Abstract
The lack of practical methods for a laboratory diagnosis of influenza C virus infections and the seemingly benign nature of the virus contribute to the fact that 50 years after its first isolation, relatively little is known about the epidemiology and the clinical impact of this virus. Reverse transcription–polymerase chain reaction (RT-PCR) was used to amplify influenza C RNA fragments from clinical specimens. Two hundred otherwise healthy adults with recent onset of a common cold were studied. Nasopharyngeal aspirates were collected at entry to the study and 1 week later. Serum samples for antibody determinations were obtained at the first visit and after 3 weeks. Influenza C was detected in 7 of the 200 patients by 2 different RT-PCR formats. All 7 patients had a significant increase in antibody titers between serum samples collected during the acute and convalescent phases of the illness. Influenza C appears to be one of the many viruses that cause acute upper respiratory tract infections in adults
To our understanding, influenza C causes mild upper respiratory tract infections in children and adolescents [1], but the virus also has been detected occasionally in patients with lower respiratory tract infections [2]. It is technically difficult to isolate the virus [3]; thus, only few laboratories provide specific diagnostic services. Therefore, little is known about the epidemiology and the clinical impact of influenza C
Reverse transcription–polymerase chain reaction (RT-PCR) for the detection of influenza C has been described elsewhere [4, 5], but the diagnostic potential of this method has not yet been established. We have used RT-PCR for the identification of influenza C RNA in nasopharyngeal aspirates (NPAs) collected from 200 adults with a common cold [6, 7]. Furthermore, we have developed an immunoperoxidase staining procedure for influenza C–infected, cultured cells, to measure virus-specific antibodies in acute- and convalescent-phase serum samples from these individuals
Materials and Methods
Fifty-nine men (mean age, 24.0 years) and 141 women (mean age, 24.1 years) were recruited from October 1994 through November 1995 for a study of the etiology of the common cold [6]. Criteria for inclusion were the presence of acute rhinorrhea, nasal congestion, and/or sore throat of <48-h duration. A recent upper respiratory tract infection, tonsillitis, a history of allergic rhinitis, any chronic illness, or use of regular medication were reasons for exclusion. An NPA was collected at entry to the study and at the first follow-up visit 1 week later. NPAs were kept frozen at −70°C for ⩽5 years before being analyzed in this study. For antibody testing, serum samples were obtained at study entry and 3 weeks later and were kept at −20°C until use
Virus propagation was done in Madin-Darby canine kidney cells (MDCK-I-N; provided by H. Nishimura, Yamagata University School of Medicine, Yamagata, Japan). Cells were grown and maintained at 36°C in Dulbecco's modified Eagle medium (DMEM) supplemented with 0.2% bovine serum albumin, 25 mM HEPES, and antibiotics (DMEM-BSA) containing 7% fetal bovine serum (FBS). Influenza C/Ann Arbor/1/50 was grown in MDCK-I-N cells, using DMEM-BSA supplemented with 4 μg/mL of 1-tosylamide-2-phenylethyl chloromethyl ketone–trypsin (TPCK-trypsin). Five days after inoculation, medium was collected, was clarified by centrifugation, and was stored in aliquots at −80°C. This virus stock served as positive control in the RT-PCR and was used in the antibody assay
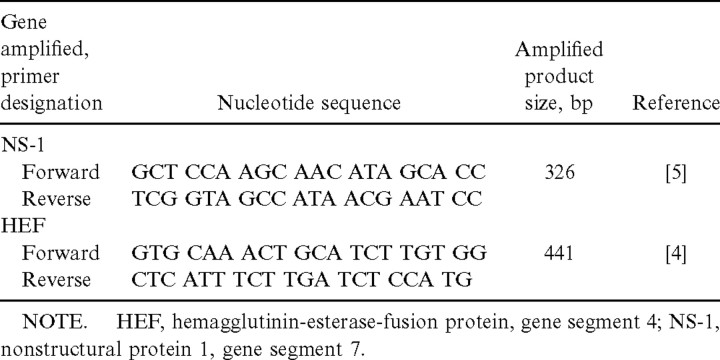
Viral RNAs were extracted from NPAs by use of a commercial kit (NucleoSpin Virus; Macherey-Nagel). Influenza C virus RNA was reverse transcribed and was amplified by 2 separate RT-PCRs, using 2 sets of primer pairs (table 1), one specific for the hemagglutinin-esterase fusion protein gene (HEF) [4] and the other for the nonstructural protein gene (NS-1) [5]. The reaction mixture contained 3 mM MgCl2, each dNTP (Finnzymes) at 0.25 mM, 9.4 pmol of the forward primer, which also primed the cDNA reaction and the reverse primer, 15 U of RNasin (Promega), 2.5 U of DNA polymerase (DyNAzyme; Finnzymes), 6 U of the enzyme avian myeloblastosis virus reverse transcriptase (Promega), and 10 μL of extracted RNA. cDNA was synthesized at 42°C for 45 min, followed by a 10-min incubation at 95°C. Thereafter, cDNA was amplified by 35 cycles consisting of denaturation at 95°C for 30 s, primer annealing at 50°C for 30 s, and primer extension at 72°C for 30 s. In the final cycle, primer extension was continued for 5 min. RNA from C/Ann Arbor/1/50 served as positive control in each run. Reaction tubes containing water instead of RNA were included as negative controls. Amplified products were detected by electrophoresis in 1% agarose gel after ethidium bromide staining
Table 1.
Nucleotide sequences of oligonucleotide primers used in reverse transcription–polymerase chain reactions
An immunoperoxidase staining assay was developed for the detection of influenza C–specific antibodies. Ninety-six–well microtiter plates (Nunc) were seeded with MDCK-I-N cells. Approximately 2.5×104 cells in 200 μL of DMEM-BSA containing 4% FBS were added to each well, and the plates were incubated at 36°C in a humidified 5% CO2 atmosphere. When the cells were confluent, 100 μL of the growth medium was replaced by 100 μL of DMEM-BSA containing C/Ann Arbor/1/50 and 8 μg/mL of TPCK-trypsin. The plates were centrifuged at 700 g at ambient temperature for 45 min and were incubated at 34°C for 2–3 days. The amount of virus was chosen so that ∼10%–20% of cells became infected. The cells then were washed with PBS, fixed with 80% acetone in PBS at room temperature for 10 min, and washed twice again with PBS, and the plates were kept at 4°C until use within 1 week
Serum samples were diluted in PBS containing 5% nonfat dry milk powder (PBS-M) and were tested in serial 4-fold dilutions starting at 1:200. A known positive serum sample and FBS as a negative control were included on each plate. Serum incubation was done at 37°C for 60 min. After the wells were washed with PBS, peroxidase-labeled rabbit antibodies to human IgG (DAKO) diluted 1:10,000 in PBS-M were added to each well, and plates were incubated at 37°C for 60 min. The wells were washed again, and 40 μL of TrueBlue substrate (Kirkegaard & Perry) was added to each well for 30 min. Wells were rinsed twice with PBS, and the plates were read with an inverted microscope at ×100 magnification. Distinct blue cytoplasmic and nuclear staining of infected cells in the absence of background staining of uninfected cells was considered to be a positive result. A ⩾4-fold increase in titer between acute and convalescent serum was considered to be a significant result
Results
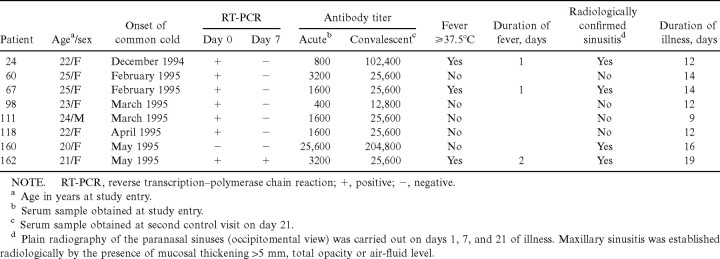
With C/Ann Arbor/1/50, the detection limit of the 2 RT-PCR protocols was ∼1 TCID50 with the NS-1 primer pair and 10 TCID50 with the HEF primers, respectively. Of the 200 NPAs collected during the first visit, 7 showed a distinct band of appropriate size with both primer pairs (table 2). Only 1 of the 7 patients from whom these samples were obtained was still positive by PCR at the first control visit 1 week later, but a signal was detected only with the NS-1 primer pair. The remaining 193 patients had negative PCR results at the initial visit. The second NPA of 107 randomly selected patients with initially negative PCR results also was tested by both PCRs, but none yielded positive signals
Table 2.
Laboratory and clinical findings for 8 study participants with evidence of acute or recent influenza C infection
Influenza C–specific antibodies were detected in all study participants. A ⩾8-fold increase between acute- and convalescent-phase serum samples was found in all 7 individuals with PCR-positive results (table 2). In addition, 1 study participant with PCR-negative results had an 8-fold increase in titer. No significant fluctuation of antibody levels was detected in the remaining 192 individuals with PCR-negative results
All 8 influenza C cases occurred during winter and spring. The duration of illness in the 7 patients with PCR-positive results varied, 3 of whom had a fever of 1 or 2 day's duration, from 9 to 19 days (table 2). Sinusitis was confirmed radiologically in 4 of the 8 individuals with laboratory evidence for a recent infection with influenza C (table 2)
Discussion
To our knowledge, this is the first successful attempt to detect influenza C RNA in clinical specimens by means of RT-PCR. Of 200 adults with a common cold of <48-h duration, 7 were diagnosed with an influenza C infection. Specimens of all 7 patients were found to be positive by both RT-PCRs, and all specimens were found to be positive on repeated examination. No other pathogen could be identified for any of these influenza C–positive patients (results not shown)
All 7 individuals with PCR-positive results had a ⩾8-fold increase in antibody titers between serum samples collected during the acute and convalescent phases of the illness. This further underlines the specificity of our PCR results. In addition, 1 study participant with PCR-negative results also had a significant increase in antibody titer (patient 160; table 2). The titer found in the first serum sample from this patient was as high as those in the convalescent-phase serum samples of the 7 patients with PCR-positive results; however, over the 3-week observation period, this patient showed an 8-fold increase in antibody titer to influenza C. This patient also had a significant titer increase to coronavirus (results not shown). This person possibly experienced an influenza C infection a few weeks earlier and entered the study because of an acute infection with a coronavirus
With laboratory-grown virus material, the sensitivity of both PCRs was relatively low. However, if a significant increase in antibody titers is accepted as definite proof of recent infection with a virus, the diagnostic sensitivity of our PCR assays is satisfactory. Prolonged storage of NPAs at −70°C and repeated freezing and thawing evidently did not affect the ability to detect virus-specific gene sequences in these specimens
NPAs obtained from the 7 patients with RT-PCR–positive results at the first control visit 1 week after study entry also were tested. Only one of these patients still had PCR-positive results, which indicates that the virus is rapidly cleared from the upper respiratory tract. Furthermore, this patient also had a longer duration of fever and illness than all other influenza C–positive patients (patient 162; table 1)
Although RT-PCR may be a rather sophisticated technique for diagnosing a relatively benign respiratory virus, the antibody assay described is easy to perform and allows for the simultaneous testing of numerous specimens. Intense nuclear staining of infected cells indicates that this assay predominantly measures antibodies to the nucleoprotein and the matrix protein, which is in contrast to the strain-specific hemagglutination inhibition (HI) test, which measures antibodies to HEF. As in influenza A and B, the nucleoprotein and matrix protein of influenza C virus are well preserved through evolution and thus offer a reliable source of antigen in the absence of more recent isolates, which could be used in HI tests [8]. In an earlier study, an EIA was shown to be more sensitive than the HI test in detecting titer increases [9]. In that study, the antigen probably consisted mainly of nucleoprotein and matrix protein
None of our 200 patients was seronegative to influenza C, which is in accordance with another study in which high seroprevalence and antibody titers were found in adults by means of EIA [9]. However, an epidemiologic study in France found a seropositivity rate of <80% in the age group reflecting our study population [10]. The authors of that study used a highly purified virus antigen in an EIA. It remains unclear why a significant fraction of their study population lacked antibodies to influenza C. It is likely that a virus causing acute upper respiratory tract infection in adults would readily and efficiently infect susceptible younger individuals; therefore, antibodies should be detectable in virtually all adults. Evidently all our influenza C patients experienced re-infection with this virus
All 7 influenza C–positive cases occurred from December through May. However, the number of positive findings is too small to draw conclusions on the epidemiology and seasonality of the virus. In a recent study from Israel, 2 cases of influenza C were identified during outbreaks of influenza A and B during the winter of 1996–1997 [11]
The common cold is an illness of viral etiology. Before this study, a viral cause was established for 148 of our 200 patients [6], whereas evidence for a bacterial infection was found in only 7 patients. Yet, a large proportion of patients with a common cold seeking medical attention receive antibiotic treatment [12]. Sinusitis, a frequent complication of the common cold, is considered to be of bacterial origin. In a recent study, though, most patients with acute sinusitis in connection with a common cold lacked laboratory parameters that are indicative of bacterial infection [7]. A better understanding of the common cold's etiology and accumulation of information about the inflammatory processes that cause the characteristic symptoms may result in a more rational use of existing drugs and in accelerated research toward new therapeutic interventions
With an incidence of 3.5% in our study, influenza C caused as many cases of common cold as did parainfluenza viruses; as many episodes as adenoviruses, respiratory syncytial virus, and enteroviruses combined; and an equal number of cases as the 4 bacterial pathogens Chlamydia pneumoniae, Mycoplasma pneumoniae, Streptococcus pneumoniae and Haemophilus influenzae [6]. Our findings indicate that influenza C is a relevant pathogen that causes acute respiratory infections
Footnotes
Presented in part: Conference on Options for the Control of Influenza IV, Hersonissos, Crete, Greece, 23–28 September 2000 (abstract P2-68)
Informed consent was obtained from each patient, and the study protocol was accepted by the Ethical Committee of the Turku University Hospital
Financial support: Foundation for Research of Infectious Disease, Finland
In memory of Professor Pekka Halonen (1927–2001)
References
- 1.Ziegler T, Cox NJ. Influenza viruses. Manual of clinical microbiology. 7th ed. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, editors. Washington, DC: American Society for Microbiology; 1999. pp. 928–35. [Google Scholar]
- 2.Moriuchi H, Katsushima N, Nishimura H, Nakamura K, Numazaki Y. Community-acquired influenza C virus infection in children. J Pediatr. 1991;118:235–8. doi: 10.1016/s0022-3476(05)80489-5. [DOI] [PubMed] [Google Scholar]
- 3.Kendal AP, Skehel JJ, Pereira MS. Atlanta: Centers for Disease Control and Prevention; 1982. Concepts and procedures for laboratory-based influenza surveillance. [Google Scholar]
- 4.Zhang W, Evans DH. Detection and identification of human influenza viruses by the polymerase chain reaction. J Virol Methods. 1991;33:165–89. doi: 10.1016/0166-0934(91)90017-t. [DOI] [PubMed] [Google Scholar]
- 5.Claas ECJ, Sprenger MJW, Kleter GEM, van Beek R, Quint WGV. Type-specific identification of influenza viruses A, B, and C by the polymerase chain reaction. J Virol Methods. 1992;39:1–13. doi: 10.1016/0166-0934(92)90120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mäkelä MJ, Puhakka T, Ruuskanen O, et al. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–42. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puhakka T, Mäkelä MJ, Alanen A, et al. Sinusitis in the common cold. J Allergy Clin Immunol. 1998;102:403–8. doi: 10.1016/S0091-6749(98)70127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buonagurio DA, Nakada S, Fitch WM, Palese P. Epidemiology of influenza C virus in man: multiple evolutionary lineages and low rate of change. Virology. 1986;153:12–21. doi: 10.1016/0042-6822(86)90003-6. [DOI] [PubMed] [Google Scholar]
- 9.Troisi CL, Monto AS. Comparison of enzyme-linked immunosorbent assay and hemagglutination inhibition in a seroepidemiological study of influenza type C infection. J Clin Microbiol. 1981;14:516–21. doi: 10.1128/jcm.14.5.516-521.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manuguerra JC, Hannoun C, Aymard M. Influenza C virus infection in France. J Infect. 1992;24:91–9. doi: 10.1016/0163-4453(92)91150-a. [DOI] [PubMed] [Google Scholar]
- 11.Greenbaum E, Morag A, Zakay-Rones Z. Isolation of influenza C virus during an outbreak of influenza A and B viruses. J Clin Microbiol. 1998;36:1441–2. doi: 10.1128/jcm.36.5.1441-1442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mainous AG, III, Hueston WJ, Clark JR. Antibiotics and upper respiratory infection: do some folks think there is a cure for the common cold. J Fam Pract. 1996;42:357–61. [PubMed] [Google Scholar]