypes and outcomes of hepatitis B virus infection in Taiwan. J Med Virol. 2004;72:17–25. doi: 10.1002/jmv.10557. [DOI] [PubMed] [Google Scholar]
- 31.Stephens HA, Klaythong R, Sirikong M, et al. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens. 2002;60:309–18. doi: 10.1034/j.1399-0039.2002.600405.x. [DOI] [PubMed] [Google Scholar]
- 32.Kilpatrick ED, Terajima M, Koster FT, Catalina MD, Cruz J, Ennis FA. Role of specific CD8+ T cells in the severity of a fulminant zoonotic viral hemorrhagic fever, hantavirus pulmonary syndrome. J Immunol. 2004;172:3297–304. doi: 10.4049/jimmunol.172.5.3297. [DOI] [PubMed] [Google Scholar]
- 33.Mustonen J, Partanen J, Kanerva M, et al. Association of HLA B27 with benign clinical course of nephropathia epidemica caused by Puumala hantavirus. Scand J Immunol. 1998;47:277–9. doi: 10.1046/j.1365-3083.1998.00302.x. [DOI] [PubMed] [Google Scholar]
- 34.Lin M, Tseng HK, Trejaut JA, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng MH, Lau KM, Li L, et al. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis. 2004;190:515–8. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bihl F, Frahm N, Di Giammarino L, et al. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol. 2006;176:4094–101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- 37.Constantin CM, Bonney EE, Altman JD, Strickland OL. Major histocompatibility complex (MHC) tetramer technology: an evaluation. Biol Res Nurs. 2002;4:115–27. doi: 10.1177/1099800402238332. [DOI] [PubMed] [Google Scholar]