Abstract
Understanding viral factors that promote cross-species transmission is important for evaluating the risk of zoonotic emergence. Weconstructed a database of viruses of domestic artiodactyls and examined the correlation between traits linked in the literature to cross-species transmission and the ability of viruses to infect humans. Among these traits-genomic material, genome segmentation, and replication without nuclear entry-the last is the strongest predictor of cross-species transmission. This finding highlights nuclear entry as a barrier to transmission and suggests that the ability to complete replication in the cytoplasm may prove to be a useful indicator of the threat of cross-species transmission.
Many emerging infections are caused by viruses that are transmitted between host species. Specific characteristics may preadapt some viral groups for cross-species transmission. If we can identify such characteristics, we can better prepare for future viral threats to humans, domestic animals, and wildlife that will emerge as interactions between potential host species change.
Previous studies have compared emerging human pathogens to nonemerging human pathogens and looked for characteristics typical of those considered to be emerging [1–3]. To ask which characteristics predict host jumps requires a different approach. Specifically, we must examine the pool of other hosts' pathogens that a target species regularly encounters. From this pool we can compare the characteristics of microbes that are able to infect the target host versus those that manifest no evidence of an ability to infect the target host. Molecular characteristics that facilitate cross-species transmission are likely to be substantially different between viruses, bacteria, and protozoa, because of large differences in the pathobiology of these different taxa.
Here, we focus on cross-species transmission of viral infections and examine the effects of 3 characteristics that are described in the literature as expected to affect the ability of a viral group to infect a novel host species: genome segmentation, genomic material, and site of replication. The ability to rapidly explore genetic state space is expected to increase the probability of a host jump, so we expect that viruses with RNA genomes will have a higher probability of jumping than viruses with DNA genomes [1, 4] and that viruses with segmented genomes will have a higher probability of jumping than viruses with nonsegmented genomes [4]. Complex interactions with a host's cellular machinery, on the other hand, are expected to decrease the probability of a host jump, so we expect that viruses that are able to complete replication in the cytoplasm will have a higher probability of jumping than viruses requiring nuclear entry [5].
To examine the effects of these characteristics, we should choose a target species that will maximize the chance that viral infection due to cross-species transmission events will have been detected; the obvious choice is humans. Likewise, we should minimize differences in exposure of the target host to infectious virions produced by the source hosts. Humans have regular contact with all potentially infectious bodily fluids of domestic food animals; we thus ensure that the target species has contact with all viral groups infecting the source hosts by analyzing the pool of viral species known to infect sheep, goats, cattle, and pigs.
Methods. We constructed a database containing taxonomic and molecular data on known viruses of domestic artiodactyls. To determine which viruses to include in the database, we searched the primary literature for references documenting infection of these species with all recognized species in all viral genera known to infect mammals. For each viral species infecting sheep, goats, pigs, or cattle, we then searched the literature to determine whether human infections have been documented (see table A 1 in appendix A, which appears only in the electronic edition of the Journal). Viruses dependent on coinfection with other viral species, known to be maintained through continuous transmission within humans (see appendix A), or for which documented instances of artiodactyl infection resulted from human-to-animal transmission or experimental infection were excluded from the database. All literature searches were performed between 10 January 2007 and 15 February 2007 using Web of Science.
Table 1.
Comparison of logistic regression models.
The database contains information on the 3 molecular characteristics hypothesized to influence the potential of a virus to cross host species: site of replication (XSR; whether replication is completed in the cytoplasm or requires nuclear entry), genomic material (XGM; RNA or DNA), and segmentation of the viral genome (XSeg; segmented or nonsegmented). These characteristics are conserved at the family level, and classifications were made on the basis of standard reference books [6, 7].
We used a combination of hypothesis testing and modelbased prediction to analyze the database. Hypothesis testing allowed us to determine how likely it was that the observed patterns were due to chance, whereas model-based prediction allowed us to determine what trait or set of traits was the best predictor of a livestock virus's ability to infect humans and to estimate the probability that a particular virus species would be able to jump host species, given knowledge of the traits of interest. Computer code and data are available at http://lalashan.mcmaster.ca/hostjumps/ or from the authors.
To determine the statistical significance of the effect of each trait on zoonotic transmission independent of the 2 other traits of interest, we performed a series of randomization tests. For a particular trait, we held the values of the other 2 traits and the ability of the viral species in the database to infect humans constant and permuted the values of the trait of interest within each subset defined by the other 2 traits (thereby preserving the full cross-correlational structure of the data with regard to the 3 viral traits) 100,000 times. The P value was given by the proportion of permutations that allowed the model to predict outcomes as well as or better than the model that was constructed using the observed data, and an α level of 5% was used to determine the statistical significance of results. We compared model fit by use of a logistic regression model that predicted the ability to infect humans as a function of replication site, genomic material, and segmentation. The logistic model was fit in the R statistics package [8], and fits were compared on the basis of likelihood.
Because the 3 traits examined are conserved at the family level for all species in our database, treating species as independent may bias our results. We therefore repeated our analysis at the genus and family levels. Permutations of the data set were constructed by permuting the values of the trait under consideration at the taxonomic level examined and assigning species within a genus (or family) the corresponding trait value after permutation. P values were calculated as in the species-level analysis.
To examine the magnitude and relative importance of the effects that the 3 molecular characteristics of interest have on the ability of the viral species in the database to infect humans, we developed a set of logistic regression models. Each model included some combination of viral traits as independent variables and the ability to infect humans as the dependent variable. Traits not having a statistically significant effect on the ability of livestock viruses to infect humans were still considered for modelbased predictions, because sample sizes were limited and small-but real-effects may not be detected via hypothesis testing. We estimated parameter values for each model in R and compared models using Akaike's information criterion adjusted for small sample size (AICc) [9].
Results. A total of 146 viral species were found to infect the livestock species of interest and meet other criteria for inclusion in the database. Of these, 141 species (representing 59 genera in 22 families) fulfilled the criteria for inclusion in the analysis.
The effect of site of replication was significant at all 3 taxonomic levels examined (P < .001, P = .018, and P = .014 for the species, genus, and family levels, respectively), with nearly half of the virus species completing replication in the cytoplasm able to infect humans. Neither genomic material nor segmentation showed a significant effect on the ability of livestock viruses to infect humans at any taxonomic level.
Logistic regression model comparisons are summarized in table 1. The models are given in order as ranked by AICc. figure 1 compares the observed data with the results of the best model. The best model included site of replication as the only variable (odds ratio, 17.4 [95% confidence interval, 3.98–75.8), and the top 4 models were the 4 that included site of replication. Each of these models showed a positive correlation between replication in the cytoplasm and the ability to infect humans, as expected. Segmentation appears in models 2, 4, 6 and 7, and all 4 models showed a positive correlation between having a segmented genome and the ability to infect humans. Genomic material appears in models 3, 4, 5, and 6. Again, all 4 models showed a correlation in the expected direction.
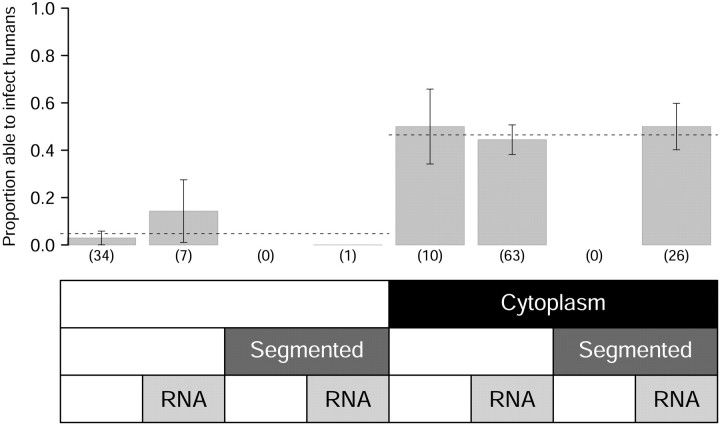
Figure 1.
Comparison of data and model predictions. Gray bars show the proportion of livestock viruses in each category that are known to infect humans. Dashed lines give the prediction of the best regression model (as determined by Akaike's information criterion adjusted for small sample size), which includes site of replication (XSR) as the only variable. Sample sizes are given in parentheses below each bar. The ability to complete replication within the cytoplasm is the single best predictor of whether livestock viruses infect humans. Error bars represent 95% exact binomial confidence intervals.
It is interesting to note that both of the viral species that caused major pandemics in humans in the 20th century (HIV and influenza virus A) require nuclear entry for replication. Because influenza virus A infects domestic artiodactyls but was excluded from our database because it is maintained through continuous transmission in humans, we confirmed the robustness of our results to this exclusion; we also confirmed that our findings were robust to the inclusion of viral species for which human infection data were based solely on serology (see table B1 in appendix B, which appears only in the electronic edition of the Journal).
Discussion. Our analyses indicate that viral species infecting domestic artiodactyls are more likely to infect humans if they complete replication in the cytoplasm without nuclear entry. The observed effect of cytoplasmic replication on host-jumping ability is not surprising given the complex molecular pathways regulating nuclear entry. Viral species that are unable to complete replication in the cytoplasm require intracellular transport from the site of penetration, targeting of the nucleus through nuclear localization signals, and importation of genetic material, proteins, and/or whole virions through the nuclear pore complex [10]. The combination of molecular mechanisms governing this chain of events is likely to be highly host specific, because of strong selective pressure against admission of foreign particles into the nucleus. To date, discussion of barriers to viral replication has largely focused on receptors for cellular entry. The concentration on this aspect of the viral life cycle exists for 2 substantive reasons. First, the inability to enter a cell obviously precludes viral replication; second, several well-documented viral host jumps have been shown to occur after point mutations that modify interactions between viral particles and cellular receptors [11–13]. The effect of nuclear entry seen in our data set emphasizes that cellular entry, while a necessary step, is insufficient for completion of the viral life cycle.
The ability to produce genetic diversity is the factor most widely discussed as expected to increase viral host-jumping ability [1, 3–5, 14]. Although the observed effects of genomic material and segmentation were not statistically significant, our data do not necessarily contradict this expectation. The hypothesized effect of segmentation, in particular, may be obscured in our data set by a combination of the small number of viral species with segmented genomes and the absence of segmented DNA viruses. On the other hand, the lack of predictive power associated with genomic material and segmentation in our data set may indicate that consideration of these traits alone is insufficient to capture the potential to generate useful genetic diversity.
The degree to which the pool of viruses infecting domestic artiodactyls is typical of all potentially zoonotic viral species is uncertain. Other pools of viral species should be examined to determine the generality of our results. Similarly, further studies should examine whether the observed patterns hold for crossspecies transmission of viruses to other target host species, including wildlife and domestic animals.
Given the rapid rates at which ecological relationships between species are changing as a result of anthropogenic landscape changes, global warming, and globalization of both human and animal populations, the development of indicators of the risk of cross-species pathogen transmission is an increasingly important goal. As humans, domestic animals, and wildlife are brought into contact with species from which they were formerly isolated, they inevitably encounter the pathogens that these species carry. The finding that the ability to complete replication in the cytoplasm is the best predictor of zoonotic transmission and that nearly half of domestic artiodactyl viruses that are able to complete replication in the cytoplasm can infect humans suggests that cytoplasmic replication will be a useful indicator of the ability of a newly encountered virus species to jump hosts, an essential prerequisite to epidemic or pandemic emergence [15]. It should be noted, however, that the present analysis focused exclusively on the ability to infect the target host, and the viral traits influencing this step in the emergence process may differ from those that predispose a virus to cause severe disease in a novel host as well as from those that facilitate transmission within a novel host species.
Acknowledgments
We thank L. Enquist for invaluable discussion and comments on the manuscript and B. Bolker, S. Bellan, M. Levy, S. Levin, A. Dobson, K. Hampson, P. Hosseini, D. Wilcove, R. Pulliam, and the Princeton Disease Cabal for comments, discussion, and support.
References
- 1.Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: pathogen characteristics, host range, and the risk of emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:991–9. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor LH, Lathan SM, Woolhouse MEJ. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:983–9. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and reemerging infectious diseases. Emerging Infect Dis. 2005;11:1842–7. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke DS. Evolvability of emerging viruses. In: Nelson A, Horsburg CJ, editors. Pathology of emerging infections. Vol. 2. Washington, DC: American Society for Microbiology Press; 1998. pp. 1–12. [Google Scholar]
- 5.Pulliam JRC. Viral host jumps: moving toward a predictive framework. EcoHealth. 2008;5:80–91. doi: 10.1007/s10393-007-0149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Berlin: Academic Press; 2005. Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. [Google Scholar]
- 7.Tidona CA, Darai G. San Diego: Springer-Verlag; 2001. The Springer index of viruses. [Google Scholar]
- 8.R Development Core Team . Vienna: R Foundation for Statistical Computing; 2007. R: a language and environment for statistical computing. [Google Scholar]
- 9.Burnham KP, Anderson DR. In: Model selection and multimodel inference: a practical information-theoretic approach. 2nd ed. Vol. 149. New York: Springer; 2002. Formal inference from more than one model: multimodel inference (MMI) p. 205. [Google Scholar]
- 10.Whittaker GR, Kahn M, Helenius A. Viral entry into the nucleus. Annu Rev Cell Dev Biol. 2000;16:627–51. doi: 10.1146/annurev.cellbio.16.1.627. [DOI] [PubMed] [Google Scholar]
- 11.Hueffer KD, Parker JSL, Weichert WS, Geisel RE, Sgro J-Y, Parrish CR. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J Virol. 2003;77:1718–26. doi: 10.1128/JVI.77.3.1718-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–8. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 13.Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–4. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webby R, Hoffman E, Webster R. Molecular constraints to interspecies transmission of viral pathogens. Nat Med. 2004;10(Suppl 12):S77–81. doi: 10.1038/nm1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–83. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]