Abstract
Transforming growth factor (TGF)-β is a pleiotropic cytokine regulating a variety of cellular processes such as cell growth, differentiation, apoptosis, migration, cell adhesion, and immune response. In the well-understood classical TGF-β signaling pathway, TGF-β activates Smad signalling via its two cell surface receptors such as TβRII and ALK5/TβRI, leading to Smad-mediated transcriptional regulation. In addition, TGF-β may also activate other signaling pathways like mitogen-activated protein kinase, PI3K, etc. The signaling of TGF-β is finely regulated at different levels. Inhibitory Smads, including Smad6 and Smad7, are key regulators of TGF-β/bone morphogenetic protein (BMP) signaling by negative feedback loops. They can form stable complexes with activated type I receptors and thereby blocking the phosphorylation of R-Smads, or recruit ubiquitin E3 ligases, such as Smurf1/2, resulting in the ubiquitination and degradation of the activated type I receptors. Besides, these inhibitory Smad proteins also inhibit TGF-β/BMP signaling in the nucleus by interacting with transcriptional repressors, such as histone deacetylases, Hoxc-8, and CtBP, or disrupting the formation of the TGF-β-induced functional Smad-DNA complexes. Smad7 is in turn regulated by different stimuli, including TGF-β, IFN-γ, TNF-α as well as ultraviolet and TPA, and mediates the crosstalk between TGF-β and other signaling pathways. Deregulation of Smad7 expression has been associated with various human diseases, such as tissue fibrosis, inflammatory disease as well as carcinogenesis. Overexpression of Smad7 has been shown to antagonize TGF-β-mediated fibrosis, carcinogenesis, and inflammation, suggesting a therapeutic potential of Smad7 to treat these diseases.
Keywords: TGF-β, signal transduction, Smad7, feedback loop, crosstalk
Introduction
Transforming growth factor (TGF)-β family cytokines have been found to play diverse roles in regulating growth, differentiation, immune response as well as development in multi-organ systems. Up to now, more than 30 factors have been discovered to belong to TGF-β superfamily, which is generally divided into two subfamilies. One of them is consisted of TGF-β, activin, Nodal, myostatin, inhibin, etc. The other one includes BMPs, anti-mullerian hormone (AMH, or MIS), as well as many growth and differentiation factors (GDFs) [1,2].
There are three species of TGF-β (TGF-β1, TGF-β2, and TGF-β3) in mammalian cells. TGF-βs are synthesized via inactive precursors, which cannot bind to their receptors until being activated. After released from cells, they associate with latency-associated protein (LAP) and form a small inactive complex. In the extracellular matrix, this complex is bound by latent TGF-β-binding protein (LTBP), a component of the extracellular matrix that is necessary for the secretion and storage of TGF-β [3]. The latent TGF-β can be activated either by enzymatic proteolysis, executed by plasmin, integrin, or thrombin, or through a conformational change [4,5].
Overview of TGF-β Signaling Pathways
Once activated, the TGF-β homodimer transduces its signal by bringing together two types of serine/threonine kinase receptors—two type I receptors and two type II receptors. ALK5 (TβRI) and TβRII are specific for TGF-β. Upon TGF-β binding, ALK5 is phosphorylated and activated by the constitutively active TβRII at the GS region, which is important for the activation of its kinase domain as well as the recruitment of R-Smads. The phosphorylation of R-Smads at the C-terminal SSXS motif by activated type I receptor is an essential step for signal transduction. Of these R-Smads, Smad2 and 3 are engaged in signal transduction of TGF-β, activin, and Nodal group cytokines, whereas Smad1, 5, and 8 are necessary for the other groups including BMPs and GDFs. Once activated, R-Smads form complexes with a common Smad (Co-Smad, Smad4), enter the nucleus and regulate transcription of target genes along with different cofactors [6]. Both R-Smads and Smad4 are characterized by two conserved regions known as MH1 domain at the N-terminus and MH2 domain at the C-terminus, respectively, which are joined together by a linker region. The MH1 domain of R-Smads and Smad4 plays important roles in cytoplasmic anchoring, nuclear import, DNA binding, and regulation of gene transcription, while the MH2 domain is responsible for Smad–receptor interaction, Smad hetero-complex formation, cytoplasmic anchoring as well as transactivation of target genes [7].
Besides the canonical Smad-mediated signaling pathway (Fig. 1), it has long been recognized that TGF-β can also regulate some cellular or physiological processes independent of Smad proteins. There is evidence showing that Smad4 is not indispensable for the development of the mammary gland, liver, or pancreas in mice [2,8]. We have demonstrated that the nucleocapsid (N) protein of severe acute respiratory syndrome-associated coronavirus can bind to Smad3, interfere with the complex formation between Smad3 and Smad4, and promote Smad3–p300 complex formation in the nucleus [9], indicating a novel mode of Smad3 effect in a Smad4-independent manner. TIF1γ is also found to selectively associate with phosphorylated Smad2/3 in hematopoietic, mesenchymal, and epithelial cells in response to TGF-β and transduce the signal independent of Smad4 to promote erythrogenesis [10]. In addition, TGF-β can also activate mitogen-activated protein kinase (MAPKs) (ERK1/2, JNK, p38), PI3K, protein phosphatase 2A, Rho family proteins as well as the epithelial polarity protein Par6, but the underlying mechanisms are not fully understood. Lee et al. [11] reported that upon TGF-β stimulation, activated TGF-β type I receptor (TβRI) recruits and directly phosphorylates ShcA protein on tyrosine and serine residues, activating ERK MAPK signaling pathway. Recently, the ubiquitin E3 ligase TRAF6 has also been reported to interact with TβRI, and this interaction is required for TGF-β-induced auto-ubiquitination of TRAF6 as well as subsequent activation of TAK1-p38 MAPK, which leads to apoptosis [12,13]. This plasticity of TGF-β signaling allows TGF-β to exert pleiotropic influences.
Fig. 1.
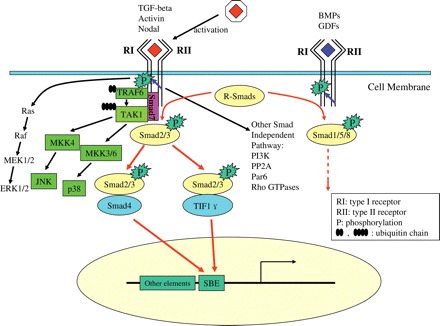
The Transforming growth factor (TGF)-β signaling pathway Once activated, TGF-β brings together two types of serine/threonine kinase receptors and propagates the signal through R-Smads phosphorylation, R-Smads/Co-Smad complex formation, and nuclear translocation, thus regulating the transcription of target genes, among which Smad7 is. Besides the canonical Smad pathway, TGF-β can also activate MAPK signaling, in which Smad7 may act as a scaffold protein. In addition, TGF-β may also activate PI3K, PP2A, Par6 as well as Rho GTPases independent of Smad signaling.
Negative Regulation of TGF-β Signaling by Smad7
As mentioned above, TGF-β is a pleiotropic cytokine that regulates embryonic development and cellular homeostasis mainly through the canonical Smad-mediated signaling pathway, which appears to be relatively simple and consists of only a few essential components. However, the signal transduction is finely regulated both temporally and spatially at different levels, including ligand activation, receptor complex formation, R-Smads activation, and translocation, as well as transcription in the nucleus. Many proteins have been identified to be associated with the receptors, R-Smads, or Co-Smad, and regulate TGF-β family signaling either in the cytoplasm or in the nucleus. In addition to R-Smads and Co-Smad, there is a third Smad protein family, namely the I-Smads (Smad6 and Smad7), which have been documented to play key roles in regulating signal transduction of TGF-β family cytokines. I-Smads are transcriptionally induced by TGF-β family cytokines and regulate these signaling pathways negatively, thus establishing an important negative feedback loop. Of these two I-Smads, Smad7 is a general antagonist of TGF-β family, while Smad6 is specific for BMP signaling. Smad6 and Smad7 also have conserved the C-terminal MH2 domain, but unlike R-Smads or Co-Smad, they lack the N-terminal MH1 domain and the phosphorylation site by the type I receptors at the C-terminal tail. The N-terminus of these two I-Smads shares a similarity of only 36.7%. Both the N-terminus and MH2 domain of Smad7 are essential for its specific inhibition of TGF-β/activin signaling [14,15]. I-Smads can also bind to DNA. For instance, we recently demonstrated that Smad7 could function in the nucleus by binding to the DNA elements containing the minimal Smad-binding element (SBE) CAGA box. By single-molecule force spectroscopy, our results revealed that Smad7 had similar binding strength to the oligonucleotides corresponding to the CAGA-containing activin responsive element (ARE) and the PAI-1 promoter, as that of Smad4 although, unlike R-Smad or Co-Smad, Smad7 also exhibits a binding activity to the mutant ARE with the CAGA sequence substituted. Interestingly, distinct from other Smad proteins, the MH2 domain, but not the N-terminal region, of Smad7 is responsible for DNA binding [16,17].
I-Smads can antagonize TGF-β family signaling through various mechanisms (Fig. 2). First, Smad7 is shown to form a stable complex with type I receptors, therefore leading to inhibition of R-Smad phosphorylation and the hetero-complex formation between R-Smads and Co-Smad [18]. Smad7 also recruits the HECT type of E3 ubiquitin ligases, Smurf1 and Smurf2. It binds to Smurfs in the nucleus and translocates into the cytoplasm in response to TGF-β and recruits the ubiquitin ligases to the activated type I receptor ALK5/TβRI, leading to the degradation of the receptor through the proteasomal pathway. Smad7 itself is also degraded in this process. Besides, the E3 ligases Nedd4-2 and WWP1/Tiul1 can also promote the degradation of type I receptor as well as R-Smads and Smad4, in which process Smad7 serves as an adaptor protein [19]. By employing similar mechanism, Smad6 interferes with BMP signaling. Moreover, Shi et al. [20] reported that the phosphatase GADD34-PP1c could be recruited to ALK5/TβRI by Smad7 and in turn dephosphorylate the receptor. Protein phosphatase 1α was also reported to have similar effect to dephosphorylate ALK1 in endothelial cells. It could be recruited by Smad7 to ALK1, and thereby controlled TGF-β/ALK1-induced Smad1/5 activation [21]. In addition, Smad6 has been shown to bind to the phosphorylated Smad1 in competition with Smad4 [22].
Fig. 2.
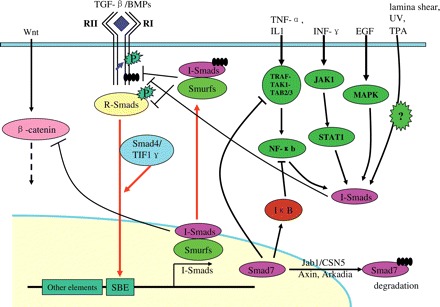
I-Smads mediate the crosstalk of the TGF-β signaling pathway with other pathways I-Smads could be transcriptionally induced by TGF-β/BMPs signaling and inhibit their signaling by negative feedback loops. Besides, they could also be induced by many other cytokines or stimuli, such as TNF-α/IL1, INF-γ, EGF, laminar shear stress, UV, TPA, etc. Smad7 has been reported to enhance the transcription of IκB, which is a key inhibitor of NF-κB signaling pathway. It may also disrupt the TRAF–TAK1–TAB2/3 complex, thus inhibiting NF-κB signaling. In addition, Smad7 was reported to down-regulate the protein level of β-catenin by recruitment of E3 ligase Smurf1. Smad7 itself could be degraded both in the nucleus and in the cytoplasm in proteasomal pathway.
It has been known that Smad6 can interfere with BMP signaling both in the cytoplasm and in the nucleus. Smad6 acts as a transcriptional repressor by interacting with Hoxc-8, or binding to DNA and recruiting transcriptional co-repressor histone deacetylases (HDACs) or CtBP to inhibit the transcription of target genes [23–26]. Recently, we reported that in some cell lines, such as Hep3B, HeLa, mink lung epithelial Mv1Lu mutant L17, and human normal lung epithelial HPL-1 cells, most Smad7 proteins retain in the nucleus even under TGF-β stimulation, and Smad7 can exert its inhibitory effect on TGF-β signaling in the nucleus [16]. Forced expression of Smad7 in the nucleus potently repressed the transcriptional activity of TGF-β, and the inhibitory effect of Smad7 can be independent of TβRI, as demonstrated in TβRI-deficient R1B/L17 cells. Furthermore, Smad7 is able to bind DNA in vivo and in vitro with the MH2 domain and disrupt the formation of functional Smad–DNA complexes [16,17].
Several proteins have been shown to interact with Smad7 and regulate TGF-β signaling (Table 1). A WD protein, STRAP, which associates with the type I and II receptors, can also interact with Smad7, stabilizing the complex between Smad7 and type I receptor, thus inhibiting TGF-β signaling synergistically with Smad7 [27]. AIP4, which is an E3 ubiquitin ligase and induces the degradation of Smad7, may negatively regulate TGF-β signaling without affecting the turnover of the type I receptor [28]. Instead, AIP4 may enhance the interaction between Smad7 and activated ALK5/TβRI.
Table 1.
Smad7-binding partners
| Smad7-binding partners | Function | References |
|---|---|---|
| AIP4 | Stabilizing the complex between ALK5 and Smad7 and inhibiting TGF-β signaling | [19] |
| Arkadia | Degradation of Smad7 | [20] |
| Axin | An adaptor between Smad7 and Arkadia | [21] |
| Cas-L (Crk-associated substrate lymphocyte type) | Facilitating TGF-β signaling by interfering with the inhibitory effect of Smad7 | [26] |
| FKBP12 | An adaptor between ALK5 and Smad7–Smurf1 | [27] |
| GADD34-PP1c | Dephosphorylation of ALK5 | [16] |
| HDACs | Deacetylation of Smad7 and inhibition of TGF-β signaling | [34,35] |
| Hic-5/ARA55 | Degradation of Smad7 | [23] |
| Jab1/CSN5 | Degradation of Smad7 | [22] |
| Nedd4-2 | Degradation of the type I receptors, R-Smads and Smad4 | [14] |
| p300/CBP | Acetylation and stabilization of Smad7 | [34,35] |
| Phosphatase 1α | Dephosphorylation of ALK1 | [17] |
| SIK (the salt-inducible kinase) | Degradation of TβRI | [25] |
| SIRT1 | Deacetylation of Smad7 and inhibition of TGF-β signaling | [33] |
| Smurf1/2 | Degradation of the type I receptors, R-Smads, Smad4 and Smad7 | [14] |
| STRAP | Stabilizing the complex between ALK5 and Smad7 and inhibiting TGF-β signaling | [18] |
| TRAFs; TAK1 | Activation of p38 MAPK signaling pathway by TGF-β with Smad7 as an adaptor; inhibition of NF-κB signaling by dissembling the TRAF–TAK1–TAB2/3 complex via Smad7 | [12,42,80] |
| Yes-associated protein (YAP65) | facilitating the recruitment of Smad7 to activated TβRI | [24] |
| WWP1/Tiul1 | Degradation of the type I receptors, R-Smads and Smad4 | [14] |
Regulation of the stability of Smad7 protein has been used as another means to influence TGF-β signaling. Arkadia facilitates TGF-β signaling by promoting the degradation of Smad7 and Axin may act as an adaptor between Arkadia and Smad7 [29,30]. Jab1/CSN5, which is a component of the COP9 signalosome complex, also regulates the stability of Smad7 and releases Smad7-mediated suppression of TGF-β signaling [31]. Hic-5/ARA55 [32], Yes-associated protein (YAP65) [33], the salt-inducible kinase (SIK) [34], and Crk-associated substrate lymphocyte type (Cas-L) [35] are identified to be binding partners of Smad7 and regulate TGF-β signaling either positively or negatively. FKBP12, which is a cytoplasmic protein that binds to the immunosuppressant drugs, Tacrolimus (FK506) and rapamycin, has been found to interact with the GS region of type I receptors and inhibit signal transduction of TGF-β family [36]. Interestingly, it can serve as an adaptor for the Smad7–Smurf1 complex and promote the ubiquitination and degradation of TβRI [37].
Post-translational modification plays a pivotal role in regulating the function of proteins. As mentioned above, ubiquitination plays an important role in the regulation of the stability of I-Smads, and TGF-β receptors. In addition, Smad7 interacts with p300, a histone acetylase, and could be acetylated at the same lysine residues where ubiquitination occurs. Smad7 can also interact with HDACs as well as SIRT1, and is deacetylated by these enzymes [38–40]. The acetylation of Smad7 inhibits its ubiquitination and proteasome-mediated degradation, so the degradation of Smad7 is regulated by the balance between acetylation, deacetylation, and ubiquitination.
Smad7 Connects Other Signaling Pathways With the TGF-β Pathway
Smad7 is an important regulator of TGF-β, activin, Nodal, and BMPs signaling via a negative feedback circuit. It is transcriptionally induced by TGF-β and BMPs. A perfect SBE has been identified in the promoter of human Smad7, and both Smad2/3 and Smad4 are involved in its transcriptional induction by TGF-β [41]. However, full induction of Smad7 by Smad proteins needs other transcriptional co-activators, such as CBP/p300, FoxH1, TFE-3 (transcription factor mE3), CBFA (PEBP2/core-binding factor A) and ATF2, and so on [42]. AP1 and SP1 were also found to promote the transcription of Smad7, indicating that other signaling pathways may be involved in the transcriptional regulation of Smad7 (Fig. 2) [43]. Similarly, Smad6 is also transcriptionally induced by BMP/Smad signaling pathway. Besides CREB, other factors, such as OAZ and Runx2, also upregulate the expression of Smad6 [44,45]. And the transcriptional co-repressor Ski was reported to inhibit the transcription of both Smad6 and Smad7 [46].
In addition to TGF-β/BMP signaling, the transcription of I-Smads can also be induced by inflammatory cytokines, such as interleukin 1, IFN-γ, and TNF-α [47]. EGF, ultraviolet irradiation, lamina shear stress as well as EGF or TPA treatment can induce Smad7 expression in a cell-line-dependent manner, but the mechanisms remain elusive [47,48]. Interestingly, Smad7 was shown to disrupt the formation of TRAF2–TAK1–TAB2/3 complex and inhibit TNF-α/NF-κB signaling [49]. Likewise, Smad6 binds to Pellino-1, an adaptor protein of mammalian interleukin 1 receptor-associated kinase 1 (IRAK1), and thereby promoting TGF-β-mediated anti-inflammatory effects [50]. Since TGF-β has an anti-inflammatory activity, and usually acts against those cytokines, I-Smads induction by one kind of cytokine may repress the signaling of another. So they mediate the balance and crosstalk of these signaling pathways.
TGF-β can activate MAPKs, including ERK, JNK, and p38 signaling in a cell-specific manner. Accumulating evidence shows that Smad7 may play roles in this process. Mazars et al. [51] reported that Smad7 could activate JNK signaling and is essential for JNK-mediated apoptosis. TGF-β induces apoptosis of prostate cancer cells by activating p38 MAPK, and Smad7 may serve as a scaffold protein in the process [52,53]. In prechondrogenic cells, Smad7 inhibits chondrocytic differentiation possibly by downregulating BMP-activated p38 MAPK pathway [52,53]. Smad7 was also reported to be involved in TGF-β-dependent activation of Rho GTPases Cdc42 and RhoA [54]. All these data indicate that I-Smads not only act as antagonists of TGF-β/BMP signaling, but also stand at the crossroad of diverse signaling pathways.
There are multiple cross-talking steps between TGF-β and Wnt signaling. β-catenin, a key signal transducer of Wnt signaling, has functional interaction with Smad7. The association of Smad7 with β-catenin regulates the transcription of c-myc, which is important for TGF-β induced apoptosis of human prostate cancer PC-3U cells [55]. In addition, Smad7 transgenic mice exhibit perturbed hair follicle morphogenesis and differentiation and accelerated sebaceous gland morphogenesis due to β-catenin degradation by the ubiquitin E3 ligase Smurf2 (recruited by Smad7) and thereby decreased Wnt signaling [56]. In contrast, a recent report showed that Smad7 stabilizes β-catenin binding to E-cadherin and modulates cell–cell adhesion [57]. Therefore, Smad7 can regulate the activity of β-catenin in a cellular context-dependent way.
The Role of Smad7 in TGF-β Mediated Physiology and Pathology
Although Smad7 is a well-documented key antagonist of TGF-β, it has been shown to promote TGF-β-mediated apoptosis of human prostatic carcinoma cells, podocytes, Mv1Lu, MDCK, and COS7 cells by activating MAPK signaling or repressing NF-κB signaling [58]. On the other hand, Smad7 can also inhibit apoptosis induced by TGF-β in some cell lines, such as B cells and gastric epithelial cells [59,60], suggesting that the function of Smad7 is context dependent.
TGF-β plays a key role in fibrosis of different tissues, such as skin, the liver, kidney, eye, lung as well as cardiovascular system. It induces the Smad3-dependent transcription of fibrillar collagens, inhibits the degradation of ECM by downregulating the expression of matrix degrading enzymes, and increases the expression of metalloproteinase (MMP) inhibitors, together leading to the accumulation of ECM. Elevated TGF-β level and decreased Smad7 level are often present in tissues where an uncontrolled fibrotic response occurs. Therefore, inhibition of Smad3 by overexpression of Smad7 dramatically reduced fibrotic responses of the kidney, lung and liver in animal models, indicating an important anti-fibrotic effect of Smad7 by antagonizing TGF-β/Smad3 signaling pathway [61–64].
In addition to pro-fibrotic activity, TGF-β also has a major role in the regulation of immune cell functions. TGF-β1 knockout mice showed a multifocal, mixed inflammatory cell response and tissue necrosis, leading to organ failure and death [65]. Consistently, blocking TGF-β signaling via specific deletion of TGF-β type II receptor in T cells leads to disruption of T-cell development as well as homeostasis, resulting in autoimmune inflammation and death at last [66]. TGF-β also regulates the differentiation and activation of many other leukocytes, including B cells, NK cells, dendritic cells, monocytes/macrophages, granulocytes, and mast cells [67]. Thus, TGF-β is a key regulator in immune system homeostasis, and dysfunction of TGF-β may results in disorders of this system, such as autoimmunity or inflammatory bowel disease. In patients with inflammatory bowel disease, the phosphorylation level of Smad3 is very low, and the formation of Smad3–Smad4 complex is also affected, so TGF-β signaling is disrupted despite the abundance of TGF-β in the inflamed gut [68,69]. Instead, Smad7 level is elevated in the tissues with inflammatory bowel disease. When Smad7 was reduced by anti-sense oligonucleotides, Smad3 activation was restored and the inflammation was subsequently inhibited by endogenous TGF-β [70]. Conversely, Smad7 may also mediate the anti-inflammatory activity of TGF-β in other situations by inhibiting NF-κB signaling upon activation by inflammatory cytokines like interleukine 1 or TNF-α. Smad7 has also been shown to disrupt the TRAF6–TAK1–TAB2/3 signaling complex or to induce the expression of IκB, which induces the degradation of NF-κB subunits by the proteasomal pathway, therefore resulting in inhibition of NF-κB signaling. In addition, Smad7 has been shown to be a key negative regulator of the renal inflammatory response [62,68,71,72]. These results suggest that Smad7 may have both anti-fibrotic and anti-inflammatory functions.
TGF-β has anti-proliferative effects in epithelial cells. However, it can also promote tumorigenesis by modulating processes such as cell invasion, metastasis, immune regulation, and microenvironment modification that cancer cells may exploit for their own good [2]. Smad7 has been shown to play a role in these processes. A genome-wide association study showed that common alleles of Smad7 influenced the risk of colorectal cancer [73]. As a key negative regulator of TGF-β signaling, Smad7 has been reported to inhibit the formation of osteolytic metastases by human breast cancer and melanoma when overexpressed [74–76]. In addition, it also inhibits endometrial carcinomas, thyroid follicular tumors, and hepatocellular carcinomas [77–79]. Azuma et al. [80] reported that overexpression of Smad7 in mouse mammary carcinoma JygMC(A) cells inhibits their metastasis via upregulation of E-cadherin and downregulation of N-cadherin, leading to a reduction of cell migration [81]. In other cases, Smad7 can promote tumorigenesis. For instance, Smad7 blocks TGF-β-mediated growth inhibition and inhibits apoptosis in pancreatic cancer as well as FET cells [82,83]. Liu et al. [84] also reported that in a xenograft model, when primary keratinocytes were co-transfected with Smad7 and H-ras, mixed with dermal fibroblasts, and grafted into nude mice, they progressed into skin squamous cell carcinomas, while control cells did not. Since Smad7 has been shown to degrade β-catenin by recruiting Smurf2, and reduced Wnt signaling leads to spontaneous skin cancer in mice [85], it would be interesting to examine the relationship among Smad7, Wnt signaling, and skin cancer. On the other hand, enhanced Wnt signaling contributes to many different types of cancers. Therefore, the role of Smad7 in the process of tumor formation is very complicated and varies depending on tumor types and their microenvironments [86].
Conclusions
TGF-β is a cytokine of crucial importance that regulates diverse cellular and physiological processes, such as cell proliferation, differentiation, apoptosis, adhesion, and migration. To maintain the cellular or/and organic homeostasis, the TGF-β signaling pathway is precisely regulated at different levels. Dysfunction or deregulation of TGF-β signaling has been associated with different human diseases, such as fibrosis, inflammatory diseases, and tumorigenesis. Smad7 regulates TGF-β signaling via a negative feedback loop and mediates the crosstalk between TGF-β and other signaling pathways. Smad7 also plays an important role in pathological processes and has both anti-fibrotic and anti-inflammatory activities, suggesting that overexpression of Smad7 may have therapeutic potential to treat fibrosis and inflammation.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (30430360, 30671033, and 30600096), the Major State Basic Research Development Program of China (2004CB720002, 2006CB943401, and 2006CB910102), and the National High Technology Research and Development Program of China (2006AA02Z172).
Acknowledgements
The authors wish to apologize to the investigators whose outstanding work was not cited here because of space limitation. The authors would also like to thank Mr Martin Ting Ma for assisting in manuscript preparation.
References
- 1.Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nat Cell Biol. 2007;9:1000–1004. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 2.Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nunes I, Gleizes PE, Metz CN, Rifkin DB. Latent transforming growth factor-beta binding protein domains involved in activation and transglutaminase-dependent cross-linking of latent transforming growth factor-beta. J Cell Biol. 1997;136:1151–1163. doi: 10.1083/jcb.136.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ten Dijke P, Arthur HM. Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol. 2007;8:857–869. doi: 10.1038/nrm2262. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 7.Brown KA, Pietenpol JA, Moses HL. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J Cell Biochem. 2007;101:9–33. doi: 10.1002/jcb.21255. [DOI] [PubMed] [Google Scholar]
- 8.Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Nicholls JM, Chen YG. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-beta signaling. J Biol Chem. 2008;283:3272–3280. doi: 10.1074/jbc.M708033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGF-beta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 11.Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, Smith SM, et al. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 2007;26:3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, Zhang S, et al. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10:1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- 13.Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell. 2008;31:918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanyu A, Ishidou Y, Ebisawa T, Shimanuki T, Imamura T, Miyazono K. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J Cell Biol. 2001;155:1017–1027. doi: 10.1083/jcb.200106023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mochizuki T, Miyazaki H, Hara T, Furuya T, Imamura T, Watabe T, Miyazono K. Roles for the MH2 domain of Smad7 in the specific inhibition of transforming growth factor-beta superfamily signaling. J Biol Chem. 2004;279:31568–31574. doi: 10.1074/jbc.M313977200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Fei T, Zhang L, Zhang R, Chen F, Ning Y, Han Y, et al. Smad7 antagonizes transforming growth factor beta signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol Cell Biol. 2007;27:4488–4499. doi: 10.1128/MCB.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi X, Chen F, Yu J, Xu Y, Zhang S, Chen YG, Fang X. Study of interaction between Smad7 and DNA by single-molecule force spectroscopy. Biochem Biophys Res Commun. 2008;377:1284–1287. doi: 10.1016/j.bbrc.2008.10.145. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, et al. The MAD-related protein Smad7 associates with the TGF-beta receptor and functions as an antagonist of TGF-beta signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 19.Itoh S, ten Dijke P. Negative regulation of TGF-beta receptor/Smad signal transduction. Curr Opin Cell Biol. 2007;19:176–184. doi: 10.1016/j.ceb.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Shi W, Sun C, He B, Xiong W, Shi X, Yao D, Cao X. GADD34-PP1c recruited by Smad7 dephosphorylates TGF-beta type I receptor. J Cell Biol. 2004;164:291–300. doi: 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valdimarsdottir G, Goumans MJ, Itoh F, Itoh S, Heldin CH, ten Dijke P. Smad7 and protein phosphatase 1alpha are critical determinants in the duration of TGF-beta/ALK1 signaling in endothelial cells. BMC Cell Biol. 2006;7:16. doi: 10.1186/1471-2121-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hata A, Lagna G, Massague J, Hemmati-Brivanlou A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai S, Cao X. A nuclear antagonistic mechanism of inhibitory Smads in transforming growth factor-beta signaling. J Biol Chem. 2002;277:4176–4182. doi: 10.1074/jbc.M105105200. [DOI] [PubMed] [Google Scholar]
- 24.Bai S, Shi X, Yang X, Cao X. Smad6 as a transcriptional corepressor. J Biol Chem. 2000;275:8267–8270. doi: 10.1074/jbc.275.12.8267. [DOI] [PubMed] [Google Scholar]
- 25.Ichijo T, Voutetakis A, Cotrim AP, Bhattachryya N, Fujii M, Chrousos GP, Kino T. The Smad6-histone deacetylase 3 complex silences the transcriptional activity of the glucocorticoid receptor: potential clinical implications. J Biol Chem. 2005;280:42067–42077. doi: 10.1074/jbc.M509338200. [DOI] [PubMed] [Google Scholar]
- 26.Lin X, Liang YY, Sun B, Liang M, Shi Y, Brunicardi FC, Feng XH. Smad6 recruits transcription corepressor CtBP to repress bone morphogenetic protein-induced transcription. Mol Cell Biol. 2003;23:9081–9093. doi: 10.1128/MCB.23.24.9081-9093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datta PK, Moses HL. STRAP and Smad7 synergize in the inhibition of transforming growth factor beta signaling. Mol Cell Biol. 2000;20:3157–3167. doi: 10.1128/mcb.20.9.3157-3167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lallemand F, Seo SR, Ferrand N, Pessah M, L'Hoste S, Rawadi G, Roman-Roman S, et al. AIP4 restricts transforming growth factor-beta signaling through a ubiquitination-independent mechanism. J Biol Chem. 2005;280:27645–27653. doi: 10.1074/jbc.M500188200. [DOI] [PubMed] [Google Scholar]
- 29.Koinuma D, Shinozaki M, Komuro A, Goto K, Saitoh M, Hanyu A, Ebina M, et al. Arkadia amplifies TGF-beta superfamily signaling through degradation of Smad7. EMBO J. 2003;22:6458–6470. doi: 10.1093/emboj/cdg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, Rui H, Wang J, Lin S, He Y, Chen M, Li Q, et al. Axin is a scaffold protein in TGF-beta signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 2006;25:1646–1658. doi: 10.1038/sj.emboj.7601057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim BC, Lee HJ, Park SH, Lee SR, Karpova TS, McNally JG, Felici A, et al. Jab1/CSN5, a component of the COP9 signalosome, regulates transforming growth factor beta signaling by binding to Smad7 and promoting its degradation. Mol Cell Biol. 2004;24:2251–2262. doi: 10.1128/MCB.24.6.2251-2262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Song K, Krebs TL, Yang J, Danielpour D. Smad7 is inactivated through a direct physical interaction with the LIM protein Hic-5/ARA55. Oncogene. 2008;27:6791–6805. doi: 10.1038/onc.2008.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrigno O, Lallemand F, Verrecchia F, L'Hoste S, Camonis J, Atfi A, Mauviel A. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene. 2002;21:4879–4884. doi: 10.1038/sj.onc.1205623. [DOI] [PubMed] [Google Scholar]
- 34.Kowanetz M, Lonn P, Vanlandewijck M, Kowanetz K, Heldin CH, Moustakas A. TGF-beta induces SIK to negatively regulate type I receptor kinase signaling. J Cell Biol. 2008;182:655–662. doi: 10.1083/jcb.200804107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inamoto S, Iwata S, Inamoto T, Nomura S, Sasaki T, Urasaki Y, Hosono O, et al. Crk-associated substrate lymphocyte type regulates transforming growth factor-beta signaling by inhibiting Smad6 and Smad7. Oncogene. 2007;26:893–904. doi: 10.1038/sj.onc.1209848. [DOI] [PubMed] [Google Scholar]
- 36.Chen YG, Liu F, Massague J. Mechanism of TGF-beta receptor inhibition by FKBP12. EMBO J. 1997;16:3866–3876. doi: 10.1093/emboj/16.13.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi T, Kurisaki A, Yamakawa N, Minakuchi K, Sugino H. FKBP12 functions as an adaptor of the Smad7-Smurf1 complex on activin type I receptor. J Mol Endocrinol. 2006;36:569–579. doi: 10.1677/jme.1.01966. [DOI] [PubMed] [Google Scholar]
- 38.Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki S, Isshiki K, Isono M, et al. SIRT1 inhibits transforming growth factor beta-induced apoptosis in glomerular mesangial cells via Smad7 deacetylation. J Biol Chem. 2007;282:151–158. doi: 10.1074/jbc.M605904200. [DOI] [PubMed] [Google Scholar]
- 39.Simonsson M, Heldin CH, Ericsson J, Gronroos E. The balance between acetylation and deacetylation controls Smad7 stability. J Biol Chem. 2005;280:21797–21803. doi: 10.1074/jbc.M503134200. [DOI] [PubMed] [Google Scholar]
- 40.Gronroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10:483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- 41.Stopa M, Anhuf D, Terstegen L, Gatsios P, Gressner AM, Dooley S. Participation of Smad2, Smad3, and Smad4 in transforming growth factor beta (TGF-beta)-induced activation of Smad7. The TGF-beta response element of the promoter requires functional Smad binding element and E-box sequences for transcriptional regulation. J Biol Chem. 2000;275:29308–29317. doi: 10.1074/jbc.M003282200. [DOI] [PubMed] [Google Scholar]
- 42.Hua X, Miller ZA, Benchabane H, Wrana JL, Lodish HF. Synergism between transcription factors TFE3 and Smad3 in transforming growth factor-beta-induced transcription of the Smad7 gene. J Biol Chem. 2000;275:33205–33208. doi: 10.1074/jbc.C000568200. [DOI] [PubMed] [Google Scholar]
- 43.Brodin G, Ahgren A, ten Dijke P, Heldin CH, Heuchel R. Efficient TGF-beta induction of the Smad7 gene requires cooperation between AP-1, Sp1, and Smad proteins on the mouse Smad7 promoter. J Biol Chem. 2000;275:29023–29030. doi: 10.1074/jbc.M002815200. [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, Wei X, Zhu T, Zhang M, Shen R, Xing L, O'Keefe RJ, et al. Bone morphogenetic protein 2 activates Smad6 gene transcription through bone-specific transcription factor Runx2. J Biol Chem. 2007;282:10742–10748. doi: 10.1074/jbc.M610997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ku M, Howard S, Ni W, Lagna G, Hata A. OAZ regulates bone morphogenetic protein signaling through Smad6 activation. J Biol Chem. 2006;281:5277–5287. doi: 10.1074/jbc.M510004200. [DOI] [PubMed] [Google Scholar]
- 46.Denissova NG, Liu F. Repression of endogenous Smad7 by Ski. J Biol Chem. 2004;279:28143–28148. doi: 10.1074/jbc.M404961200. [DOI] [PubMed] [Google Scholar]
- 47.Park SH. Fine tuning and cross-talking of TGF-beta signal by inhibitory Smads. J Biochem Mol Biol. 2005;38:9–16. doi: 10.5483/bmbrep.2005.38.1.009. [DOI] [PubMed] [Google Scholar]
- 48.Quan T, He T, Voorhees JJ, Fisher GJ. Ultraviolet irradiation induces Smad7 via induction of transcription factor AP-1 in human skin fibroblasts. J Biol Chem. 2005;280:8079–8085. doi: 10.1074/jbc.M409647200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong S, Lim S, Li AG, Lee C, Lee YS, Lee EK, Park SH, et al. Smad7 binds to the adaptors TAB2 and TAB3 to block recruitment of the kinase TAK1 to the adaptor TRAF2. Nat Immunol. 2007;8:504–513. doi: 10.1038/ni1451. [DOI] [PubMed] [Google Scholar]
- 50.Choi KC, Lee YS, Lim S, Choi HK, Lee CH, Lee EK, Hong S, et al. Smad6 negatively regulates interleukin 1-receptor-Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat Immunol. 2006;7:1057–1065. doi: 10.1038/ni1383. [DOI] [PubMed] [Google Scholar]
- 51.Mazars A, Lallemand F, Prunier C, Marais J, Ferrand N, Pessah M, Cherqui G, et al. Evidence for a role of the JNK cascade in Smad7-mediated apoptosis. J Biol Chem. 2001;276:36797–36803. doi: 10.1074/jbc.M101672200. [DOI] [PubMed] [Google Scholar]
- 52.Edlund S, Bu S, Schuster N, Aspenstrom P, Heuchel R, Heldin NE, ten Dijke P, et al. Transforming growth factor-beta1 (TGF-beta1)-induced apoptosis of prostate cancer cells involves Smad7-dependent activation of p38 by TGF-beta-activated kinase 1 and mitogen-activated protein kinase kinase 3. Mol Biol Cell. 2003;14:529–544. doi: 10.1091/mbc.02-03-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwai T, Murai J, Yoshikawa H, Tsumaki N. Smad7 inhibits chondrocyte differentiation at multiple steps during endochondral bone formation and down-regulates p38 MAPK pathways. J Biol Chem. 2008;283:27154–27164. doi: 10.1074/jbc.M801175200. [DOI] [PubMed] [Google Scholar]
- 54.Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Smad7 is required for TGF-beta-induced activation of the small GTPase Cdc42. J Cell Sci. 2004;117:1835–1847. doi: 10.1242/jcs.01036. [DOI] [PubMed] [Google Scholar]
- 55.Edlund S, Lee SY, Grimsby S, Zhang S, Aspenstrom P, Heldin CH, Landstrom M. Interaction between Smad7 and beta-catenin: importance for transforming growth factor beta-induced apoptosis. Mol Cell Biol. 2005;25:1475–1488. doi: 10.1128/MCB.25.4.1475-1488.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han G, Li AG, Liang YY, Owens P, He W, Lu S, Yoshimatsu Y, et al. Smad7-induced beta-catenin degradation alters epidermal appendage development. Dev Cell. 2006;11:301–312. doi: 10.1016/j.devcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 57.Tang Y, Liu Z, Zhao L, Clemens TL, Cao X. Smad7 stabilizes beta-catenin binding to E-cadherin complex and promotes cell-cell adhesion. J Biol Chem. 2008;283:23956–23963. doi: 10.1074/jbc.M800351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez-Capelo A. Dual role for TGF-beta1 in apoptosis. Cytokine Growth Factor Rev. 2005;16:15–34. doi: 10.1016/j.cytogfr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Ohgushi M, Kuroki S, Fukamachi H, O'Reilly LA, Kuida K, Strasser A, Yonehara S. Transforming growth factor beta-dependent sequential activation of Smad, Bim, and caspase-9 mediates physiological apoptosis in gastric epithelial cells. Mol Cell Biol. 2005;25:10017–10028. doi: 10.1128/MCB.25.22.10017-10028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schuster N, Krieglstein K. Mechanisms of TGF-beta-mediated apoptosis. Cell Tissue Res. 2002;307:1–14. doi: 10.1007/s00441-001-0479-6. [DOI] [PubMed] [Google Scholar]
- 61.Flanders KC. Smad3 as a mediator of the fibrotic response. Int J Exp Pathol. 2004;85:47–64. doi: 10.1111/j.0959-9673.2004.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W, Koka V, Lan HY. Transforming growth factor-beta and Smad signalling in kidney diseases. Nephrology (Carlton) 2005;10:48–56. doi: 10.1111/j.1440-1797.2005.00334.x. [DOI] [PubMed] [Google Scholar]
- 63.Lan HY. Smad7 as a therapeutic agent for chronic kidney diseases. Front Biosci. 2008;13:4984–4992. doi: 10.2741/3057. [DOI] [PubMed] [Google Scholar]
- 64.Wang B, Omar A, Angelovska T, Drobic V, Rattan SG, Jones SC, Dixon IM. Regulation of collagen synthesis by inhibitory Smad7 in cardiac myofibroblasts. Am J Physiol Heart Circ Physiol. 2007;293:H1282–H1290. doi: 10.1152/ajpheart.00910.2006. [DOI] [PubMed] [Google Scholar]
- 65.Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 67.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 68.Monteleone G, Pallone F, MacDonald TT. Smad7 in TGF-beta-mediated negative regulation of gut inflammation. Trends Immunol. 2004;25:513–517. doi: 10.1016/j.it.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 69.Monteleone G, Kumberova A, Croft NM, McKenzie C, Steer HW, MacDonald TT. Blocking Smad7 restores TGF-beta1 signaling in chronic inflammatory bowel disease. J Clin Invest. 2001;108:601–609. doi: 10.1172/JCI12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monteleone G, Mann J, Monteleone I, Vavassori P, Bremner R, Fantini M, Del Vecchio Blanco G, et al. A failure of transforming growth factor-beta1 negative regulation maintains sustained NF-kappaB activation in gut inflammation. J Biol Chem. 2004;279:3925–3932. doi: 10.1074/jbc.M303654200. [DOI] [PubMed] [Google Scholar]
- 71.Wang W, Huang XR, Li AG, Liu F, Li JH, Truong LD, Wang XJ, et al. Signaling mechanism of TGF-beta1 in prevention of renal inflammation: role of Smad7. J Am Soc Nephrol. 2005;16:1371–1383. doi: 10.1681/ASN.2004121070. [DOI] [PubMed] [Google Scholar]
- 72.Ng YY, Hou CC, Wang W, Huang XR, Lan HY. Blockade of NF-kappaB activation and renal inflammation by ultrasound-mediated gene transfer of Smad7 in rat remnant kidney. Kidney Int Suppl. 2005;94:S83–S91. doi: 10.1111/j.1523-1755.2005.09421.x. [DOI] [PubMed] [Google Scholar]
- 73.Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, Lubbe S, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 74.Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massague J, et al. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Javelaud D, Mohammad KS, McKenna CR, Fournier P, Luciani F, Niewolna M, Andre J, et al. Stable overexpression of Smad7 in human melanoma cells impairs bone metastasis. Cancer Res. 2007;67:2317–2324. doi: 10.1158/0008-5472.CAN-06-3950. [DOI] [PubMed] [Google Scholar]
- 76.Javelaud D, Delmas V, Moller M, Sextius P, Andre J, Menashi S, Larue L, et al. Stable overexpression of Smad7 in human melanoma cells inhibits their tumorigenicity in vitro and in vivo. Oncogene. 2005;24:7624–7629. doi: 10.1038/sj.onc.1208900. [DOI] [PubMed] [Google Scholar]
- 77.Cerutti JM, Ebina KN, Matsuo SE, Martins L, Maciel RM, Kimura ET. Expression of Smad4 and Smad7 in human thyroid follicular carcinoma cell lines. J Endocrinol Invest. 2003;26:516–521. doi: 10.1007/BF03345213. [DOI] [PubMed] [Google Scholar]
- 78.Dowdy SC, Mariani A, Reinholz MM, Keeney GL, Spelsberg TC, Podratz KC, Janknecht R. Overexpression of the TGF-beta antagonist Smad7 in endometrial cancer. Gynecol Oncol. 2005;96:368–373. doi: 10.1016/j.ygyno.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 79.Mikula M, Proell V, Fischer AN, Mikulits W. Activated hepatic stellate cells induce tumor progression of neoplastic hepatocytes in a TGF-beta dependent fashion. J Cell Physiol. 2006;209:560–567. doi: 10.1002/jcp.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Azuma H, Ehata S, Miyazaki H, Watabe T, Maruyama O, Imamura T, Sakamoto T, et al. Effect of Smad7 expression on metastasis of mouse mammary carcinoma JygMC(A) cells. J Natl Cancer Inst. 2005;97:1734–1746. doi: 10.1093/jnci/dji399. [DOI] [PubMed] [Google Scholar]
- 81.Fidler IJ. Blockade of the TGF-beta superfamily by Smad7 breaking a link in the metastatic chain. J Natl Cancer Inst. 2005;97:1714–1715. doi: 10.1093/jnci/dji437. [DOI] [PubMed] [Google Scholar]
- 82.Halder SK, Beauchamp RD, Datta PK. Smad7 induces tumorigenicity by blocking TGF-beta-induced growth inhibition and apoptosis. Exp Cell Res. 2005;307:231–246. doi: 10.1016/j.yexcr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 83.Kleeff J, Ishiwata T, Maruyama H, Friess H, Truong P, Buchler MW, Falb D, et al. The TGF-beta signaling inhibitor Smad7 enhances tumorigenicity in pancreatic cancer. Oncogene. 1999;18:5363–5372. doi: 10.1038/sj.onc.1202909. [DOI] [PubMed] [Google Scholar]
- 84.Liu X, Lee J, Cooley M, Bhogte E, Hartley S, Glick A. Smad7 but not Smad6 cooperates with oncogenic ras to cause malignant conversion in a mouse model for squamous cell carcinoma. Cancer Res. 2003;63:7760–7768. [PubMed] [Google Scholar]
- 85.Niemann C, Owens DM, Hulsken J, Birchmeier W, Watt FM. Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development. 2002;129:95–109. doi: 10.1242/dev.129.1.95. [DOI] [PubMed] [Google Scholar]
- 86.Bornstein S, Hoot K, Han GW, Lu SL, Wang XJ. Distinct roles of individual Smads in skin carcinogenesis. Mol Carcinog. 2007;46:660–664. doi: 10.1002/mc.20336. [DOI] [PubMed] [Google Scholar]


