Abstract
Objectives: To investigate the etiology of viral respiratory tract infections mainly in hospitalized children and adults over a 12-month consecutive period after implementation of a 14-virus multiplex nucleic acid amplification test.
Methods: From January 2014 to January 2015, a total of 2,237 respiratory samples were analyzed with the US Food and Drug Administration–cleared eSensor Respiratory Viral Panel (GenMark Diagnostics, Carlsbad, CA).
Results: Of the 2,237 specimens tested, 788 specimens were positive for at least one virus, giving a positivity rate of 35.2%, and because of viral codetection, a total of 862 viral targets were identified. The age groups with the highest positivity rates were the 0- to 1-year (73.5%) and 2- to 6-year (78.4%) age groups. The overall viral codetection rate was 9.1%. Human rhinovirus (HRV) was the most prevalent respiratory virus found in children and adults. The peak of HRV seen in September 2014 represented a combination of HRV and enterovirus D68, 2014 epidemic respiratory infections.
Conclusion: The ability to detect a wider range of respiratory viruses gave us a better understanding of the etiology of respiratory infections in our population, particularly for HRV and enhanced our ability to detect viral coinfection.
Keywords: Respiratory viruses, Multiplex nucleic acid amplification, Etiology, Epidemiology, Viral codetection
Viral infections of the respiratory tract are the most frequent illnesses seen in humans, and annually, millions of people are afflicted worldwide.1‐3 They affect all age groups but particularly the very young, the elderly, and those with chronic medical conditions.2‐6 In these specific risk groups, respiratory viral infections may predispose them to secondary bacterial infections, including pneumonia, otitis media, and sinusitis, which can lead to severe outcomes, such as hospitalization and death.7,8 Emergence of new respiratory viral pathogens, such as severe acute respiratory syndrome coronavirus, avian H5N1 influenza, pandemic (h1N1) 2009 influenza, and the discovery of novel viruses known to cause respiratory tract infections in humans, such as human metapneumovirus (hMPV), have increased our understanding of life-threatening illness associated with respiratory infections.2,9
The current pharmacologic interventions for respiratory viral infections are largely limited, and no vaccines or therapeutics of proven value are currently available for respiratory viruses other than for influenza. To develop effective vaccines and antiviral therapies, a better understanding of disease etiology and epidemiology is necessary. This has driven some clinical diagnostic laboratories to move from traditional viral cultures that were time-consuming and had a limited repertoire to new molecular diagnostic technologies that are relatively rapid and permit the detection of a wider spectrum of viral respiratory pathogens.
The US Food and Drug Administration (FDA) has cleared several nucleic acid amplification respiratory viral panels able to detect multiple respiratory viruses in a single test. In September 2012, GenMark Diagnostics (Carlsbad, CA) received clearance from the FDA for its eSensor Respiratory Viral Panel that is able to detect up to 14 different viral targets in a single test on nasopharyngeal specimens.
The objective of this investigational study was to study the etiology and epidemiology of respiratory virus infections and, in particular, to determine the virus-specific positivity rates, age and sex distribution, viral codetection, and seasonality, mainly in hospitalized children and adults in a Midwest university medical center after implementation of the eSensor Respiratory Viral Panel over a 12-month consecutive period.
Materials and Methods
Samples
From January 7, 2014, through January 1, 2015, prospectively, for each specimen tested (nasopharyngeal swabs, nasal washes, and bronchoalveolar lavages) on the eSensor Respiratory Viral Panel, the age, the sex of the patient, the specimen type, the date of collection, and the test result were recorded. Most of the respiratory specimens tested were from hospitalized patients from the University of Minnesota Medical Center and the University of Minnesota Children’s Hospital. Ordering guidelines stating that the assay was to be ordered only on hospitalized patients in respiratory distress and for the evaluation of severely immunocompromised or critically ill patients with respiratory symptoms were put in place to avoid misuse of the assay. The assay was discouraged in patients seen in the emergency department (ED) or in clinics, and respiratory viral cultures and/or rapid antigen tests for respiratory viruses were recommended as alternative testing. Despite this, some specimens were submitted from the ED and outpatient clinics per physician orders.
The eSensor Respiratory Viral Panel is FDA approved for nasopharyngeal swabs only, and thus the laboratory performed validations of the bronchoalveolar lavages and nasal wash specimens to assess the performance characteristics of the assay in detecting the intended viral targets in these specimen types. The validation showed that the performance characteristics of the assay for these specimen types were acceptable and in agreement with published data.10 For any other respiratory specimen types than nasopharyngeal swabs, bronchoalveolar lavages, and nasal wash, a disclaimer, stating that this particular specimen type had not been validated and needed to be interpreted accordingly, was attached with the result.
To determine the period of time necessary to differentiate between a new infection and prolonged virus presence/persistence of a previous infection, a review of the literature was performed and indicated that, in most cases, viral nucleic acid was not detected beyond 30 days after initial viral infection.11,12 It was decided for this study that patients with repeat specimens sent for testing in a 4-week period were excluded for analysis if they were repeatedly negative or repeatedly positive with the same virus.
Detection of Respiratory Viruses
The eSensor Respiratory Viral Panel is a qualitative nucleic acid multiplex in vitro diagnostic test that uses the eSensor XT-8 system for the simultaneous detection and identification of the following virus types and subtypes: influenza A (all subtypes), influenza A H1 seasonal subtype, influenza A H3 seasonal subtype, influenza A 2009 H1N1 subtype, influenza B, respiratory syncytial virus subtype A (RSV A), respiratory syncytial virus subtype B (RSV B), parainfluenza virus 1 (PIV-1), parainfluenza virus 2 (PIV-2), parainfluenza virus 3 (PIV-3), hMPV, human rhinovirus (HRV), adenovirus species B/E (ADV B/E), and adenovirus species C (ADV C).
The NucliSENS EasyMAG automated extractor (bioMerieux, Durham, NC) was used for the isolation of viral nucleic acid to be compliant with the FDA-approved extraction method for the eSensor Respiratory Viral Panel.
Statistical Analysis
Categorical variables were compared using χ2. All tests were two-tailed, and P values .05 or less were considered significant.
Ethical Approval
An institutional review board application (study 1510E78822), submitted to the University of Minnesota Human Research Protection Program, was reviewed and approved.
Results
From January 2014 through January 2015, a total of 2,237 respiratory samples consisting of 1,634 (73.0%) nasopharyngeal swabs, 579 (25.9%) bronchoalveolar lavages, and 24 (1.1%) nasal washes were analyzed with the eSensor Respiratory Viral Panel. Of the 2,237 specimens tested, 752 (33.6%) were from pediatric patients (aged 1 day to 19 years), and 1,485 (66.4%) were from adult patients (aged 20-94 years).
There were 788 positive specimens, giving a positivity rate of 35.2%, and because some specimens were positive for more than one virus, a total of 862 viruses were detected, and the distribution among the total viruses was as follows: 449 (52.0%) HRV; 102 (11.8%) RSV A and RSV B; 89 (10.3%) PIV-1, PIV-2, and PIV-3; 85 (9.9%) influenza A (all subtypes); 73 (8.5%) hMPV; 50 (5.8%) ADV B/E and ADV C; and 14 (1.6%) influenza B Table 1. The age groups with the highest positivity rates (total positive viruses per age group/total tested specimens per age group) were 0 to 1 years (73.5%) and 2 to 6 years (78.4%), followed by 7 to 12 years (56.6%) Table 2.
Table 1.
Positivity Rate per Viral Target and Virus Type (n = 862)
| Characteristic | Value |
|---|---|
| Viral target positivity rate, No. (%) | |
| Influenza A H3 | 71 (8.2) |
| Influenza A H1N1 2009 | 14 (1.6) |
| Influenza B | 14 (1.6) |
| RSV A | 64 (7.4) |
| RSV B | 38 (4.4) |
| PIV-1 | 7 (0.8) |
| PIV-2 | 20 (2.3) |
| PIV-3 | 62 (7.2) |
| hMPV | 73 (8.4) |
| ADV B/E | 8 (0.9) |
| ADV C | 42 (4.8) |
| HRV | 449 (52.0) |
| Virus-specific positivity rate, No. (%) | |
| Influenza Aa | 85 (9.9) |
| Influenza B | 14 (1.6) |
| RSV A and RSV B | 102 (11.8) |
| PIV-1, PIV-2, and PIV-3 | 89 (10.3) |
| hMPV | 73 (8.5) |
| ADV B/E-C | 50 (5.8) |
| HRV | 449 (52.0) |
ADV B/E, adenovirus species B/E; ADV C, adenovirus species C; hMPV, human metapneumovirus; HRV, human rhinovirus; PIV-1, parainfluenza virus 1; PIV-2, parainfluenza virus 2; PIV-3, parainfluenza virus 3; RSV A, respiratory syncytial virus subtype A; RSV B, respiratory syncytial virus subtype B.
aInfluenza A seasonal subtype H1 not detected during study period.
Table 2.
Viral Targets Detected According to Age Groupa
| Age, y |
||||||||
|---|---|---|---|---|---|---|---|---|
| Viral Target | 0-1 | 2-6 | 7-12 | 13-19 | 20-39 | 40-59 | 60-79 | >80 |
| Influenza A H3 | 5 | 6 | 11 | 5 | 16 | 10 | 12 | 6 |
| Influenza A H1N1 2009 | 0 | 1 | 0 | 2 | 5 | 3 | 3 | 0 |
| Influenza B | 1 | 1 | 0 | 1 | 3 | 3 | 5 | 0 |
| RSV A | 36 | 14 | 0 | 0 | 1 | 6 | 6 | 1 |
| RSV B | 11 | 8 | 2 | 1 | 2 | 8 | 5 | 1 |
| PIV-1 | 2 | 0 | 2 | 0 | 2 | 0 | 1 | 0 |
| PIV-2 | 4 | 1 | 2 | 0 | 5 | 5 | 3 | 0 |
| PIV-3 | 26 | 7 | 1 | 0 | 6 | 13 | 9 | 0 |
| hMPV | 20 | 13 | 6 | 3 | 5 | 9 | 14 | 3 |
| ADV B/E | 2 | 3 | 0 | 0 | 1 | 2 | 0 | 0 |
| ADV C | 23 | 8 | 1 | 0 | 3 | 5 | 2 | 0 |
| HRV | 123 | 91 | 35 | 17 | 56 | 59 | 62 | 6 |
| Total detected viruses | 253 | 153 | 60 | 29 | 105 | 123 | 122 | 17 |
| Total specimens tested | 344 | 195 | 106 | 107 | 322 | 549 | 538 | 76 |
| Viral targets detected per total specimens tested, % | 73.5 | 78.4 | 56.6 | 27.1 | 32.6 | 22.4 | 22.7 | 22.3 |
ADV B/E, adenovirus species B/E; ADV C, adenovirus species C; hMPV, human metapneumovirus; HRV, human rhinovirus; PIV-1, parainfluenza virus 1; PIV-2, parainfluenza virus 2; PIV-3, parainfluenza virus 3; RSV A, respiratory syncytial virus subtype A; RSV B, respiratory syncytial virus subtype B.
aValues are presented as numbers unless otherwise indicated.
The number of pediatric specimens tested (752 specimens, 33.6%) was smaller than those from adults (1,485 specimens, 66.4%), but they had the highest number of detected viruses, 495 detected viruses vs 367 detected, respectively.
HRV was the most common virus detected in all age groups except for those 80 years or older. The highest HRV prevalence was seen in the 0- to 1-year age group (123 positive for HRV). RSV A was the most common virus isolated after HRV in the 0- to 1-year age group (36 positive for RSV A) and 2- to 6-years (14 positive for RSV A) age group. Influenza A was the main cause of respiratory infection after HRV, for the remaining age groups, and was equivalent to HRV in the age group 80 years or older, although the numbers were small (Table 2). The influenza A subtypes detected were H3 and H1N1 2009. The influenza A seasonal subtype H1 was not detected during the study period.
ADV C was more prevalent than ADV B/E (42 detected vs 8) (Table 1). The highest number of positive ADV C (23) was seen in the 0- to 1-year age group (Table 2).
PIV-3 was the most prevalent of the PIV subtypes with 62 positive specimens vs 20 positives for PIV-2 and 7 positives for PIV-1 (Table 1).
Of the 2,237 specimens tested, 1,042 (46.6%) were from female patients and 1,195 (53.4%) were from male patients. The total positivity rate for the female patients was 37.1% (total female positive viral targets [387]/total female specimens tested) and 39.7% for the male patients (total male positive viral targets [475]/total male specimens tested) (P = .39). The numbers of positive viral targets in males were higher than in females for all age groups, except for the 40- to 59-year and the 80-year or older age groups, where females had a higher number of positive viral targets than the males. The age group with the greatest difference between male and female positivity rates was the 13- to 19-year age group with a positivity rate of 34.0% and 20.3%, respectively (P = .11).
Seventy-two specimens had more than one virus detected, thus giving codetection rates of 9.1% for the total 788 positive specimens or 3.2% for the total 2,237 specimens tested Table 3. The infant (0-1 year) and young children (2-6 years) age groups had the highest codetection rates, 9.9% (34/344) and 7.2% (14/195), respectively (Table 3). The most frequent codetection combinations were HRV with ADV C and HRV with PIV-3.
Table 3.
Codetection Rate of Viral Targets per Age Group
| Age, y |
||||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | 0-1 | 2-6 | 7-12 | 13-19 | 20-39 | 40-59 | 60-79 | >80 |
| Total specimens tested | 344 | 195 | 106 | 107 | 322 | 549 | 538 | 76 |
| Total specimens with codetection of virusesb | 34 | 14 | 3 | 0 | 6 | 8 | 7 | 0 |
| Codetection rate, % | 9.9 | 7.2 | 2.8 | 0 | 1.8 | 1.4 | 1.3 | 0 |
aValues are presented as numbers unless otherwise indicated.
bTwo or more positive viral targets.
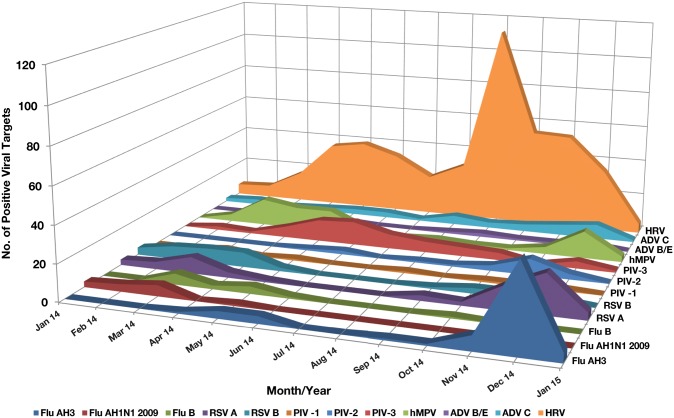
RSV and influenza were more prevalent during the colder months of the year. PIV-2 showed a peak in November, and PIV-3 was present most of the year with peaks in the spring months. Adenovirus C was seen all year around with peaks in the late fall into the winter. hMPV was seen in all age groups, all year around, with the highest number of positives in December, March, April, and May Figure 1. HRV was seen throughout the year, with major peaks in the spring and in the fall months of September, October, and November and was the most prevalent respiratory virus detected in children and adults with 449 positive targets. The hierarchy of virus isolation in our patients likely represented what was circulating in the community. Of the 449 patients with positive HRV, 290 were hospitalized, of whom 100 (34.4%) were children aged 1 year and younger.
Figure 1.
Virus detected according to month and year from January 2014 to January 2015. Observe the peaks of influenza A H3 and respiratory syncytial virus (RSV) A during November and December. Human metapneumovirus (hMPV) had two separate peaks of activity in March and December. Parainfluenza virus (PIV) 3 had a peak of activity in May and June. Human rhinovirus (HRV) had the highest activity in September, October, and November. ADV, adenovirus.
Discussion
The introduction of nucleic acid testing in our clinical virology laboratories has enhanced our ability to detect viruses that we were not able to recover with traditional viral culture methods and thus permitted us to identify the “hidden” burden of viral respiratory infections in our patient population.
HRV was the most prevalent respiratory virus in our population, with 449 specimens positive for HRV, which represented 20% of all specimen tested. Of these, 290 were from hospitalized patients that included 100 children aged 1 year and younger. HRV, previously considered a respiratory virus with minimal pathogenicity, has emerged as a cause of severe respiratory illness in healthy individuals, those with underlying asthma, and in vulnerable populations (very young, elderly, and immunocompromised).13,14 HRV is responsible for most common colds and more likely to be symptomatic in children than in adults, as shown by the Better Identification of Germs Longitudinal Viral Epidemiology study.13 The HRV family includes three species: HRV-A, HRV-B, and HRV-C, and within each species, there are multiple serotypes, types, and strains. The most recent described species, HRV-C, has been associated with serious lower respiratory disease in infants and young children, elderly persons, and immunocompromised patients.14
In our study, the large peak of positive HRV detection seen in September 2014 raised questions, particularly because it coincided with the enterovirus D68 outbreak.15 Upon further investigation, it was demonstrated that the GenMark Diagnostics HRV primers and probe used in the eSensor Respiratory Viral Panel were cross-reacting with the enterovirus D68.16 Thus, the high positive HRV peak seen in September represented a combination of HRV and epidemic enterovirus D68.16
We also demonstrated that the pediatric population had a higher burden of viral respiratory infections compared with adults; 33.6% (752) of the tested specimens were from pediatric patients, but they had the highest number of detected viruses, with 495 detected viruses compared with 367 detected viruses in the 1,485 tested adult specimens (P < .05).
RSV was the most common virus detected in the young children (0- to 1-year and 2- to 6-year age groups) after HRV. RSV can cause severe infections in infants and young children and is the most important etiologic agent of acute lower respiratory tract infection in infants and young children in the United States and worldwide.17
In our study, hMPV was more prevalent in infants and young children and in the older population (60- to 79-year age group), which is consistent with previous studies.18 hMPV has a high prevalence in bronchiolitis and pneumonia in hospitalized infants and young children. Advanced age and underlying cardiopulmonary diseases are two risk factors for severe hMPV infection in the older population.19
Of all the parainfluenza virus subtypes, PIV-3 is the subtype that is the most commonly associated with bronchiolitis, bronchitis, and pneumonia and thus often requires hospital admission for clinical management. In our study, most of the respiratory specimens were from hospitalized patients, and PIV-3 was the most prevalent of the PIV subtypes. PIV-1 and PIV-2 cause cold-like symptoms in infected people, and PIV-4 may cause mild to severe respiratory tract illnesses in infected patients. We could not assess the prevalence of PIV-4 in our study as this target was not included in the eSensor Respiratory Viral Panel.
ADV, an important pathogen in infants, was also detected frequently and is responsible for approximately 7% to 8% of reported childhood viral respiratory infections globally and causes a broad spectrum of clinical disease, including respiratory tract infection.20 In an immunocompromised host, ADV can cause severe localized disease or disseminated disease with multiorgan failure.21
Viral codetection, which is defined as the detection of more than one viral pathogen in the same specimen, is a phenomenon that was underestimated when using viral cultures. Since the introduction of new technologies that have moved diagnosis abilities forward, the capability to detect more than one virus per specimen has greatly increased our understanding of the true incidence of viral coinfection. Viral codetection was seen in 72 (9.1%) of the 788 positive specimens in our study. HRV was the virus most commonly associated with other viruses. The clinical significance of such viral codetection and thus possible coinfection is unclear, and review of the literature is conflicting. Some studies link viral coinfection with more severe clinical outcomes, and some show no overall difference between single respiratory virus infection and viral coinfections.3,22‐27 Also, as shown in our study (Table 3), viral codetection appeared to be more commonly seen in pediatric patients than in adults.24 The reasons for this difference are not known. Even though the impact of viral coinfection on severity of disease is unclear, the need for rapid and accurate detection of a wide spectrum of viral respiratory pathogens is desirable to understand their correlation with clinical illness and for epidemiologic and hospital infection control reasons.
The field of clinical virology has changed dramatically since introduction of new diagnostic techniques in the clinical laboratory. Newer assays are still being developed to expand the number of detected targets to include viral and bacterial respiratory pathogens such as PIV-4, coronavirus, and Bordetella species with decreased turnaround time. This will further increase our understanding of the circulating respiratory pathogens during different seasons of the year and in different health care settings (hospital based vs community based) and potentially help in the development of new vaccines.
Continued advances in diagnostic technology have enabled diagnostic microbiology laboratories to expand their capability of detecting respiratory viruses and thus help understand the epidemiology of respiratory viruses. It is evident that many unanswered questions still need to be addressed: for example, what is the scientific reason behind the seasonality of the different respiratory viruses, and which antiviral treatments should be used, if any, to treat these viral respiratory tract infections? Despite progress made in the 20th and 21st centuries, there is still no common oral antiviral therapy available for the treatment of these respiratory viruses except for influenza. Further studies need to be conducted to answer these questions.
The study was limited by the fact that no clinical data were associated with test results; thus, we could not assess symptoms associated with viral infection or coinfection disease progression, treatment plans, and response to treatment.
Note: Nineteen specimens composed of tracheal aspirates, sputum, and throat swabs were excluded from this analysis. The performance characteristics of these specimens types had not been validated on the eSensor Respiratory Viral Panel, and thus the accuracy of the results could not be verified.
Acknowledgments
Acknowledgment: We are grateful for the excellent technical expertise of our molecular staff in the Infectious Diseases Diagnostic Laboratory.
References
- 1. Monto AS. Epidemiology of viral respiratory infections. Am J Med. 2002;112:4S-12S. [DOI] [PubMed] [Google Scholar]
- 2. Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23:74-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stefanska I, Romanowska M, Donevski S, et al. Co-infections with influenza and other respiratory viruses. Adv Exp Med Biol. 2013;756:291-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Louie JK, Schnurr DP, Pan CY, et al. A summer outbreak of human metapneumovirus infection in a long-term-care facility. J Infect Dis. 2007;196:705-708. [DOI] [PubMed] [Google Scholar]
- 5. Whimbey E, Englund JA, Couch RB. Community respiratory virus infections in immunocompromised patients with cancer. Am J Med. 1997;102:10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hicks KL, Chemaly RF, Kontoyiannis DP. Common community respiratory viruses in patients with cancer: more than just “common colds.” Cancer. 2003;97:2576-2587. [DOI] [PubMed] [Google Scholar]
- 7. Peltola VT, McCullers JA. Respiratory viruses predisposing to bacterial infections: role of neuraminidase. Pediatr Infect Dis J. 2004;23:S87-S97. [DOI] [PubMed] [Google Scholar]
- 8. Chonmaitree T, Revai K, Grady JJ, et al. Viral upper respiratory tract infection and otitis media complication in young children. Clin Infect Dis. 2008;46:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Osterhaus ADME. New respiratory viruses of humans. Pediatr Infect Dis J. 2008;27:S71-S74. [DOI] [PubMed] [Google Scholar]
- 10. Ruggiero P, McMillen T, Tang YW, et al. Evaluation of the BioFire FilmArray respiratory panel and the GenMark eSensor respiratory viral panel on lower respiratory tract specimens. J Clin Microbiol. 2014;52:288-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loeffelholz MJ, Trujillo R, Pyles RB, et al. Duration of rhinovirus shedding in the upper respiratory tract in the first year of life. Pediatrics. 2014;134:1144-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chu CM, Leung WS, Cheng VCC, et al. Duration of RT-PCR positivity in severe acute respiratory syndrome. Eur Respir J. 2005;25:12-14. [DOI] [PubMed] [Google Scholar]
- 13. Byington CL, Ampofo K, Stockmann C, et al. Community surveillance of respiratory viruses among families in the Utah Better Identification of Germs-Longitudinal Viral Epidemiology (BIG-LoVE) Study. Clin Infect Dis. 2015;61:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jacobs SE, Lamson DM, St George K, et al. Human rhinoviruses. Clin Microbiol Rev. 2013;26:135-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Midgley CM, Jackson MA, Selvarangan R, et al. Severe respiratory illness associated with enterovirus D68—Missouri and Illinois, 2014. MMWR Morb Mortal Wkly Rep. 2014;63:798-799. [PMC free article] [PubMed] [Google Scholar]
- 16. McAllister SC, Schleiss MR, Arbefeville S, et al. Epidemic 2014 enterovirus D68 cross-reacts with human rhinovirus on a respiratory molecular diagnostic platform. PLoS One. 2015;10:e0118529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haynes AK, Prill MM, Iwane MK, et al. ; Centers for Disease Control and Prevention (CDC). Respiratory syncytial virus—United States, July 2012–June 2014. MMWR Morb Mortal Wkly Rep. 2014;63:1133-1136. [PMC free article] [PubMed] [Google Scholar]
- 18. Panda S, Mohakud NK, Pena L, Kumar S. Human metapneumovirus: review of an important respiratory pathogen. Int Infect Dis. 2014;25:45-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Centers for Disease Control and Prevention (CDC). Outbreaks of human metapneumovirus in two skilled nursing facilities—West Virginia and Idaho, 2011-2012. MMWR Morb Mortal Wkly Rep. 2013;62:909-913. [PMC free article] [PubMed] [Google Scholar]
- 20. Li H, Wei Q, Tan A, et al. Epidemiological analysis of respiratory viral etiology for influenza-like illness during 2010 in Zhuhai, China. Virol J. 2013;10:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Echavarría M. Adenoviruses in immunocompromised hosts. Clin Microbiol Rev. 2008;21:704-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mengelle C, Mansuy JM, Pierre A, et al. The use of a multiplex real-time PCR assay for diagnosing acute respiratory viral infections in children attending an emergency unit. J Clin Virol. 2014;61:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Debiaggi M, Canducci F, Ceresola ER, et al. The role of infections and coinfections with newly identified and emerging respiratory viruses in children. Virol J. 2012;9:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zimmerman RK, Rinaldo CR, Nowalk MP, et al. Viral infections in outpatients with medically attended acute respiratory illness during the 2012-2013 influenza season. BMC Infect Dis. 2015;15:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Asner SA, Science ME, Tran D, et al. Clinical disease severity of respiratory viral co-infection versus single viral infection: a systematic review and meta-analysis. PLoS One. 2014;9:e99392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nitsch-Osuch A, Kuchar E, Topczewska-Cabanek A, et al. Incidence and clinical course of respiratory viral coinfections in children aged 0-59 months. Adv Exp Med Biol. 2016;905:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crotty MP, Meyers S, Hampton N, et al. Epidemiology, co-infections, and outcomes of viral pneumonia in adults: an observational cohort study. Medicine (Baltimore). 2015;94:e2332. [DOI] [PMC free article] [PubMed] [Google Scholar]