Abstract
Acute respiratory infection is a significant cause of morbidity and mortality in children worldwide. Accurate identification of causative agents is critical to case management and to prioritization in vaccine development. Sensitive multiplex diagnostics provide us with an opportunity to investigate the relative contributions of individual agents andmayalso facilitate the discovery of new pathogens. Recently, application of MassTag polymerase chain reaction (PCR) to undiagnosed infuenza-like illness in New York State led to the discovery of a novel rhinovirus genotype. Here we report the investigation, by MassTag PCR, of pediatric respiratory-tract infections in Germany, studying 97 cases for which no pathogen was identified through routine laboratory evaluation. Respiratory viruses were identified in 49 cases (51%); of the 55 identified viruses, 41 (75%) were rhinoviruses. The novel genotype represented 73% of rhinoviruses and 55% of all identified viruses. Infections with the novel genotype were associated with upper-respiratory-tract symptoms but, more frequently, with bronchitis, bronchiolitis, and pneumonia.
Human rhinoviruses (HRVs) are the most frequent cause of acute respiratory illness worldwide. Although HRVs are most commonly associated with mild upperrespiratory- tract disease [1–3], infection of lower airways does occur [4–7]. Lower-respiratory-tract infections (LRTIs) related to HRV are increasingly being reported in infants, elderly persons, and immunocompromised patients [8]. HRVs are also implicated in exacerbations of asthma [9, 10], chronic bronchitis [11], and acute bronchiolitis [12].
Taxonomically, HRVs are currently grouped into 2 species, human rhinovirus A (HRV-A) and human rhinovirus B (HRV-B), in the genus Rhinovirus of the family Picornaviridae ([13, 14]). These nonenveloped, positive-sense, single-stranded RNA viruses have been classified serologically [15, 16] and on the basis of their antiviral susceptibility profiles [17, 18], their nucleotidesequence relatedness [19, 20], and their use of receptors (intercellular adhesion molecule 1, low-density lipoprotein receptor, and decay-accelerating factor) [21–23]. Phylogenetic analyses of the VP4/VP2 and VP1 coding regions have indicated the presence of 76 serotypes in genetic group A and 25 serotypes genetic group B [18– 20, 24].
An agent is commonly not implicated in up to 50% of cases of severe respiratory disease, despite the application of polymerase-chain-reaction (PCR) assays as well as classical diagnostic methods, including antigen tests, serology, and culture methods. Broad-range molecular assay systems, such as multiplex PCR (hexaplex [25], GeneScan [26], and MassTag [27]), or microarrays (ViroChip [28] and panmicrobial GreeneChips [29]) may therefore allow us to gain new insights into epidemiology and clinical associations [30, 31]. With respect to HRV, recent studies employing sensitive PCR systems for these difficult-to-isolate organisms have shown an increased detection rate, compared with culture methods [1, 32–35]. Applying a multiplex MassTag PCR platform, we recently detected numerous agents of respiratory illness in samples that had been submitted for laboratory diagnosis but that had tested negative during routine diagnostic assessment [30]. HRVs were identified at high frequency in this set of samples. Detailed genetic analysis indicated that a large fraction of these viruses represent a previously uncharacterized genotype of rhinovirus, one that diverges from either HRV-A or HRV-B.
In an attempt to gather additional information on the potential pathogenicity, as well as temporal and geographic distribution, of rhinoviruses, including the recently identified genotype, we evaluated specimens collected, during the 2003–2006 seasons in Bad Kreuznach, Germany, from children hospitalized because of severe LRTI.
Materials and Methods
Clinical specimens and sample collection. Nasopharyngeal aspirates were obtained from children admitted, because of acute respiratory-tract infection, to the Kreuznacher Diakonie Hospital (Bad Kreuznach, Germany) during the interval of 2003–2006. Individuals ranged in age from 2 weeks to 5 years (mean age, 5 months; median age, 10 months); 46% were male, 54% female. Specimens collected at the time of admission were forwarded undiluted to the Robert Koch Institute (Berlin, Germany) for laboratory evaluation.RNAextraction was performed by use of QIAamp Viral RNA Kits (Qiagen). The 97 samples for which no pathogen had been diagnosed after assessment by realtime reverse-transcription (RT)-PCR assay for influenza virus [36] and respiratory syncytial virus infection were stored at −70°C (2003–2004 season, n = 30; 2004–2005 season, n = 27; 2005–2006 season, n = 40 [assay details available on request]).
Assay procedures. The 97 RNA samples representing cases of undiagnosed respiratory diseases were employed as a template for cDNA synthesis by use of Superscript II kits with random hexamer priming (Invitrogen), and they were analyzed by MassTag PCR by using a viral primer panel [27] that targeted influenza virus A and B (FLUAV and FLUBV, respectively), respiratory syncytial virus A and B (RSV-A and RSV-B, respectively), human parainfluenza virus 1, 2, 3, and 4 (HPIV-1, HPIV-2, HPIV-3, and HPIV-4, respectively), human coronavirus 229E and OC-43 (HCoV-229E and HCoV-OC43, respectively), human metapneumovirus, entero- and rhinoviruses, and adenoviruses. The fidelity of the MassTag PCR signal was verified by reamplification of products and by sequence analysis for all positive specimens. In instances in which MassTag PCR indicated the presence of a picornavirus, the VP4/VP2 region was amplified [37]. Amplification products were purified from agarose gels (Qia Gel Extraction Kit; Qiagen), and nucleotidesequencing reactions were performed on both strands by use of the ABI Prism Big Dye cycle sequencing kits and the ABI Prism Genetic Analyzer systems (Applied Biosytems). Identical results were obtained with duplicate aliquots processed at the New York and Berlin laboratories. Sequence analyses, alignments, and phylogenetic reconstructions were performed by use of programs from the Wisconsin GCG Package (Accelrys) and by MEGA3.1 software [38]. Nucleic-acid sequences generated during this work are available at GenBank, under the accession numbers EU081778–EU081816.
Results
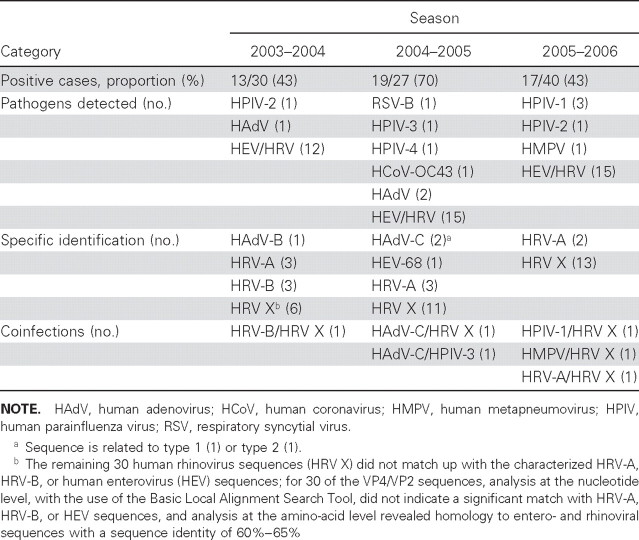
Identification of pathogens. We used MassTag PCR to investigate 97 nasopharyngeal aspirates from children hospitalized because of acute respiratory illness for which no pathogen was identified through routine laboratory testing. MassTag PCR identified at least 1 candidate respiratory-viral pathogen in 49 specimens. Although there was variability across the 3 seasons included in this study, 43% of specimens were positive in the 2003–2004 season, 70% of specimens were positive in the 2004–2005 season, and 43% of specimens were positive in the 2005–2006 season. Picornaviruses represented the majority of identified viruses in each season (table 1). For purposes of molecular identification, nucleic-acid sequences were obtained from all specimens that MassTag PCR had identified as being positive for virus. We identified 3 cases of human adenovirus (HAdV) infection—1 HAdV-B and 2 HAdV-C (table 1). In the case of the picornavirus-positive specimens, we identified, by use of molecular typing, 1 human enterovirus 68 (HEV-68), 8 HRV-A, and 3 HRV-B infections; the remaining 30 HRV sequences (HRV X) did not match with known HRV-A, HRV-B, or HEV sequences.
Table 1.
Viral pathogens detected by MassTag polymerase chain reaction, in children hospitalized with respiratory disease.
Clinical associations. HRVs were the viruses most frequently detected in our set of samples, representing 75% (41/55) of the identified viruses; coinfection with another virus was observed in only 12% (5/41) of these cases (table 2). The frequency of fever or cough in infections with HRV (82%) was comparable to that in infections with the other viruses (89%); the frequency of rhinitis or pharyngitis with HRV (79%) was comparable to that with other viruses; and the frequency of LRTI symptoms (bronchitis, bronchiolitis, and pneumonia) with HRV (71%) was comparable to that with the other viruses (67%). Whereas pneumonia was more common in infections with HRV A/B (56%) than in infections with HRV X (36%), the frequency of bronchiolitis with HRV A/B (11%) was comparable to that with HRV X (12%), and the frequency of bronchitis with HRV A/B (67%) was comparable to that with HRV X (60%). LRTI was recorded in 72% of HRV X infections; however, some cases were related to milder disease (table 2).
Table 2.
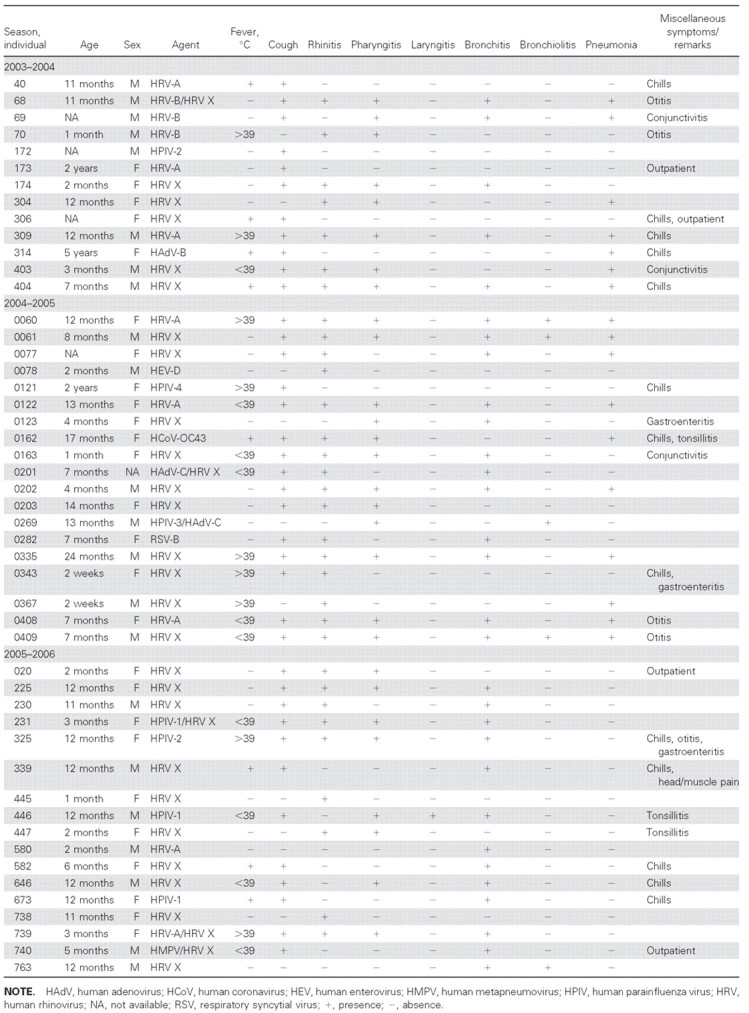
Patient and clinical data.
Molecular epidemiology of identified picornaviruses. MassTag PCR targets conserved sequences in the 5′-untranslated region of entero- and rhinoviruses; thus, to facilitate phylogenetic analysis of HEV and HRV, we amplified and sequenced the VP4/VP2 gene region. However, when we used the Basic Local Alignment Search Tool for analysis at the nucleotide level, we did not find, for 30 of the VP4/VP2 sequences, a significant match with HRV-A, HRV-B, or HEV sequences; analysis at the amino-acid level revealed homology to enteroand rhinoviral sequences, with a sequence identity of 60%–65%. High similarity at both the nucleotide- and amino-acid levels was evident when sequences were aligned with an unclassified genetic clade of picornaviruses recently identified in New York State [30]. However, detailed phylogenetic analysis indicated significant sequence diversity among the 30 viruses (figure 1). Temporal analysis over 3 seasons indicated a lower frequency of the novel genotype during the 2003 season (20% [6/30]) compared with the 2004 (41% [11/27]) and 2005 (33% [13/40]) seasons; phylogenetic clustering by season was not obvious. No significant relationship between the HRV genotypes and clinical diagnoses was observed (figure 1).
Figure 1.
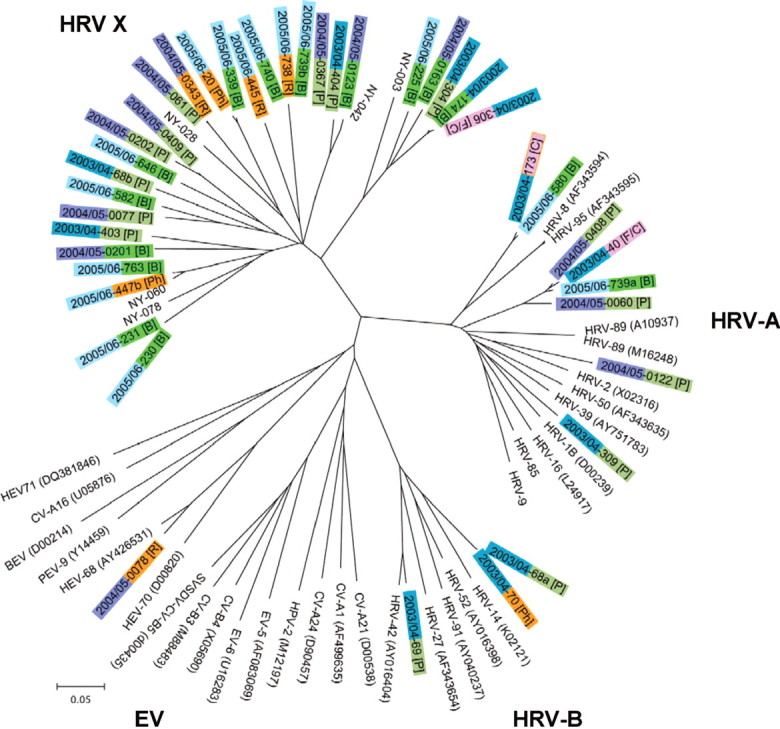
Phylogenetic analysis of the VP4/VP2 coding region of viruses identified in association with pediatric respiratory disease in Germany. Neighbor-joining analysis of the VP4/VP2 nucleotide sequence was performed by applying the Kimura 2-parameter model; the scale bar indicates nucleotide substitutions per site. Included for comparison are sequences belonging to the novel genotype recently identified in New York State (NY-003, -028, -042, -060, and -078); selected human rhinovirus (HRV)-A serotypes (the GenBank accession numbers for all reference sequences are indicated in parentheses); HRV-B serotypes; human enterovirus (HEV)-C viruses human coxsackievirus A1, A21, and A24 (CV-A1, CV-A21, and CV-A24, respectively); human poliovirus 2 (HPV-2); HEV-B viruses human echovirus 5 and 6 (HEV-5 and HEV-6, respectively); human coxsackievirus B4 (CV-B4), and swine vesicular disease virus (CV-B5); HEV-D viruses human enterovirus 68 and 70 (HEV-68 and HEV-70, respectively); Porcine enterovirus B virus porcine enterovirus 9 (PEV-9); Bovine enterovirus virus bovine enterovirus 1 (BEV-1); and HEV-A viruses human coxsackievirus A16 (CV-A16), and human enterovirus 71 (HEV-71). Major clinical symptoms associated with the specimen in which the respective virus was detected are indicated in square brackets: bronchitis/bronchiolitis [B], cough [C], fever [F], pharyngitis [Ph], pneumonia [P], and rhinitis [R].
Discussion
In this study of samples collected, during a 3-year interval, from hospitalized children with severe undiagnosed respiratory infection, MassTag PCR allowed us to detect viral pathogens in 49 (51%) of 97 cases. The pathogens most commonly identified were HRVs. These findings are consistent with other studies, which have indicated that rhinoviruses or picornaviruses account for 20%–80% of acute respiratory infections [1, 32, 33, 39–41]—exceeding, in some instances, even the frequency of RSV infection in pediatric-patient populations [34, 41–43].
The presence of HRV is not sufficient to prove causation. Asymptomatic HRV infection has been described; however, the extent to which infection without disease represents carriage, incubation, or convalescence is unknown [35, 39, 40, 42, 44]. Although we did not have samples to test for the presence of HRV in the lower respiratory tract, the high frequency at which HRV was identified as being the sole virus detected suggests a correlation between the agent and the observed LRTI symptoms. Support for the plausibility of HRV being pathogenic in LRTI comes from the facts that (1) in situ hybridization has demonstrated that HRVs exist in lower airways and (2) HRVs have been shown to trigger inflammatory processes in the infected cells and tissues [5–7, 45–48].
Among the HRVs identified in the present study were representatives of the novel genetic clade recently discovered in New York State [30]; indeed, these viruses comprised the majority of HRVs detected. HRV-A and HRV-B have been implicated in common colds as well as in severe LRTI. In our patients, viruses of the novel genetic clade were also associated with a wide range of diseases, ranging from rhinitis to bronchitis to severe pneumonia, necessitating supplemental oxygen in ∼50% of cases. A seasonal pattern of HRV infections has been described [2, 3, 43]; however, data regarding either serotype- or genotype-specific patterns of seasonality or disease symptoms are limited [49–51]. A temporal trend of sequence diversity or of correlation between genotype (within the novel HRV clade) and clinical diagnosis was not apparent in our data (figure 1).
No detailed information is available yet concerning the history of the novel HRV clade; nonetheless, the sequence diversity observed within it (figure 1) is not consistent with a recent introduction. This clade may account, in part, for earlier reports of nontypeable rhinoviruses [41, 44]. Indeed, its discovery may reflect the implementation of new technologies rather than novelty of the agent itself. We anticipate that future work will define more than 1 serogroup. Our findings reinforce other groups' recent work indicating the significance of HRVs in pediatric LRTI. The presence of novel HRVs in 2 disparate geographic locations, in association with serious respiratory disease in children as well as in adults, mandates further work in epidemiology and pathogenesis.
Acknowledgments
We thank Ingrid Zadow, Bettina Bauer, Manuela Friedrich, Marlies Hartwig, and Sven Tietze for their excellent technical assistance.
Footnotes
Potential conflicts of interest: none reported.
Financial support: College of American Pathology Foundation (scholars research award to N.R.); National Institutes of Health (awards UC1 AI062705, AI51292, and U54 AI5715803, all to the Northeast Biodefense Center-Lipkin); Ellison Medical Foundation (SS-0030-01 to W.I.L.).
References
- 1.Arruda E, Pitkaranta A, Witek TJ Jr, Doyle CA, Hayden FG. Frequency and natural history of rhinovirus infections in adults during autumn. J Clin Microbiol. 1997;35:2864–8. doi: 10.1128/jcm.35.11.2864-2868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mäkelä MJ, Puhakka T, Ruuskanen O, et al. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–42. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monto AS. The seasonality of rhinovirus infections and its implications for clinical recognition. Clin Ther. 2002;24:1987–97. doi: 10.1016/S0149-2918(02)80093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ketler A, Hamparian VV, Hilleman MR. Characterization and classification of ECHO 28-rhinovirus-coryzavirus agents. Proc Soc Exp Biol Med. 1962;110:821–31. doi: 10.3181/00379727-110-27662. [DOI] [PubMed] [Google Scholar]
- 5.Gern JE, Galagan DM, Jarjour NN, Dick EC, Busse WW. Detection of rhinovirus RNA in lower airway cells during experimentally induced infection. Am J Respir Crit Care Med. 1997;155:1159–61. doi: 10.1164/ajrccm.155.3.9117003. [DOI] [PubMed] [Google Scholar]
- 6.Papadopoulos NG, Bates PJ, Bardin PG, et al. Rhinoviruses infect the lower airways. J Infect Dis. 2000;181:1875–84. doi: 10.1086/315513. [DOI] [PubMed] [Google Scholar]
- 7.Mosser AG, Vrtis R, Burchell L, et al. Quantitative and qualitative analysis of rhinovirus infection in bronchial tissues. Am J Respir Crit Care Med. 2005;171:645–51. doi: 10.1164/rccm.200407-970OC. [DOI] [PubMed] [Google Scholar]
- 8.Hayden FG. Rhinovirus and the lower respiratory tract. Rev Med Virol. 2004;14:17–31. doi: 10.1002/rmv.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grissell TV, Powell H, Shafren DR, et al. Interleukin-10 gene expression in acute virus-induced asthma. Am J Respir Crit Care Med. 2005;172:433–9. doi: 10.1164/rccm.200412-1621OC. [DOI] [PubMed] [Google Scholar]
- 10.Xatzipsalti M, Kyrana S, Tsolia M, et al. Rhinovirus viremia in children with respiratory infections. Am J Respir Crit Care Med. 2005;172:1037–40. doi: 10.1164/rccm.200502-315OC. [DOI] [PubMed] [Google Scholar]
- 11.Gwaltney JM., Jr . Rhinoviruses. In: Evans AS, editor. Viral infections of human. New York: Plenum Medical Book; 1989. pp. 593–615. [Google Scholar]
- 12.Papadopoulos NG, Moustaki M, Tsolia M, et al. Association of rhinovirus infection with increased disease severity in acute bronchiolitis. Am J Respir Crit Care Med. 2002;165:1285–9. doi: 10.1164/rccm.200112-118BC. [DOI] [PubMed] [Google Scholar]
- 13.International Committee on Taxonomy of Viruses. [Accessed 17 October 2007];Universal Database of the International Committee on Taxonomy of Viruses. Available at: http://phene.cpmc.columbia.edu.
- 14.Fauquet MF, Mayo MA, Maniloff J, Desselberger U, Ball LA. Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. London: Elsevier Academic Press; 2005. pp. 757–78. [Google Scholar]
- 15.Cooney MK, Fox JP, Kenny GE. Antigenic groupings of 90 rhinovirus serotypes. Infect Immun. 1982;37:642–7. doi: 10.1128/iai.37.2.642-647.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamparian VV, Colonno RJ, Cooney MK, et al. A collaborative report: rhinoviruses—extension of the numbering system from 89 to 100. Virology. 1987;159:191–2. doi: 10.1016/0042-6822(87)90367-9. [DOI] [PubMed] [Google Scholar]
- 17.Andries K, Dewindt B, Snoeks J, et al. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J Virol. 1990;64:1117–23. doi: 10.1128/jvi.64.3.1117-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laine P, Blomqvist S, Savolainen C, Andries K, Hovi T. Alignment of capsid protein VP1 sequences of all human rhinovirus prototype strains: conserved motifs and functional domains. J Gen Virol. 2006;87:129–38. doi: 10.1099/vir.0.81137-0. [DOI] [PubMed] [Google Scholar]
- 19.Horsnell C, Gama RE, Hughes PJ, Stanway G. Molecular relationships between 21 human rhinovirus serotypes. J Gen Virol. 1995;76((Pt 10)):2549–55. doi: 10.1099/0022-1317-76-10-2549. [DOI] [PubMed] [Google Scholar]
- 20.Savolainen C, Blomqvist S, Mulders MN, Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J Gen Virol. 2002;83:333–40. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- 21.Abraham G, Colonno RJ. Many rhinovirus serotypes share the same cellular receptor. J Virol. 1984;51:340–5. doi: 10.1128/jvi.51.2.340-345.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uncapher CR, DeWitt CM, Colonno RJ. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology. 1991;180:814–7. doi: 10.1016/0042-6822(91)90098-v. [DOI] [PubMed] [Google Scholar]
- 23.Blomqvist S, Savolainen C, Raman L, Roivainen M, Hovi T. Human rhinovirus 87 and enterovirus 68 represent a unique serotype with rhinovirus and enterovirus features. J Clin Microbiol. 2002;40:4218–23. doi: 10.1128/JCM.40.11.4218-4223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ledford RM, Patel NR, Demenczuk TM, et al. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J Virol. 2004;78:3663–74. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan J, Henrickson KJ, Savatski LL. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex) Clin Infect Dis. 1998;26:1397–402. doi: 10.1086/516357. [DOI] [PubMed] [Google Scholar]
- 26.Erdman DD, Weinberg GA, Edwards KM, et al. GeneScan reverse transcription-PCR assay for detection of six common respiratory viruses in young children hospitalized with acute respiratory illness. J Clin Microbiol. 2003;41:4298–303. doi: 10.1128/JCM.41.9.4298-4303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briese T, Palacios G, Kokoris M, et al. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg Infect Dis. 2005;11:310–3. doi: 10.3201/eid1102.040492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Coscoy L, Zylberberg M, et al. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA. 2002;99:15687–92. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palacios G, Quan P-L, Jabado OJ, et al. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis. 2007;13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamson D, Renwick N, Kapoor V, et al. MassTag polymerase-chainreaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influeza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiu CY, Rouskin S, Koshy A, et al. Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clin Infect Dis. 2006;43:e71–6. doi: 10.1086/507896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ireland DC, Kent J, Nicholson KG. Improved detection of rhinoviruses in nasal and throat swabs by seminested RT-PCR. J Med Virol. 1993;40:96–101. doi: 10.1002/jmv.1890400204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyypiä T, Puhakka T, Ruuskanen O, Mäkelä M, Arola A, Arstila P. Molecular diagnosis of human rhinovirus infections: comparison with virus isolation. J Clin Microbiol. 1998;36:2081–3. doi: 10.1128/jcm.36.7.2081-2083.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loens K, Goossens H, de Laat C, et al. Detection of rhinoviruses by tissue culture and two independent amplification techniques, nucleic acid sequence-based amplification and reverse transcription-PCR, in children with acute respiratory infections during a winter season. J Clin Microbiol. 2006;44:166–71. doi: 10.1128/JCM.44.1.166-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winther B, Hayden FG, Hendley JO. Picornavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: association with symptomatic illness and effect of season. J Med Virol. 2006;78:644–50. doi: 10.1002/jmv.20588. [DOI] [PubMed] [Google Scholar]
- 36.Schweiger B, Zadow I, Heckler R, Timm H, Pauli G. Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J Clin Microbiol. 2000;38:1552–8. doi: 10.1128/jcm.38.4.1552-1558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coiras MT, Aguilar JC, Garcia ML, Casas I, Perez-Brena P. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J Med Virol. 2004;72:484–95. doi: 10.1002/jmv.20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 39.Johnston SL, Sanderson G, Pattemore PK, et al. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. J Clin Microbiol. 1993;31:111–7. doi: 10.1128/jcm.31.1.111-117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nokso-Koivisto J, Kinnari TJ, Lindahl P, Hovi T, Pitkaranta A. Human picornavirus and coronavirus RNA in nasopharynx of children without concurrent respiratory symptoms. J Med Virol. 2002;66:417–20. doi: 10.1002/jmv.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jartti T, Lehtinen P, Vuorinen T, et al. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis. 2004;10:1095–101. doi: 10.3201/eid1006.030629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rakes GP, Arruda E, Ingram JM, et al. Rhinovirus and respiratory syncytial virus in wheezing children requiring emergency care: IgE and eosinophil analyses. Am J Respir Crit Care Med. 1999;159:785–90. doi: 10.1164/ajrccm.159.3.9801052. [DOI] [PubMed] [Google Scholar]
- 43.Miller EK, Lu X, Erdman DD, et al. Rhinovirus-associated hospitalizations in young children. J Infect Dis. 2007;195:773–81. doi: 10.1086/511821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jartti T, Lehtinen P, Vuorinen T, Koskenvuo M, Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol. 2004;72:695–9. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- 45.Fraenkel DJ, Bardin PG, Sanderson G, Lampe F, Johnston SL, Holgate ST. Lower airways inflammation during rhinovirus colds in normal and in asthmatic subjects. Am J Respir Crit Care Med. 1995;151:879–86. doi: 10.1164/ajrccm/151.3_Pt_1.879. [DOI] [PubMed] [Google Scholar]
- 46.Johnston SL, Papi A, Bates PJ, Mastronarde JG, Monick MM, Hunninghake GW. Low grade rhinovirus infection induces a prolonged release of IL-8 in pulmonary epithelium. J Immunol. 1998;160:6172–81. [PubMed] [Google Scholar]
- 47.Schroth MK, Grimm E, Frindt P, et al. Rhinovirus replication causes RANTES production in primary bronchial epithelial cells. Am J Respir Cell Mol Biol. 1999;20:1220–8. doi: 10.1165/ajrcmb.20.6.3261. [DOI] [PubMed] [Google Scholar]
- 48.Seymour ML, Gilby N, Bardin PG, et al. Rhinovirus infection increases 5-lipoxygenase and cyclooxygenase-2 in bronchial biopsy specimens from nonatopic subjects. J Infect Dis. 2002;185:540–4. doi: 10.1086/338570. [DOI] [PubMed] [Google Scholar]
- 49.Calhoun AM, Jordan WS, Jr, Gwaltney JM., Jr Rhinovirus infections in an industrial population. V. Change in distribution of serotypes. Am J Epidemiol. 1974;99:58–64. doi: 10.1093/oxfordjournals.aje.a121585. [DOI] [PubMed] [Google Scholar]
- 50.Fox JP, Cooney MK, Hall CE. The Seattle virus watch. V. Epidemiologic observations of rhinovirus infections, 1965–1969, in families with young children. Am J Epidemiol. 1975;101:122–43. doi: 10.1093/oxfordjournals.aje.a112078. [DOI] [PubMed] [Google Scholar]
- 51.Roebuck MO. Rhinoviruses in Britain 1963–1973. J Hyg (Lond) 1976;76:137–46. doi: 10.1017/s0022172400055029. [DOI] [PMC free article] [PubMed] [Google Scholar]