Abstract
Deposition of H2A.Z in chromatin is known to be mediated by a conserved SWR1 chromatin‐remodeling complex in eukaryotes. However, little is known about whether and how the SWR1 complex cooperates with other chromatin regulators. Using immunoprecipitation followed by mass spectrometry, we found all known components of the Arabidopsis thaliana SWR1 complex and additionally identified the following three classes of previously uncharacterized plant‐specific SWR1 components: MBD9, a methyl‐CpG‐binding domain‐containing protein; CHR11 and CHR17 (CHR11/17), ISWI chromatin remodelers responsible for nucleosome sliding; and TRA1a and TRA1b, accessory subunits of the conserved NuA4 histone acetyltransferase complex. MBD9 directly interacts with CHR11/17 and the SWR1 catalytic subunit PIE1, and is responsible for the association of CHR11/17 with the SWR1 complex. MBD9, TRA1a, and TRA1b function as canonical components of the SWR1 complex to mediate H2A.Z deposition. CHR11/17 are not only responsible for nucleosome sliding but also involved in H2A.Z deposition. These results indicate that the association of the SWR1 complex with CHR11/17 may facilitate the coupling of H2A.Z deposition with nucleosome sliding, thereby co‐regulating gene expression, development, and flowering time.
Keywords: chromatin remodeling, H2A.Z, ISWI, MBD9, SWR1
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Plant Biology
Proteomic analysis of Arabidopsis thaliana SWR1 complex components implicates nucleosome‐sliding ISWI remodeler subunits CHR11 and CHR17 also in histone deposition.

Introduction
In eukaryotes, genomic DNA is packaged into chromatin within the nucleus. The fundamental packaging unit is the nucleosome, which consists of a histone octamer wrapped by 147 base pairs of DNA. Although the formation of the nucleosome facilitates packaging of genomic DNA into the limited space of the nucleus, it blocks accessibility of nuclear factors to DNA, thus reducing the ability of these factors to regulate transcription, DNA replication, and repair (Khorasanizadeh, 2004; Luger et al, 2012; Li et al, 2017). To counteract the repressive effect of the nucleosome, eukaryotic organisms have evolved histone variants, DNA and histone modifiers, and chromatin remodelers (Clapier & Cairns, 2009; March‐Díaz & Reyes, 2009; Han et al, 2015). H2A.Z is a highly conserved histone variant and is required for the regulation of gene expression, DNA repair, and recombination (Choi et al, 2013; Rosa et al, 2013; Jarillo & Piñeiro, 2015). In Arabidopsis thaliana, H2A.Z is involved in the regulation of development, flowering time, meiotic crossover, DNA repair, and biotic and abiotic stress responses (March‐Díaz & Reyes, 2009; Kumar & Wigge, 2010; Coleman‐Derr & Zilberman, 2012; Jarillo & Piñeiro, 2015). Therefore, investigation of the mechanism of H2A.Z deposition in chromatin should increase our understanding of how H2A.Z is involved in the regulation of diverse biological processes.
In eukaryotes, the ATPase domain‐containing chromatin‐remodeling factors are classified into four families: SWI/SNF, ISWI, CHD, and INO80 (Clapier & Cairns, 2009). In yeast, SWR1, a member of the INO80 family, interacts with accessory subunits and forms a multi‐subunit SWR1 complex (Mizuguchi et al, 2004; Clapier & Cairns, 2009; Kapoor & Shen, 2014). The SWR1 complex is responsible for exchanging the H2A/H2B dimer of the nucleosome with the free H2A.Z/H2B dimer, thereby depositing H2A.Z into the nucleosome (March‐Díaz & Reyes, 2009; Bao & Shen, 2011). In genic regions, H2A.Z is deposited by the SWR1 complex predominantly in the +1 nucleosome near the transcription start site and to a lesser extent in the downstream nucleosomes (Raisner et al, 2005; Albert et al, 2007; Zilberman et al, 2008). H2A.Z in the +1 nucleosome is known to poise repressive genes for transcriptional activation (Zhang et al, 2005; Talbert & Henikoff, 2014; Sura et al, 2017). Conserved multi‐subunit SWR1/SRCAP complexes have been identified and characterized in yeast and metazoans (Clapier & Cairns, 2009; Kapoor & Shen, 2014). Most of the conserved SWR1 components identified in yeast and metazoans have also been found in Arabidopsis (March‐Díaz & Reyes, 2009; Jarillo & Piñeiro, 2015), suggesting that the SWR1 complexes are conserved in eukaryotes.
In the Arabidopsis SWR1 complex, PIE1 is the ATPase domain‐containing chromatin‐remodeling factor (Noh & Amasino, 2003). Loss‐of‐function mutation of PIE1 leads to pleiotropic developmental abnormalities, which include smaller plants, short siliques, reduced fertility, and earlier flowering (Choi et al, 2007; Deal et al, 2007; March‐Díaz et al, 2007). Depletion of accessory subunits of the SWR1 complex causes similar (although weaker) developmental defects (Kandasamy et al, 2005; Choi et al, 2007; Deal et al, 2007; March‐Díaz et al, 2007), suggesting that, as in other eukaryotes, the accessory subunits are required for the function of the SWR1 complex in Arabidopsis. According to protein–protein interaction assays, the C terminus of PIE1 binds to the accessory subunits SWC2, SWC6, and ARP6, while the N terminus of PIE1 binds to H2A.Z (Choi et al, 2007; March‐Díaz et al, 2007). The interaction of these SWR1 components in Arabidopsis is comparable to that in yeast (Wu et al, 2009; Nguyen et al, 2013), suggesting that the architecture of the SWR1 complexes is probably conserved in eukaryotes. Although most of the SWR1 components are conserved, some SWR1 components do differ between yeast and metazoans (Clapier & Cairns, 2009; Kapoor & Shen, 2014). Research is needed to determine whether there are non‐conserved SWR1 components that are exclusively present in plants.
ISWI, a class of conserved ATP‐dependent chromatin remodelers, mediates nucleosome sliding and thus enables dynamic accessibility of packaged DNA in eukaryotes (Gkikopoulos et al, 2011; Li et al, 2017). An ISWI protein typically interacts with 1–3 accessory subunits and forms an ISWI complex (Yadon & Tsukiyama, 2011). In Arabidopsis, CHR11 and CHR17 (CHR11/17), two closely related ISWI chromatin remodelers, function redundantly and are required for gametic and zygotic development (Huanca‐Mamani et al, 2005; Li et al, 2012; Smaczniak et al, 2012). A family of DTT domain‐containing proteins interact with CHR11/17 and thus form ISWI complexes (Li et al, 2012; Dong et al, 2013). The closely related DTT domain‐containing proteins RLT1 and RLT2 function together with CHR11/17 and are required for the regulation of development and flowering time (Li et al, 2012). Loss‐of‐function mutations of CHR11/17 result in loss of the evenly spaced nucleosome pattern in the gene body, suggesting that CHR11/17 function in positioning nucleosomes in an evenly spaced array (Li et al, 2014). The evenly spaced nucleosome array starts from the +1 nucleosome, in which H2A.Z is predominantly deposited by the SWR1 complex (Raisner et al, 2005; Albert et al, 2007; Zilberman et al, 2008; Dai et al, 2017). However, it is largely unknown how H2A.Z deposition by the SWR1 complex and nucleosome positioning by the ISWI complex are coordinated.
Here, we identified all components of the Arabidopsis SWR1 complex by immunoprecipitation followed by mass spectrometry (IP‐MS). We found not only conserved SWR1 components but also three classes of previously uncharacterized plant‐specific SWR1 components including the methyl‐CpG‐binding domain‐containing protein MBD9, the ISWI chromatin‐remodeling factors CHR11/17, and the conserved subunits of the NuA4 complex TRA1a and TRA1b (TRA1a/1b). MBD9 and TRA1a/1b promote H2A.Z deposition at the whole‐genome level, whereas CHR11/17 are involved in H2A.Z deposition at a subset of H2A.Z target genes. As determined by RNA‐seq, MBD9, CHR11/17, and other SWR1 components co‐regulate the expression of a number of genes, supporting the notion that these components function together in the SWR1 complex. Furthermore, we demonstrated that CHR11/17 but not PIE1 are required for nucleosome sliding in the gene body. These results suggest that association of the SWR1 complex with CHR11/17 may facilitate the coordination between H2A.Z deposition and nucleosome sliding and thus ensure proper expression of their common target genes.
Results
Identification and characterization of the Arabidopsis SWR1 complex
To investigate how H2A.Z is incorporated into chromatin in Arabidopsis, we generated transgenic plants expressing the H2A.Z variant HTA9 tagged by a Flag epitope at its C terminus, and performed IP‐MS to identify H2A.Z‐associated proteins. In addition to detecting all previously known SWR1 components, we identified the following previously unknown H2A.Z‐associated proteins: MBD9, a methyl‐CpG‐binding domain‐containing protein; CHR11 and CHR17, ISWI chromatin remodelers responsible for nucleosome sliding; and TRA1a and TRA1b, accessory subunits of the conserved NuA4 histone acetyltransferase complex (Fig 1A; Dataset EV1). To determine whether these newly identified proteins are components of the Arabidopsis SWR1 complex, we generated transgenic plants expressing the known SWR1 components PIE1, RIN1 (i.e., RVB1), RIN2 (i.e., RVB2A), SWC2, and GAS41 (i.e., YAF9A) tagged by an epitope. The developmental defect in the pie1 mutant was complemented by the PIE1 transgene tagged by Flag, suggesting that the PIE1‐Flag transgene functions in vivo (Fig EV1A). By subjecting the transgenic plants to IP‐MS, we found that MBD9, CHR11, CHR17, TRA1a, and TRA1b were co‐precipitated with the known SWR1 components (Fig 1A and B; Appendix Fig S1). We then generated transgenic plants expressing MBD9, CHR11, and CHR17 tagged by Flag for IP‐MS. The biological function of the MBD9‐Flag and CHR11‐Flag transgenes was confirmed by complementation testing (Fig EV1B–E). Our IP‐MS result indicated that the known SWR1 components were also co‐precipitated with MBD9, CHR11, and CHR17 (Fig 1A and B; Appendix Fig S1), suggesting that these proteins are new components of the SWR1 complex. The interaction of MBD9 with H2A.Z, PIE1, and CHR11/17 was demonstrated by co‐immunoprecipitation (Fig 1C). These results indicate that MBD9, CHR11/17, and TRA1a/1b are subunits of the SWR1 complex in Arabidopsis. Considering that homologs of these proteins are not components of the SWR1 complexes in yeast and metazoans, we suspect that they function as plant‐specific components of the SWR1 complex.
Figure 1. Identification of all components of the SWR1 complex in Arabidopsis .

- The list of co‐precipitated proteins as determined by immunoprecipitation followed by mass spectrometry (IP‐MS). The SWR1 subunits not shared by the ISWI complexes are shown in blue; the DDT domain‐containing ISWI‐specific subunits are shown in orange; CHR11 and CHR17 are shared subunits of the SWR1 and ISWI complexes and are shown in green.
- Schematic representation of the protein–protein interaction as determined by IP‐MS. Red lines represent the interaction among the SWR1 components; gray lines indicate the interaction among the ISWI components. Arrows point to the interaction proteins that were identified by IP‐MS.
- Determination of the interaction of MBD9 with HTA9, HTA11, PIE1, CHR11, and CHR17 by co‐immunoprecipitation.
Figure EV1. The complementation test of the SWR1 complex mutants mbd9‐3, pie1, and chr11/17 .
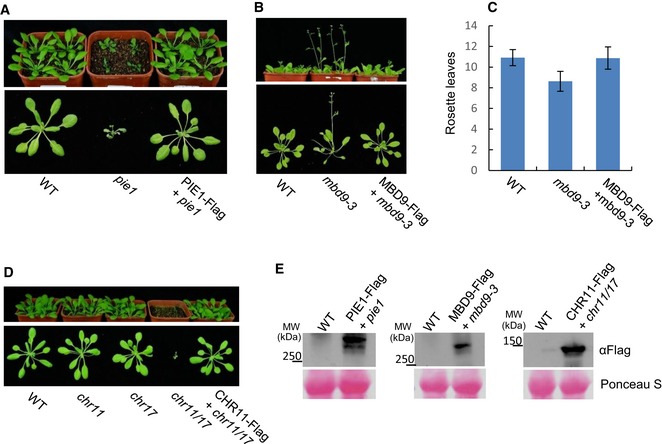
- The complementation test of the pie1 mutant. The top and bottom panels show morphological phenotypes of 24‐day‐old plants in the soil and on the black cloth, respectively.
- The complementation test of the mbd9‐3 mutant. Shown are morphological phenotypes of 24‐day‐old plants in the soil (the top panel) and on the black cloth (the bottom panel).
- Numbers of rosette leaves of the WT, mbd9‐3, and MBD9‐Flag transgenic plants in the mbd9‐3 background. At least 20 plants of each genotype were counted. Values are mean ± SD.
- The complementation test of the chr11/17 mutant. The developmental defect of the chr11/17 mutant was complemented by CHR11‐Flag. The top and bottom panels show the phenotypes in the soil and on the black cloth, respectively.
- Verification of the expression of MBD9‐Flag, PIE1‐Flag, and CHR11‐Flag in each transgenic line. The top panel is the Western blot results of the expression of these three transgenes. The bottom is the corresponding Ponceau S staining loading control.
In yeast and metazoans, TRA1a/1b homologs are subunits of the NuA4/TIP60 histone acetyltransferase complexes but not of the SWR1/SRCAP complexes even though several subunits are shared by the two types of complexes (Clapier & Cairns, 2009; Kapoor & Shen, 2014), suggesting that TRA1a/1b function as subunits of the SWR1 complex exclusively in plants. CHR11/17 are catalytic subunits of the ATP‐dependent ISWI chromatin‐remodeling complexes (Li et al, 2017). A family of DDT domain‐containing proteins interact with CHR11/17 and function as accessory subunits of the ISWI complexes in Arabidopsis (Dong et al, 2013). We found that, while the DDT domain‐containing proteins were co‐precipitated with CHR11 and CHR17, none of the DDT domain‐containing proteins were co‐precipitated with MBD9 or the other SWR1 components (Fig 1A and B). We also performed IP‐MS for RLT2, a well‐known DDT domain‐containing ISWI complex subunit, and found that CHR11/17 but not the other SWR1 components were co‐precipitated with RLT2 (Fig 1A and B). These results suggest that, in the ISWI complex, the catalytic subunits CHR11 and CHR17 but not the DDT domain‐containing accessory subunits function as components of the SWR1 complex in Arabidopsis.
MBD9 interacts with PIE1 and CHR11/17 and is responsible for the association of the SWR1 complex with CHR11/17
We performed yeast two‐hybrid (Y2H) assays to determine how MBD9 and CHR11/17 interact with known SWR1 components. A number of protein–protein interactions were identified among the known SWR1 components (Fig 2A; Appendix Figs S2–S4). Although some of the interactions, such as PIE1‐SWC6, PIE1‐HTA9, SWC6‐SWC2, SWC6‐ARP6, and SWC6‐SWC4, were previously identified (Choi et al, 2007; March‐Díaz et al, 2007; Gómez‐Zambrano et al, 2018), most of the interactions identified in this study had not been previously reported. In particular, we identified the interaction of PIE1 with SWC2, ARP6, RIN1, and RIN2 (Fig 2A; Appendix Figs S3 and S4), supporting the notion that the SWR1 catalytic subunit functions as a scaffold for the assembly of the SWR1 complex not only in yeast and metazoans but also in plants (Wu et al, 2009; Jarillo & Piñeiro, 2015). Different from our study reporting the PIE1–SWC2 interaction, a previous study was unable to detect the interaction because of some uncertain reasons (Choi et al, 2007). More importantly, we observed that MBD9 interacts with PIE1 and CHR11/17 but not with the other SWR1 components, whereas CHR11/17 interact only with MBD9 (Fig 2A; Appendix Figs S3–S5). We then expressed and purified epitope‐tagged MBD9, PIE1, and CHR11 and performed in vitro pull‐down assays to assess how these proteins interact with each other (Fig 2B and C; Appendix Fig S6). We found that MBD9 directly interacts with PIE1 and CHR11, whereas PIE1 and CHR11 do not directly interact (Fig 2B and C). The interaction between PIE1 and CHR11 was observed when MBD9 was added to the pull‐down system (Fig 2B). Based on these analyses, we predict that MBD9 acts as a bridge protein between PIE1 and CHR11/17 in the SWR1 complex.
Figure 2. CHR11 and CHR17 are recruited to the SWR1 complex by MBD9.

-
ASchematic representation of the interaction of SWR1 components as determined by Y2H assays. Proteins shown in blue are the SWR1 subunits that are not shared by the ISWI complex; the ISWI catalytic subunits CHR11 and CHR17 are shown in green.
-
B, CDetermination of the interaction of PIE1‐Flag, MBD9‐Myc, and CHR11‐HA by in vitro pull‐down assays. PIE1‐Flag, MBD9‐Myc, and CHR11‐HA were expressed from yeast and subjected to the pull‐down assays. Anti‐Flag agarose (B) and anti‐HA (C) agarose were independently used in the pull‐down assays.
-
DThe effect of the pie1 and mbd9‐1 mutations on the formation of the SWR1 complex as determined by IP‐MS. Transgenic plants expressing Flag‐tagged MBD9, PIE1, and CHR11 in the WT and mutant backgrounds were subjected to the IP‐MS experiment with anti‐Flag agarose.
-
EDiagrams showing the interaction regions of CHR11, MBD9, and PIE1 as determined by Y2H assays. The interaction regions are labeled by red frames. Conserved domains are shown.
We introduced epitope‐tagged MBD9 into the pie1 mutant plants and determined whether the pie1 mutation affects the interaction of MBD9 with the conserved SWR1 components and with CHR11/17 by IP‐MS. We found that, although the pie1 mutation did not affect the interaction of MBD9 with CHR11/17, it eliminated the interaction of MBD9 with the conserved SWR1 components (Fig 2D), suggesting that the interaction of MBD9 with PIE1 is necessary for the incorporation of MBD9 into the SWR1 complex. We then transformed epitope‐tagged PIE1 and CHR11 into the mbd9‐1 mutant to determine whether the mbd9 mutation affects the association of the SWR1 complex with CHR11/17. In the wild‐type (WT) background, the SWR1 components and CHR11/17 co‐precipitated with each other (Figs 1A and 2D), indicating that the SWR1 components interact with CHR11/17. In the mbd9‐1 mutant background, however, the interaction between the SWR1 components and CHR11/17 was abrogated (Fig 2D). These results strongly suggest that MBD9 is responsible for the association of the SWR1 complex with CHR11/17.
MBD9 contains a putative methyl‐CpG‐binding domain (MBD) in the N‐terminal region (Appendix Fig S7). However, the MBD did not bind to methylated and unmethylated DNA in an electrophoretic mobility shift assay (EMSA) (Appendix Fig S8). In agreement with this observation, the MBD was found in dicotyledons but not in monocotyledons (Appendix Fig S7), suggesting that the MBD is not conserved among angiosperms. Thus, the putative methylated DNA‐binding ability is unlikely to be required for the function of MBD9 in plants. The DDT, BROMO, and the second PHD are conserved, while the first PHD and WSD are quite divergent (Appendix Fig S7). We generated a series of truncated MBD9 for Y2H assays and found that a long MBD9 C‐terminal region harboring the DDT, BROMO, PHD, and WSD domains is required for the interaction with both CHR11/17 and PIE1 (Fig 2E; Appendix Figs S3, S5, S9, and S10). Similarly, by using a series of truncated PIE1 and CHR11 in Y2H assays, we demonstrated that a SANT domain of PIE1 and a SLIDE domain of CHR11 are responsible for interaction with MBD9 (Fig 2E; Appendix Figs S9 and S10). Interestingly, the SLIDE domain of CHR11 is also responsible for interaction with the canonical DDT domain‐containing ISWI subunit RLT1 (Dong et al, 2013). Compared with the canonical DDT domain‐containing ISWI subunits, MBD9 has evolved the additional ability to interact with the SWR1 complex while maintaining the ability to interact with the ISWI catalytic subunits.
Like known SWR1 components, MBD9, CHR11/17, and TRA1a regulate flowering time
In the Arabidopsis SWR1 complex, PIE1 is homologous to the ATPase‐containing catalytic subunit of the yeast SWR1 complex (March‐Díaz & Reyes, 2009; Jarillo & Piñeiro, 2015). Consistent with previous studies reporting the pleiotropic developmental defects in the pie1 mutant (Choi et al, 2007; Deal et al, 2007; March‐Díaz et al, 2007), we observed that loss‐of‐function mutation of PIE1 resulted in severe developmental defects, such as smaller plant size, reduced fertility, and shorter siliques (Figs 3A and B, and EV2A–C; Appendix Figs S11 and S12). The developmental defects were also observed in the mutants of the accessory SWR1 subunits SWC6, ARP4, and ARP6, even though the developmental defects in these mutants were weaker than in the pie1 mutant (Kandasamy et al, 2005; Choi et al, 2007; Deal et al, 2007; March‐Díaz et al, 2007; Gómez‐Zambrano et al, 2018; Figs 3E and EV2A–C; Appendix Figs S11 and S12). The development defects in the chr11 chr17 (chr11/17) double mutant were more serious than those in the pie1 mutant (Fig 3A). Considering that CHR11/17 are shared by the ISWI complex and the SWR1 complex (Fig 1A and B), the severe developmental defects in the chr11/17 mutant may be caused by disruption of both the ISWI and SWR1 complexes.
Figure 3. The effect of the SWR1 mutations on development and flowering time.
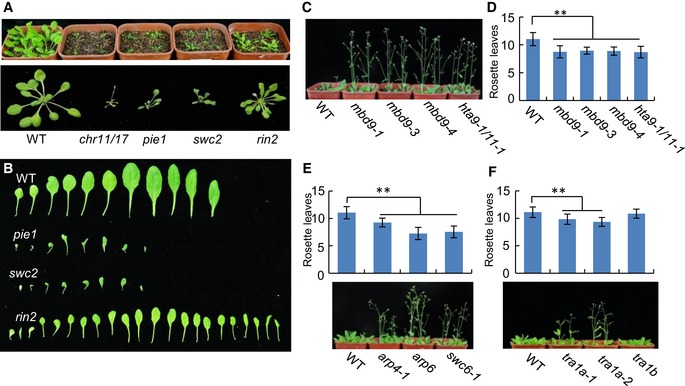
- Morphological phenotypes of the WT, chr11/17, pie1, swc2, and rin2 mutant plants. Shown are 24‐day‐old plants. The top panel shows the phenotypes of plants in the soil; the bottom panel shows the phenotypes of plants on the black cloth.
- Shown are rosette leaves of the WT, pie1, swc2, and rin2 mutant plants. Rosette leaves were cut from bolting plants.
- The early‐flowering phenotype of the mbd9‐1, mbd9‐3, mbd9‐4, and hta9‐1/11‐1 mutants compared with the WT. 24‐day‐old plants in soil are shown.
- Numbers of rosette leaves of the WT and mutant plants shortly after bolting. Rosette leaves from at least 20 plants were counted for each genotype. Values are mean ± SD. **P < 0.01, Student's t‐test.
- Determination of the effect of the arp4‐1, arp6, and swc6‐1 mutations on flowering time. The top panel shows the number of rosette leaves in the WT, arp4‐1, arp6, and swc6‐1 mutant plants; the bottom panel shows a photograph of the plants. Rosette leaves from at least 20 plants were counted for each genotype. Values are mean ± SD. **P < 0.01, Student's t‐test.
- Determination of the effect of the tra1a and tra1b mutations on flowering time. The top panel shows numbers of rosette leaves of the WT, tra1a‐1, tra1a‐2, and tra1b mutant plants; the bottom panel shows a photograph of the plants. Rosette leaves from at least 20 plants were counted for each genotype. Values are mean ± SD. **P < 0.01, Student's t‐test.
Figure EV2. The morphological phenotypes of reproductive organs in the WT and SWR1 complex mutants.
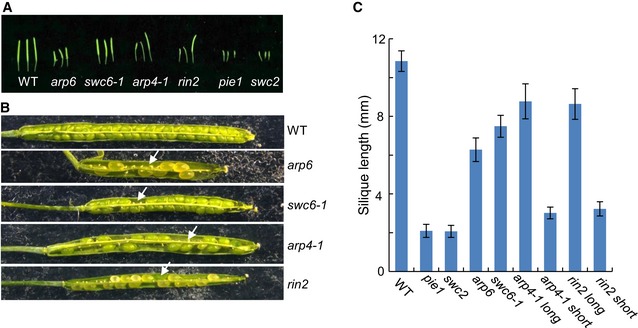
- Siliques of WT, arp6, swc6‐1, arp4‐1, rin2, pie1, and swc2. In the arp4‐1 and rin2 mutants, there are two different types of siliques. One is similar to that in the WT and the other is shorter than that in the WT. Both types of siliques are shown.
- Open siliques of WT, arp6, swc6‐1, arp4‐1, and rin2. Arrowheads indicate aborted ovules. Long siliques of arp4‐1 and rin2 are shown.
- Average silique length of WT, arp6, swc6‐1, arp4‐1, and rin2. At least 20 siliques were calculated for each mutant. Values are mean ± SD. “arp6 long” and “rin2 long” stand for the long siliques of arp6 and rin2 mutants, while “arp6 short” and “rin2 short” stand for the short siliques of these two mutants.
In addition to the developmental defects, an early‐flowering phenotype was evident in the mutants of the conserved SWR1 components PIE1, SWC4, SWC6, ARP4, and ARP6 as well as in the H2A.Z mutant hta9‐1 hta11‐1 (hta9‐1/11‐1) (March‐Díaz & Reyes, 2009; Coleman‐Derr & Zilberman, 2012; Jarillo & Piñeiro, 2015; Fig 3A–E; Appendix Fig S11). Unlike the previous study reporting an early‐flowering phenotype of the pie1‐5 mutant (Deal et al, 2007), our result indicated that the pie1 mutant allele (SALK_096434) showed severe growth retardation (Fig 3A and B; Appendix Fig S11), suggesting that the pie1‐5 mutant might be a weak mutant allele. The similar growth retardation was also observed in the swc2 mutant (Fig 3A and B; Appendix Fig S11). These results suggest that PIE1 as well as the SWR1 complex not only regulates flowering time but also plays important roles in plant growth and development. Therefore, the early‐flowering phenotype may only be observed in weak mutant alleles of core SWR1 subunits and in loss‐of‐function mutants of accessory SWR1 subunits (Fig 3A–E, Appendix Fig S11).
In Arabidopsis, there are three H2A.Z variants: HTA8, HTA9, and HTA11 (Choi et al, 2007). The hta9‐1/11‐1 double mutants showed a significant early‐flowering phenotype, but the developmental defects were weak (Fig 3C and D; Appendix Figs S11 and S12). Severe developmental defects were previously observed in the hta8/9/11 triple mutant (Coleman‐Derr & Zilberman, 2012), suggesting that the three H2A.Z variants are partially redundant. All three independent mbd9 mutant alleles showed an obvious early‐flowering phenotype, whereas the developmental defect in the mbd9‐1 mutant plants was weak and was comparable to that in the hta9‐1/11‐1 double mutant (Fig 3C and D; Appendix Figs S11 and S12).
We crossed the mbd9‐1 mutant with the swc2 mutant and obtained the mbd9‐1 swc2 double mutant. The developmental defect in the swc2 mutant was not enhanced in the mbd9‐1 swc2 double mutant even though a weak but significant developmental defect was observed in the mbd9‐1 mutant compared with the WT (Fig EV3A and B). These results demonstrate that MBD9 and the conserved SWR1 component SWC2 function in the same pathway, supporting the notion that MBD9 is a subunit of the SWR1 complex. The genetic interaction between MBD9 and ARP6 was analyzed by two recently published studies (Potok et al, 2019; Sijacic et al, 2019), indicating that the arp6‐1 mbd9‐3 double mutants displayed a more severe developmental defect than either of the single mutants. However, regarding flowering time, ARP6 is epistatic to MBD9 (Potok et al, 2019). As shown by our results (Fig EV2A–C; Appendix Fig S11), the developmental defect in the arp6 mutant is weaker than that in the mutant of the core SWR1 component PIE1, suggesting that ARP6 functions as an accessory component. It is not surprising to find an additive effect on development between the mutants of two accessory SWR1 complex components. In contrast, the developmental defect in the swc2 mutant is comparable with that in the pie1 mutant (Fig EV2A–C; Appendix Fig S11). Therefore, the swc2 mutant rather than the arp6 mutant is more suitable to be used as a representative of the SWR1 complex mutants to determine the relationship between MBD9 and the SWR1 complex.
Figure EV3. The developmental defect of the swc2 mutant is comparable to that of the mbd9‐1 swc2 double mutant.
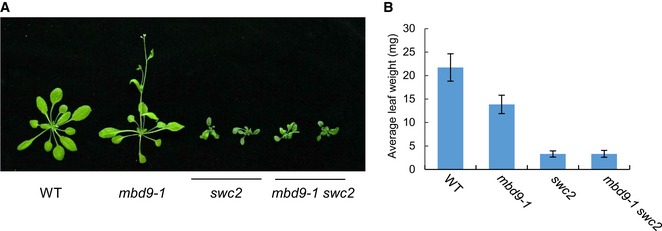
- The morphological phenotype of WT, mbd9‐1, swc2, and mbd9‐1 swc2. Shown are 24‐day‐old plants.
- Average leaf weight of WT, mbd9‐1, swc2, and mbd9‐1 swc2. For each plant, the total leaf weight and leaf number were recorded and the average leaf weight of each plant was calculated. Only rosette leaves were used for the analysis. At least 20 plants were used for each sample. Values are mean ± SD.
The tra1a‐1 and tra1a‐2 single mutants showed an early‐flowering phenotype but no obvious developmental defects, whereas the tra1b mutant did not show visible changes in either flowering time or development (Fig 3F; Appendix Figs S11 and S12). We crossed tra1a‐2 with tra1b to generate their F2 segregation group but did not obtain the tra1a‐2 tra1b (tra1a‐2/1b) double mutant (Appendix Fig S13), indicating that the double mutant is lethal. These results suggest that TRA1a and TRA1b are partially redundant in the SWR1 complex.
While the rin1 mutant was unavailable because of lethality, the rin2 mutant showed an obvious developmental defect, but the defect was much weaker than that in the pie1 and swc2 mutants (Figs 3A and B, and EV2A–C; Appendix Figs S11 and S12). Although the growth of the rin2 mutant was unlikely to be severely retarded, the flowering time was markedly delayed (Fig 3A and B; Appendix Fig S11). In yeast and metazoans, homologs of RIN1 and RIN2 are shared subunits of the SWR1 complexes and the INO80 chromatin‐remodeling complexes (Clapier & Cairns, 2009). Considering that the INO80 complexes are conserved in eukaryotes, we predict that RIN1 and RIN2 may also function as subunits of the Arabidopsis INO80 complex. The distinct developmental phenotypes of the rin2 mutant may be caused by disruption of both the SWR1 and INO80 complexes in Arabidopsis.
MBD9, CHR11/17, and other SWR1 components co‐regulate the expression of a number of genes
To investigate how MBD9 and CHR11/17 function in the SWR1 complex, we performed RNA sequencing (RNA‐seq) in order to compare the effect of pie1, hta9‐1/11‐1, mbd9‐1, and chr11/17 on gene expression. The RNA‐seq analysis identified a number of differentially expressed genes (DEGs) (log2 (fold change) > 1 or < −1; P < 0.01, Cufflinks) in the mutants relative to the WT (Fig 4A; Dataset EV2 and Dataset EV3). The numbers of DEGs identified in the mutants generally corresponded to the severity of their developmental defects (Fig 3A–D, 4A; Appendix Figs S11 and S12). The number of DEGs in the mbd9‐1 mutant (393) was comparable to that in the hta9‐1/11‐1 double mutant (519) and was fewer than in the pie1 mutant (2273) (Fig 4A; Dataset EV2 and Dataset EV3). Although a small number of DEGs were identified in the mbd9‐1 mutant, these genes significantly overlapped (P < 0.01, hypergeometric test) with those in the pie1 and hta9‐1/11‐1 mutants (Fig 4A and B; Dataset EV2 and Dataset EV3). In detail, 69% (160/233) and 41% (96/233) of mbd9‐1 up‐regulated genes were up‐regulated in the pie1 and hta9‐1/11‐1 mutants, respectively; 51% (81/160) and 41% (66/160) of mbd9‐1 down‐regulated genes were down‐regulated in the pie1 and hta9‐1/11‐1 mutants, respectively. As indicated by scatter plotting, the expression of the DEGs identified in the hta9‐1/11‐1 mutant was positively correlated with that in the pie1 and mbd9‐1 mutants, and the correlation coefficients of both were 0.73 (Fig 4C; Appendix Fig S14). These analyses support the notion that MBD9 functions as a component of the SWR1 complex to regulate gene expression.
Figure 4. The effect of chr11/17, hta9‐1/11‐1, mbd9‐1, and pie1 on gene expression as determined by RNA‐seq.

- Venn diagrams showing overlaps of differentially expressed genes in the mutants relative to the WT. Up‐ and down‐regulated genes were independently analyzed. The data were generated from two biological replicates. Differentially regulated genes were called when log2 (fold change) > 1 or < −1; P < 0.01.
- Heat maps showing the effect of indicated mutations on the expression of differentially expressed genes identified in the mbd9‐1 mutant. The color bar shows log2 (fold change). Up‐regulation and down‐regulation are shown by red and blue, respectively.
- Scatter plots showing that the expression pattern in the hta9‐1/11‐1 mutant is correlated with that in the chr11/17, mbd9‐1, and pie1 mutants. The differentially expressed genes identified in the hta9‐1/11‐1 mutant were subjected to the analysis.
A large number of DEGs (7,412) were identified in the chr11/17 mutant (Fig 4A; Dataset EV2 and Dataset EV3). These genes significantly overlapped (P < 0.01, hypergeometric test) with the DEGs identified in the pie1, mbd9‐1, and hta9‐1/11‐1 mutants (Fig 4A and B), supporting the notion that CHR11/17 can function in the SWR1 complex. However, the number of DEGs was markedly higher in the chr11/17 mutant than in the other SWR1 mutants (Fig 4A). This observation is consistent with the fact that the developmental defects are more serious in the chr11/17 mutant than in the other SWR1 mutants (Fig 3A). Given that CHR11/17 are shared subunits of the SWR1 and ISWI complexes as indicated above (Fig 1A and B), the severe developmental defects observed in the chr11/17 mutant may result from disruption of both complexes. Although the effect of chr11/17 on gene expression is correlated with that of pie1, hta9‐1/11‐1, and mbd9‐1, the correlations are generally lower than the correlations among pie1, hta9‐1/11‐1, and mbd9‐1 (Fig 4C; Appendix Fig S14). We predict that, unlike unique subunits of the SWR1 complex, CHR11/17 are shared by the ISWI and SWR1 complexes and have dual role in the regulation of gene expression.
Newly identified SWR1 components are involved in H2A.Z deposition
To determine whether the newly identified SWR1 components MBD9, CHR11/17, and TRA1a/1b are required for deposition of H2A.Z into chromatin, we extracted chromatin‐associated proteins and determined whether chromatin‐associated H2A.Z levels were affected by mutations of the SWR1 components. Immunoblotting assays showed that the chromatin‐associated H2A.Z level was markedly lower in the hta9‐1/11‐1 mutant than in the WT (Fig 5A), indicating that the H2A.Z antibody used in the immunoblotting assays was specific. In the mutants of the known SWR1 components PIE1, SWC2, SWC6, and ARP6, the chromatin‐associated H2A.Z levels were also markedly lower than in the WT even though the transcript levels of the H2A.Z genes were not reduced (Fig 5A; Appendix Fig S15), supporting the notion that the SWR1 complex is responsible for the deposition of H2A.Z into chromatin. In the mutants of the known SWR1 components ARP4 and RIN2, however, the chromatin‐associated H2A.Z levels were not affected (Fig 5A). In yeast and metazoans, the homologs of PIE1, SWC2, SWC6, and ARP6 are specific components of the SWR1 complexes, whereas the homologs of SWC4, ARP4, and RIN2 are components not only in the SWR1 complexes but also in the INO80 chromatin‐remodeling complexes and in the NuA4/TIP60 histone acetyltransferase complexes (Kobor et al, 2004; Kapoor & Shen, 2014). Therefore, we predict that the SWR1 components that are shared by the other chromatin‐related complexes are more likely to play a regulatory role rather than an essential role in H2A.Z deposition.
Figure 5. Determination of the function of the SWR1 complex components in H2A.Z deposition.
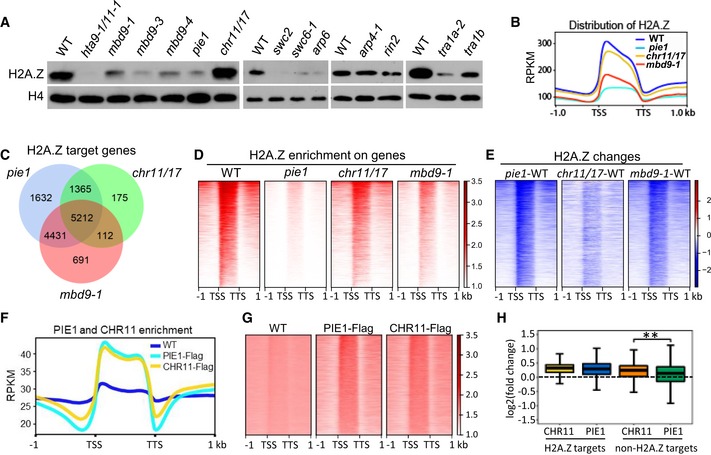
- The effect of indicated mutations on H2A.Z deposition as determined by immunoblotting. Chromatin‐associated proteins were extracted and subjected to immunoblotting by the anti‐H2A.Z antibody. The histone H4 level was detected by the anti‐H4 antibody and indicated as a loading control.
- Meta plots showing H2A.Z occupancy on H2A.Z target genes. TSS, transcription start site; TTS, transcription termination site. RPKM, reads per kilobase per million. The H2A.Z ChIP‐seq results were generated from two independent biological replicates.
- Venn diagrams showing overlaps of genes with lower H2A.Z levels in the pie1, mbd9‐1, and chr11/17 mutant than in the WT. The significance of the overlaps between each pair of samples was determined by hypergeometric test (P→0).
- Heat maps showing H2A.Z enrichment in the WT, pie1, chr11/17, and mbd9‐1 mutants over the PIE1‐dependent H2A.Z target genes. The peaks were sorted by H2A.Z density in the WT.
- Shown are H2A.Z changes in the pie1, chr11/17, and mbd9‐1 mutants relative to the WT. PIE1‐dependent H2A.Z target genes were subjected to the analysis and sorted by H2A.Z density in the WT. The color bar shows log2 (fold change).
- Meta plots showing CHR11 and PIE1 occupancy on PIE1‐dependent H2A.Z target genes. TSS, transcription start site; TTS, transcription termination site. The CHR11‐Flag and PIE1‐Flag ChIP‐seq data are from two independent biological replicates.
- Heat maps showing the enrichment of PIE1‐Flag and CHR11‐Flag over the PIE1‐dependent H2A.Z target genes. The peaks were sorted by H2A.Z density in the WT.
- Box plot showing the enrichment of CHR11 and PIE1 over H2A.Z target genes and non‐H2A.Z target genes. The log2 (fold change) values of RPKM between the CHR11‐Flag or PIE1‐Flag signals and the WT signals are shown. Center lines and box edges represent medians and the interquartile range (IQR), respectively. Whiskers extend to minimum and maximum values within 1.5 times of the IQR. The data shown are from two independent biological replicates. The difference between the CHR11‐Flag and PIE1‐Flag ChIP‐seq levels was determined by Student's t‐test. **P < 0.01.
In the mbd9‐1, tra1a‐2, and tra1b mutants, the chromatin‐associated H2A.Z levels were markedly lower than in the WT even though the transcript levels of the H2A.Z genes were not reduced (Fig 5A; Appendix Fig S15), indicating that MBD9, TRA1a, and TRA1b are involved in the deposition of H2A.Z into chromatin. In the chr11/17 mutant, however, the overall H2A.Z level was not affected (Fig 5A), suggesting that CHR11/17 are dispensable for H2A.Z deposition at the whole‐genome level. Given that CHR11/17 are shared subunits of the SWR1 complex and of the ISWI complex as indicated above (Fig 1A and B), this observation is consistent with the prediction that the SWR1 subunits that are shared by the other chromatin‐related complexes are dispensable for H2A.Z deposition at the whole‐genome level and may play a regulatory role in H2A.Z deposition.
We next performed chromatin immunoprecipitation followed by deep sequencing (ChIP‐seq) to determine how H2A.Z deposition is affected by the mutations of PIE1, MBD9, and CHR11/17. Based on the ChIP‐seq analysis, we identified 13,742 H2A.Z peaks showing reduced H2A.Z enrichment in the pie1 mutant relative to the WT (Appendix Fig S16). The PIE1‐dependent H2A.Z peaks identified in this study are highly overlapped with the peaks identified in a previous study (Carter et al, 2018; Appendix Fig S16), suggesting that our ChIP‐seq data are reliable. The H2A.Z peaks are predominantly located in the gene body and especially at the 5′‐end of genes (Fig 5B; Appendix Fig S17). We subsequently focused our analysis on the occupancy of H2A.Z in the genic region. We identified 12,640, 10,446, and 6,864 genes showing reduced H2A.Z occupancy in the pie1, mbd9‐1, and chr11/17 mutants, respectively (Fig 5C; Dataset EV2 and Dataset EV4). MBD9‐ and CHR11/17‐dependent H2A.Z target genes are significantly overlapped (P < 0.01, hypergeometric test) with the PIE1‐dependent H2A.Z target genes (Fig 5C). In the PIE1‐dependent H2A.Z target genes, the H2A.Z enrichment was markedly reduced in the mbd9‐1 mutant even though the reduction was less than that in the pie1 mutant (Fig 5B, D, and E), while the H3 enrichment was not reduced in either mbd9‐1 or pie1 mutants (Appendix Fig S18), suggesting that MBD9 functions in the SWR1 complex to specifically promote the incorporation of H2A.Z into chromatin. Although the H2A.Z enrichment was also reduced in the chr11/17 mutant, the reduction was weaker than in the pie1 and mbd9‐1 mutants (Fig 5B, D, and E), suggesting that CHR11/17 play an auxiliary role in the incorporation of H2A.Z into chromatin.
We performed ChIP‐seq to determine the enrichment of CHR11 and PIE1 at the whole‐genome level. The ChIP‐seq results indicated that both CHR11 and PIE1 signals are enriched in PIE1‐dependent H2A.Z target genes at the whole‐genome level (Fig 5F and G; Dataset EV5), which is comparable to the enrichment of H2A.Z (Fig 5B). As indicated by box plots, while the enrichment of CHR11 and PIE1 is comparable at H2A.Z target genes, the enrichment of CHR11 is significantly higher than that of PIE1 at non‐H2A.Z target genes (Fig 5H). This finding suggests that, while CHR11/17 can function in the SWR1 complex to mediate the coupling of nucleosome sliding and H2A.Z deposition, it can also function at non‐H2A.Z target genes in a SWR1‐independent manner, which is consistent with the idea that CHR11/17 can function in the ISWI complex to mediate nucleosome sliding at non‐H2A.Z target genes. As indicated by Venn diagram, H2A.Z target genes are significantly overlapped with CHR11‐enriched genes, supporting the notion that CHR11 is involved in H2A.Z deposition (Appendix Fig S19; Dataset EV5 and Dataset EV6). Meanwhile, a part of H2A.Z target genes are not overlapped with CHR11‐enriched genes (Appendix Fig S19). It appears that the overlap between H2A.Z target genes and CHR11‐enriched genes was underestimated due to the use of stringent parameters in the identification of H2A.Z target genes and CHR11‐enriched genes. Alternatively, it may also be possible that CHR11 can only function at a part of H2A.Z target genes.
We also tried to test the association of MBD9 with chromatin by CHIP‐seq but failed due to technical difficulties. A recent study showed that the conserved BROMO of MBD9 can bind to acetylated histone peptides as determined by an in vitro pull‐down assay (Nie et al, 2019), suggesting that MBD9 may mediate the association of the SWR1 complex with histone acetylation on chromatin. We mutated critical residues in the BROMO and the second PHD of MBD9 and performed a complementation assay to test whether the BROMO and PHD domains are required for the regulation of flowering time by MBD9. We found that, like the WT MBD9 construct, the MBD9 constructs harboring mutations in both the BROMO and PHD domains can completely complement the early‐flowering phenotype of the mbd9‐1 mutant (Fig EV4A–D). Therefore, the BROMO and PHD domains of MBD9 are unlikely to be necessary for the association of MBD9 with chromatin and for the regulation of flowering time by MBD9.
Figure EV4. Mutations of conserved residues in the BROMO and the second PHD domains of MBD9 do not affect the biological function of MBD9.

- Shown are mutation sites in the BROMO domain and the second PHD domain of MBD9. The alignment of MBD9 and its plant orthologs is shown.
- Shown are MBD9 mutation sites in each of the complementary transgenic lines in the mbd9‐1 mutant background.
- The flowering‐time phenotypes of WT, mbd9‐1, and the complementary transgenic lines harboring WT and mutated MBD9 transgenes.
- The number of rosette leaves of WT, mbd9‐1, and the complementary transgenic lines harboring WT and mutated MBD9 transgenes. Values are mean ± SD. **P < 0.01, Student's t‐test.
Coordination of H2A.Z deposition, nucleosome positioning, and gene expression
In investigating how H2A.Z deposition affects gene expression, we found that the DEGs identified in the pie1, hta9‐1/11‐1, and mbd9‐1 mutants are highly enriched in H2A.Z target genes (Fig 4A; Appendix Fig S20), confirming the involvement of H2A.Z deposition in the regulation of gene expression. In each of the three mutants, both up‐ and down‐regulated H2A.Z target genes were present even though the number of up‐regulated H2A.Z target genes was higher than the number of down‐regulated ones, which is consistent with the previous finding that H2A.Z has dual role in transcriptional regulation (Sura et al, 2017). The DEGs in the chr11/17 mutant are also enriched in H2A.Z target genes (Fig 4A; Appendix Fig S20), suggesting that H2A.Z is involved in the regulation of gene expression by CHR11/17. Given that CHR11/17 are shared subunits of the SWR1 and ISWI complexes, the DEGs in the chr11/17 mutant may be caused by disruption of both the SWR1 and ISWI complexes.
The early‐flowering phenotype was reported not only in previously reported SWR1 mutants but also in the mbd9 and chr11/17 mutants; reduced expression of the core flowering repressor gene FLC was known to play a critical role in the early‐flowering phenotype of previously reported SWR1 mutants and the chr11/17 mutant (Kandasamy et al, 2005; Peng et al, 2006; Choi et al, 2007; Deal et al, 2007; March‐Díaz et al, 2007; Li et al, 2012; Gómez‐Zambrano et al, 2018). Our RNA‐seq data confirmed the reduced expression of FLC in the pie1 and chr11/17 mutant (Fig EV5; Dataset EV3). Our RNA‐seq data showed that the expression of FLC was also reduced in the mbd9‐1 mutant even though the reduction was not significant (Fig EV5A). Our RT–qPCR analysis demonstrated that the expression of FLC was significantly reduced in the two independent mbd9 mutant alleles (Fig EV5B), which is consistent with the previous study (Peng et al, 2006). Given the finding that MBD9 and CHR11/17 function as subunits of the SWR1 complex, we expected that the reduced expression of FLC in the mbd9 and chr11/17 mutants may be caused by reduction in H2A.Z enrichment at the FLC chromatin. Our H2A.Z ChIP‐seq data indicated that H2A.Z was enriched at the transcriptional start site of FLC in the WT and the enrichment was reduced not only in the pie1 mutant but also in the mbd9‐1 and chr11/17 mutants (Fig EV5A). The result demonstrates that the regulation of FLC expression by MBD9 and CHR11/17 is at least partially attributed to their function in H2A.Z deposition, further supporting the inference that MBD9 and CHR11/17 function as subunits of the SWR1 complex.
Figure EV5. The expression and H2A.Z enrichment of FLC in the WT, mbd9‐1, pie1, and chr11/17 mutants.
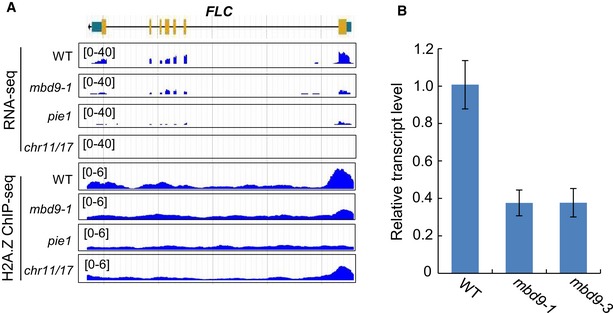
- Snapshots showing RNA‐seq and H2A.Z ChIP‐seq signals of FLC in WT, mbd9‐1, pie1, and chr11/17.
- Determination of the transcript levels of FLC in WT, mbd9‐1, and mbd9‐3 by RT‐qPCR. Results shown are mean ± SD from three independent biological replicates.
Because CHR11/17 were previously found to be required for nucleosome sliding in the gene body (Li et al, 2014), we investigated whether the CHR11/17‐interacting chromatin remodeler PIE1 is also required for nucleosome sliding. Using an in vitro nucleosome sliding assay, we observed that, while CHR11 slides the nucleosome from the end to the middle of the 245‐bp DNA, PIE1 fails to slide the nucleosome (Fig 6A). A previous work indicated that some of the ISWI family proteins can slide the nucleosome from the middle to the end of the DNA fragment (Eberharter et al, 2001). Our results indicated that both PIE1 and CHR11 cannot slide the nucleosome from the middle to the end (Fig 6A). These results suggest that, unlike CHR11/17, PIE1 is not a nucleosome sliding enzyme. We then performed micrococcal nuclease digestion followed by deep sequencing (MNase‐seq) to determine whether the genome‐wide nucleosome pattern differs in the chr11/17, pie1, and mbd9‐1 mutants relative to the WT. Consistent with a previous report (Li et al, 2014), the nucleosomes in the gene body were evenly spaced in the WT, whereas the evenly spaced pattern was markedly disrupted in the chr11/17 mutant (Fig 6B). In the pie1 mutant, the evenly spaced nucleosome pattern was similar to that in the WT, not only at H2A.Z target genes but also at non‐H2A.Z target genes (Fig 6B; Appendix Fig S21). The same is for the mbd9‐1 mutant, suggesting that PIE1 and MBD9 are dispensable for nucleosome sliding.
Figure 6. Determination of the function of CHR11/17 and PIE1 in nucleosome sliding.

- Increasing amounts of CHR11 (left panel) or PIE1 (right panel) were incubated with end‐positioned (lanes 1–4 and 9–12) or center‐positioned (lanes 5–8 and 13–16) mono‐nucleosomes in the presence of ATP. The sliding results were then visualized on native PAGE gels by EB staining.
- Distribution of nucleosomes over TSS of all protein‐coding genes in the WT, chr11/17, pie1, and mbd9‐1 mutants. The MNase‐seq results were generated from two independent biological replicates.
- The proposed model for the function of SWR1‐MBD9‐CHR11/17 complex in nucleosome regulation. NFR, nucleosome‐free region.
Based on the results shown in this study, we proposed a working model for the association of the SWR1 complex with CHR11/17 (Fig 6C). CHR11/17 mediate nucleosome sliding in the gene body independently of the SWR1 complex, while they function as subunits of the SWR1 complex to mediate H2A.Z deposition. MBD9 functions as a bridge protein to recruit the SWR1 complex to CHR11/17. It is well known that the conserved core catalytic subunits (i.e., CHR11/17 homologs) of the ISWI complexes are sufficient to mediate nucleosome sliding even when accessory subunits of the ISWI complexes are absent (Corona et al, 1999; Li et al, 2017; Fig 6A). Therefore, it is reasonable to conclude that CHR11/17 can mediate nucleosome sliding not only in the ISWI complex but also in the SWR1 complex. Although we cannot exclude the possibility that the CHR11/17‐containing ISWI complex may mediate nucleosome sliding at H2A.Z target loci, the results shown in our study strongly support the notion that the CHR11/17‐containing SWR1 complex can mediate nucleosome sliding and facilitate the coupling between nucleosome sliding and H2A.Z deposition, thereby co‐regulating gene expression, development, and flowering time.
Discussion
In eukaryotes, H2A.Z is incorporated into chromatin by a conserved SWR1/SRCAP complex (Mizuguchi et al, 2004; Clapier & Cairns, 2009; Kapoor & Shen, 2014). Components of the SWR1/SRCAP complexes were previously identified and characterized in yeast and metazoans (Clapier & Cairns, 2009; Kapoor & Shen, 2014). Although several Arabidopsis SWR1 components have been identified, all of them are conserved in yeast and metazoans (Jarillo & Piñeiro, 2015; Gómez‐Zambrano et al, 2018). Before the current study, whether the Arabidopsis SWR1 complex has non‐conserved components was unknown. In the current study, we used IP‐MS to identify all components of the Arabidopsis SWR1 complex. Intriguingly, we identified not only conserved components but also the following previously uncharacterized components in the Arabidopsis SWR1 complex: MBD9, CHR11/17, and TRA1a/1b. Considering that the homologs of MBD9, CHR11/17, and TRA1a/1b have not been detected in the SWR1 complexes of eukaryotes other than plants, we suggest that MBD9, CHR11/17, and TRA1a/1b are plant‐specific components of the SWR1 complex.
Pleiotropic developmental phenotypes including early flowering and developmental abnormalities were previously observed in mutants or knockdown lines of conserved SWR1 components (March‐Díaz & Reyes, 2009; Jarillo & Piñeiro, 2015; Gómez‐Zambrano et al, 2018). Like the mutants of known SWR1 components, the mbd9 mutant also showed early flowering and developmental defects (Peng et al, 2006). However, the molecular mechanism underlying the function of MBD9 has been elusive (Yaish et al, 2009; Hale et al, 2016). Our study indicated that H2A.Z deposition was markedly reduced by loss‐of‐function mutations of MBD9, confirming that MBD9 is a canonical component of the Arabidopsis SWR1 complex. The developmental defect in the chr11/17 mutant is more serious than that in the SWR1 mutant pie1, suggesting that CHR11 and CHR17 can function independently of the SWR1 complex. CHR11 and CHR17 interact with the DDT domain‐containing proteins RLT1 and RLT2 and thus form an ISWI chromatin‐remodeling complex that is responsible for nucleosome positioning in the gene body (Li et al, 2014). Thus, CHR11 and CHR17 are shared subunits of the SWR1 and ISWI complexes in Arabidopsis.
In yeast and metazoans, TRA1/TRRAP is a conserved subunit of the NuA4/Tip60 complexes but not of the SWR1/SRCAP complexes (Clapier & Cairns, 2009; Kapoor & Shen, 2014). The finding of the TRA1 homologs TRA1a/1b in the Arabidopsis SWR1 complex suggests that TRA1a/1b can function as subunits of the SWR1 complex in plants. Our previous IP‐MS analysis identified TRA1b as a subunit of the Arabidopsis NuA4 complex using the histone acetyltransferase HAM1 at a bait protein (Tan et al, 2018). However, the SWR1 unique components and HAM1 cannot co‐purify each other in the IP‐MS experiment (Tan et al, 2018; Fig 1A; Dataset EV1), suggesting that the components of the SWR1 and NuA4 complexes form two separated complexes and cannot interact to form a larger one in Arabidopsis. Therefore, TRA1a/1b are likely to be shared subunits of the SWR1 and NuA4 complexes. Reduced deposition of H2A.Z was observed in both the tra1a‐2 and tra1b single mutants (Fig 5A), supporting the notion that TRA1a/1b function as subunits of the SWR1 complex. The early‐flowering phenotype of TRA1a further confirmed that TRA1a/1b are functionally related to the SWR1 complex. Although either of the tra1a‐2 and tra1b single mutants did not show severe developmental defects, the tra1a‐2/1b double mutant is unavailable by genetic crossing between tra1a‐2 and tra1b, suggesting TRA1a and TRA1b function redundantly and are required for the viability of either gametophytes or embryos. Given that TRA1a/1b are shared subunits of the SWR1 and NuA4 complexes, the tra1a‐2/1b mutant may disrupt both the SWR1 and NuA4 complexes and thus lead to more serious developmental defects than the SWR1 mutant pie1. GAS41/YAF9A is another component shared by the SWR1 and NuA4 complexes in Arabidopsis (Gómez‐Zambrano et al, 2018; Tan et al, 2018). A recent study reported that GAS41/YAF9A can function redundantly with its homolog, YAF9B, and mediate acetylation of H2A.Z and H4, thereby regulating development and flowering time in parallel to the SWR1 complex (Crevillen et al, 2019). The share of subunits by the SWR1 complex and the other chromatin‐related complexes may add a layer of complexity to regulate development and flowering time.
DDT domain‐containing proteins, such as Itc1 in yeast and ACF1, BPTF, and WSTF in humans, function as conserved subunits of the ISWI chromatin‐remodeling complexes (Bartholomew, 2014). In Arabidopsis, a family of DDT domain‐containing proteins interact with the ISWI catalytic subunits CHR11 and CHR17 (Li et al, 2012; Dong et al, 2013). Here, we found that most of the DDT domain‐containing proteins were co‐precipitated with CHR11/17 but not with any conserved SWR1 components (Fig 1A), suggesting that the DDT domain‐containing proteins exclusively function as subunits of the ISWI complexes in Arabidopsis. The DDT domain‐containing ISWI subunits contain not only the DDT domain but also other conserved domains including DDT, BROMO, PHD, and/or WSD (Bartholomew, 2014; Li et al, 2017). The BROMO and PHD domains of the human ISWI subunit BPTF were previously found to bind acetylated histone and methylated histone H3K4, respectively, thereby enhancing the association of the ISWI complex with chromatin (Ruthenburg et al, 2011). Although the BROMO of MBD9 was recently shown to bind to acetylated histone peptides as determined by an in vitro assay (Nie et al, 2019), our genetic analysis indicated that disruption of the BROMO and PHD domains of MBD9 did not affect the function of MBD9 in the regulation of flowering time (Fig EV4A–D). This finding excludes the possibility that the BROMO and PHD domains of MBD9 are necessary for the association of the SWR1 complex with chromatin. Future research should determine how MBD9 functions in the SWR1 complex to promote H2A.Z deposition.
Two recent studies reported the identification of MBD9 as a subunit of the Arabidopsis SWR1 complex during the revision of our paper (Potok et al, 2019; Sijacic et al, 2019). One of the studies detected the interaction between MBD9 and CHR11/17 but did not detect the interaction between ARP6 and CHR11/17, thus concluding that MBD9 interacts with two separate complexes (Potok et al, 2019). Different from the published studies, our study reported the association of the SWR1 complex with the ISWI chromatin‐remodeling regulators CHR11/17, and demonstrated that MBD9 functions as a bridge protein between the known SWR1 components and the ISWI chromatin‐remodeling regulators, thus revealing a distinct mechanism of chromatin regulation. MBD9 shares conserved domains (i.e., DTT, PHD, WSD) with the canonical DDT domain‐containing subunits of the ISWI complexes, which is consistent with the interaction of MBD9 with CHR11/17. However, unlike the DDT domain‐containing ISWI subunits that interact with CHR11/17 but not with the SWR1 components, MBD9 interacts with both CHR11/17 and the SWR1 components (Fig 1A). The dual recognition of CHR11/17 and PIE1 by MBD9 mediates the association of CHR11/17 with the SWR1 complex and thereby facilitates the coupling of H2A.Z deposition with nucleosome sliding.
Materials and Methods
Plant materials and constructs
Arabidopsis thaliana seedlings were grown on MS (Murashige and Skoog) medium plates under long‐day conditions (16‐h light, 8‐h dark, 22°C). All of the Arabidopsis T‐DNA insertion lines used in this study were obtained from the Arabidopsis Biological Resource Center (ABRC); these lines included mbd9‐1 (SALK_054659), mbd9‐3 (SALK_039302C), mbd9‐4 (SALK_027780), pie1 (SALK_096434), chr11 (GK_424F01), chr17 (SALK_080144), rin2 (SALK_147720), swc2 (SK40839), swc6‐1 (SAIL_1142_C03), arp4‐1 (SAIL_760_H04), arp6 (SAIL_236_C07), hta9‐1 (SALK_054814), hta11‐1 (SALK_017235), tra1a‐1 (SAIL_655_C11), tra1a‐2 (SALK_087015), and tra1b (SALK_053301). Double mutants used in this study were generated by crossing single mutants. Because of the severely reduced fertility, pie1, swc2, and chr11/17 mutants were maintained in a heterozygous state.
The full‐length MBD9 driven by its native promoter was introduced into the modified pRI909 vector with 3 × Flag or 5 × Myc tagged at its 3′‐terminus. The full‐length PIE1, CHR11, CHR17, HTA9, RIN1, RIN2, SWC2, GAS41, and RLT2 driven by their native promoters were introduced into the modified pCAMBIA1305 vector with 3 × Flag tagged at their 3′‐ends. These constructs were introduced into the Agrobacterium strain GV3101 and transformed into the WT or mutants via the floral‐dipping method. In detail, the PIE1, CHR11, CHR17, HTA9, RIN1, RIN2, SWC2, GAS41, and RLT2 constructs were transformed into WT for IP‐MS experiments (Fig 1A), and the MBD9, CHR11, CHR17, PIE1 constructs were also transformed into their corresponding mutants for IP‐MS experiments (Fig 1A, Appendix Fig S1) and complementation testing (Fig EV1). The constructs were obtained according to the In‐Fusion method (Vazyme, C112‐01) or the restriction endonuclease and T4 DNA ligation method. Mutation sites were introduced by site‐directed mutagenesis. All constructs were sequenced for verification, and the primers are listed in Dataset EV7.
Immunoprecipitation, mass spectrometry, and co‐immunoprecipitation
A 3 g quantity of flowers from transgenic and WT plants was used for immunoprecipitation followed by mass spectrometry. Tissues were ground in liquid nitrogen and homogenized in plant lysis buffer (50 mM Tris–HCl, pH 7.6, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% NP‐40, 5 mM DTT, 0.1 mM PMSF, and proteinase inhibitor cocktail, Roche). After centrifugation, the supernatant was incubated with anti‐Flag M2 agarose (Sigma, A2220) at 4°C for 3 h, and the resins were washed four times with plant lysis buffer. The agarose‐bound proteins were eluted with 3 × Flag peptide (Sigma, F4799) and were subjected to SDS–PAGE followed by silver staining (Sigma, PROT‐SIL2). The silver‐stained proteins were de‐stained and digested in‐gel with trypsin at 37°C overnight. The digested peptides were eluted on a capillary column and sprayed into a Q Exactive Mass Spectrometer (Thermo Fisher Scientific, USA) equipped with a nano‐ESI ion source. Database searches were performed on an in‐house Mascot Server (Matrix Science, UK) against IPI (International Protein Index) Arabidopsis protein database.
For co‐immunoprecipitation analysis, the MBD9‐Myc transgenic plants were crossed with the plants harboring HTA9‐Flag, HTA11‐Flag, PIE1‐Flag, CHR11‐Flag, and CHR17‐Flag transgenes. A 1 g quantity of tissue from F1 plants and parental lines was ground in liquid nitrogen and homogenized in 5 ml of plant lysis buffer. After centrifugation, the supernatant was incubated with anti‐Flag M2 agarose (Sigma, A2220) for 3 h. The resins were washed four times with plant lysis buffer and eluted with 3 × Flag peptides (Sigma, F4799). The input and elution samples were boiled and subjected to SDS–PAGE followed by Western blotting.
Yeast two‐hybrid assay
For Y2H assays, the full‐length cDNA sequences of the genes were cloned into the pGADT7 and pGBKT7 vectors in frame with the 3′‐terminus of GAL4‐AD and GAL4‐BD, respectively. Series of truncations of MBD9, PIE1, CHR11, and CHR17 were also constructed. Primers used for these constructs are listed in Dataset EV7. The yeast strains AH109 and Y187 were transformed by the pGADT7 and pGBKT7 constructs and were grown on synthetic dropout medium lacking Leu (SD‐L) and Trp (SD‐W), respectively. The positive clones from these two media were mated for 16–20 h in YPDA (yeast peptone dextrose adenine) liquid medium. After mating, the mixtures were spotted on synthetic dropout medium lacking both Trp and Leu (SD‐LW). Positive clones on SD‐LW were resuspended in ddH2O and spotted on SD‐LW and synthetic dropout medium lacking Leu, Trp, and His (SD‐LWH). 3‐AT was added to SD‐LWH to inhibit the background growth.
Protein purification
The cDNA sequence of the truncated MBD9 encoding 1‐750 aa was cloned into the pET28a vector to generate a construct expressing the His‐tagged MBD9 fragment. The construct was then transformed into Escherichia coli strain BL21 (DE3) for protein expression. Protein purification was performed as previously described (Luo et al, 2018). In brief, the bacteria were precipitated and resuspended in His‐tag lysis buffer (20 mM Tris–HCl, pH 8.0, 500 mM NaCl, 20 mM imidazole, 1 mM DTT, and 1 mM PMSF), followed by sonication and centrifugation at 38,000 g for 1 h. The supernatant was incubated with Ni‐NTA Resin (Millipore, 70666‐4) for 2 h at 4°C with rotation. The resin was then washed for several times by His‐tag lysis buffer. Proteins were eluted by His‐tag lysis buffer containing 250 mM imidazole.
The full‐length cDNAs of PIE1 and MBD9 were inserted into the pAT423 vector tagged with 3 × Flag and 3 × Myc, respectively, in frame with their 3′‐termini. The full‐length cDNA of CHR11 was inserted into the pAT425 vector tagged with 3 × HA in frame with its 3′‐terminus. These constructs were transformed into the yeast strain YPH499 for protein purification (Ishii et al, 2014). The yeast pellets were ground in liquid nitrogen and resuspended in yeast lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.05% NP‐40, 1 mM DTT, 1 mM PMSF, and cocktail, Roche). After centrifugation, the supernatants were incubated with anti‐Flag M2 agarose (Sigma, A2220), anti‐c‐Myc agarose (Sigma, A7470), or anti‐HA agarose (DiNing, SA068001). The resins were washed six times with yeast lysis buffer. Flag‐, Myc‐, and HA‐tagged proteins were eluted by 3 × Flag peptides, 0.1 M NH3·H2O, and HA peptides (SMART Lifesciences), respectively. Eluted proteins were concentrated and stored at −80°C.
Pull‐down assay
For the Flag pull‐down assay, yeast strains expressing PIE1‐Flag, MBD9‐Myc, and CHR11‐HA proteins were ground in liquid nitrogen and resuspended in yeast lysis buffer. After centrifugation, the supernatants of PIE1‐Flag, MBD9‐Myc, and CHR11‐HA were mixed in different combinations and incubated with anti‐Flag M2 agarose (Sigma, A2220) for 2 h. After washing for several times, the proteins were eluted with 3 × Flag peptides. For the HA pull‐down assay, yeast strains expressing MBD9‐Myc and CHR11‐HA proteins were ground and centrifuged. Supernatants were mixed and incubated with anti‐HA agarose. Proteins were eluted with HA peptides. Input and elution samples were analyzed by immunoblotting.
Chromatin isolation and H2A.Z detection
Chromatin was isolated as previously described (Zhang et al, 2016). In brief, a 1 g quantity of 11‐day‐old seedlings of each mutant was ground in liquid nitrogen and homogenized in 5 ml of Honda buffer (20 mM HEPES–KOH, pH 7.4, 0.44 M sucrose, 1.25% Ficoll, 2.5% Dextran T40, 10 mM MgCl2, 0.5% Triton X‐100, 5 mM DTT, 1 mM PMSF, and proteinase inhibitor cocktail, Roche). The homogenate was filtered through Miracloth (Merck Millipore, 475855) and then centrifuged at 1,500 g. The pellets were washed three times with Honda buffer and one time with 1 × PBS (1 mM EDTA). The pellets were then resuspended in 0.5 ml of prechilled glycerol buffer (20 mM Tris–HCl, pH 7.9, 75 mM NaCl, 0.5 mM EDTA, 50% glycerol, 0.85 mM DTT, 0.125 mM PMSF, proteinase inhibitor cocktail, and 10 mM β‐mercaptoethanol) followed by addition of 0.5 ml of cold nuclei lysis buffer (10 mM HEPES, pH 7.6, 7.5 mM MgCl2, 0.2 mM EDTA, 0.3 M NaCl, 1 M urea, 1 mM DTT, 1% NP‐40, 0.5 mM PMSF, proteinase inhibitor cocktail, and 10 mM β‐mercaptoethanol). The samples were gently mixed two times, each for 2 s, with a vortex mixer, and then incubated on ice for 2 min followed by centrifugation at 13,000 g at 4°C for 2 min to separate the chromatin fraction and the nucleoplasmic fraction. The proteins extracted from the chromatin fraction were analyzed by immunoblotting to detect the amount of H2A.Z in the WT and mutant plants. H2A.Z antibody was produced as previously described (Deal et al, 2007).
RNA analysis and RNA‐seq
Total RNA was extracted from 11‐day‐old seedlings with TRIzol reagent (Ambion, 15596018) according to the manual. A 2 μg quantity of total RNA of each sample was subjected to reverse transcription using All‐In‐One MasterMix (abm, G492) followed by quantitative PCR. For RNA‐seq analysis, library preparation and sequencing were performed by Vazyme Biotech (Illumina HiSeq X10; 150‐bp pair‐end sequencing). HISAT2 version 2.1.0 (Kim et al, 2015) was used to map clean reads to the Arabidopsis reference genome Araport11 (Cheng et al, 2017). Reads mapped to each transcript were counted using featureCounts version 1.6.0 (Liao et al, 2013). Expression quantification and differential expression analysis were performed using the edgeR package in R (Nikolayeva & Robinson, 2014).
ChIP‐seq
A 2 g quantity of 11‐day‐old seedlings was ground in liquid nitrogen and suspended in 15 ml of ChIP lysis buffer (20 mM Tris–HCl, pH 7.5, 20 mM KCl, 2 mM EDTA, pH 8.0, 2.5 mM MgCl2, 25% glycerol, 250 mM sucrose, 5 mM DTT, 1 mM PMSF, and cocktail, Roche). The suspension was cross‐linked with 1% formaldehyde (Sigma, F8775), and the reaction was stopped by addition of 0.125 mM glycine. After the preparation was passed through two layers of Miracloth and centrifuged, the pellet with nuclei was washed four times using nuclear resuspension buffer (NRBT, 20 mM Tris–HCl, pH 7.5, 2.5 mM MgCl2, 25% glycerol, and 0.2% Triton X‐100) followed by sonication in sonication buffer (20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 4 mM MgCl2, 0.2% NP‐40, 0.1 mM PMSF, and cocktail, Roche). The chromatin was then incubated with HTA9 antibody and Dynabeads Protein G (Thermo, 10004D) or anti‐Flag M2 magnetic beads (Sigma, M8823) overnight with rotation at 4°C. The ChIP DNA was purified using a standard phenol–chloroform method and was sent to Novogene (Tianjin, China) for library generation and sequencing (Illumina HiSeq X10; 150‐bp pair‐end sequencing). ChIP‐seq reads were mapped to the Arabidopsis reference genome Araport11 using bowtie2 version 2.3.4 (Langdon, 2015). SICER was used to determine ChIP‐enriched regions and to assess regions of differential enrichment between the WT and mutant plants (Xu et al, 2014). DeepTools was used for generating metaplots (Ramírez et al, 2014). HOMER was used for identifying genes with nearby peaks (Heinz et al, 2010). For H2A.Z ChIP‐seq analysis, previously published data for the WT were used as an additional biological control (Carter et al, 2018).
MNase‐seq
A 1 g quantity of 11‐day‐old seedlings was ground in liquid nitrogen and resuspended in Honda buffer. After the preparation was passed through Miracloth and centrifuged, the pellet was washed three times with Honda buffer and one time with MNase buffer (20 mM Tris–HCl, pH 8.0, 5 mM CaCl2, 1 mM PMSF, cocktail, Roche). After centrifugation, the pellet was resuspended in 800 μl of MNase buffer and digested with 5 μl of MNase (NEB, M0247S) at 37°C for 15 min, and the digestion was stopped by addition of 10 mM EDTA. The digested DNA was treated with RNase and Proteinase K and was then purified using a standard phenol–chloroform method. The DNA samples were sent to Novogene (Tianjin, China) for library preparation and sequencing (Illumina HiSeq X10; 150‐bp pair‐end sequencing). Bowtie 1.1.1 (Langmead et al, 2009) was used to align the MNase‐seq reads to TAIR10 genome with ‐v 2 and ‐m 1 parameters. The insertion size was limited from 140 bp to 160 bp. The midpoint of insertion segment was employed in the follow‐up analysis. The 1‐kb upstream and downstream regions of TSS of all protein‐coding genes were assigned to 200 bins for counting MNase‐seq reads. Normalization was conducted by gene counts and mapped reads.
Electrophoretic mobility shift assay
The cDNA sequence encoding the MBD domain of MBD9 was amplified and cloned into the pET28a vector. The construct was transformed into E. coli strain BL21 (DE3) for protein purification. The purified proteins were incubated with 1.25 μM methylated or unmethylated double‐stranded DNA in binding buffer (25 mM HEPES–KOH, pH 7.6, 50 mM KCl, 0.1 mM EDTA, 12.5 mM MgCl2, 0.5% (w/v) BSA, 5% glycerol, and 1 mM DTT) at room temperature for 30 min. The binding reaction mixture was resolved with 5% native PAGE in 0.5× TBE at 4°C for 150 min at 80 V. The interaction between proteins and DNA was detected by ethidium bromide staining.
Nucleosome assembly
Arabidopsis proteins H2A (HTA10, AT1G51060), H2B (HTB1, AT1G07790), H3 (AT1G09200), and H4 (AT2G28740) were chosen for nucleosome assembly. Coding sequences of these proteins were inserted into pET28a vector without any tags or excess amino acids. The coding sequence of H4 was optimized according to the preference of E. coli to increase the protein expression level. All of these plasmids were transformed into E. coli strain BL21 (DE3), which was grown in LB medium at 37°C until an OD600 of 0.6 was obtained. Protein expression was induced by the addition of 0.6 mM IPTG for 4 h at 37°C, and the histones were purified as the inclusion body. Cells were harvested, and pellets were washed twice with 10 ml of cold PBS buffer. Pellets from 1 liter of culture were then resuspended in 10 ml of cold lysis buffer (50 mM Tris–HCl, pH 7.6, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, cocktail, Roche), sonicated, and centrifuged at 30,000 g for 30 min at 4°C. The inclusion body pellet was washed twice with cold lysis buffer with 1% Triton X‐100 and once with cold lysis buffer without Triton X‐100. The inclusion body pellets from 1 liter of culture were resuspended in 10 ml of the resuspension buffer (20 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 6 M guanidine hydrochloride, and 1 mM DTT) and incubated at 4°C for 2 h with rotation. After centrifugation at 30,000 g for 30 min, the supernatants were transferred to SnakeSkin Dialysis Tubing (Thermo, 68035) and were dialyzed against low‐salt urea buffer (10 mM Tris, pH 7.5, 100 mM NaCl, 1 mM EDTA, 7 M urea, and 1 mM DTT) overnight. Samples were then loaded onto a HiTrap SP HP 5‐ml column (GE Healthcare, 17‐1152‐01) equilibrated with low‐salt urea buffer (filtered and degassed), and were eluted from the SP column by a linear salt gradient from low‐salt urea buffer to high‐salt urea buffer (10 mM Tris, pH 7.5, 1 M NaCl, 1 mM EDTA, 7 M urea, and 1 mM DTT). Histone fractions were collected (Dann et al, 2017).
For octamer assembly, equal amounts of the histones H2A, H2B, H3, and H4 were mixed and dialyzed against refolding buffer (10 mM Tris–HCl, pH 7.5, 2 M NaCl, 1 mM EDTA, and 1 mM DTT) at 4°C three times, with the last time overnight. After centrifugation, the supernatant was loaded onto a HiLoad 16/600 Superdex‐200 pg (GE Healthcare, 28‐9893‐35) column equilibrated with refolding buffer to collect the octamer. For nucleosome assembly, proper amounts of octamer and 601 DNA (245 bp produced by PCR) were mixed and dialyzed against buffers (20 mM Tris–HCl, pH 8.0) with a series of decreasing concentrations of NaCl (2.0, 1.6, 1.2, 1.0, 0.8, and 0.6 M), each for 1 h at 4°C. The mixture was finally dialyzed against TE buffer overnight to obtain nucleosomes.
Nucleosome sliding assay
The nucleosome sliding assay was performed as previously described (Watanabe et al, 2015). In brief, a 2 μg quantity of mono‐nucleosomes with 245‐bp 601 DNA was incubated with chromatin‐remodeling enzymes (CHR11 or PIE1) and 2 mM ATP in sliding buffer (10 mM Tris–HCl, pH 8.0, 70 mM NaCl, 5 mM MgCl2, 0.1 mg/ml BSA, and 1 mM DTT) for 1 h at 30°C. The reactions were quenched with 2 mg of λDNA and 5% glycerol, and then incubated for another 5 min at 30°C. Samples were resolved on 5% native PAGE in 0.5× TBE buffer and were run at 80 V for 150 min at 4°C. Nucleosomes were visualized by ethidium bromide staining.
Author contributions
Y‐XL, X‐MH, and X‐JH designed the research; Y‐XL, X‐MH, L‐MT, C‐JZ, C‐RS, LL, and SC performed the research; R‐NL, Y‐NS, and X‐WC performed the bioinformatic analysis; and Y‐XL and X‐JH wrote the paper. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Dataset EV7
Review Process File
Acknowledgments
We thank Chang Huang and Dr. Bing Zhu for their technical assistance in the assembly of nucleosomes. This work was supported by the National Key Research and Development Program of China (2016YFA0500801) from the Chinese Ministry of Science and Technology.
The EMBO Journal (2020) 39: e102008
Data availability
Raw RNA‐seq, ChIP‐seq, and MNase‐seq data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE139465; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139465).
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF (2007) Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572 [DOI] [PubMed] [Google Scholar]
- Bao Y, Shen X (2011) SnapShot: chromatin remodeling: INO80 and SWR1. Cell 144: 158.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B (2014) ISWI chromatin remodeling: one primary actor or a coordinated effort? Curr Opin Struct Biol 24: 150–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B, Bishop B, Ho KK, Huang R, Jia W, Zhang H, Pascuzzi PE, Deal RB, Ogas J (2018) The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in Arabidopsis . Plant Cell 30: 1337–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud‐Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Choi K, Park C, Lee J, Oh M, Noh B, Lee I (2007) Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134: 1931–1941 [DOI] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FCH (2013) Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet 45: 1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Coleman‐Derr D, Zilberman D (2012) Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet 8: e1002988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Längst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB (1999) ISWI is an ATP‐dependent nucleosome remodeling factor. Mol Cell 3: 239–245 [DOI] [PubMed] [Google Scholar]
- Crevillen P, Gomez‐Zambrano A, Lopez JA, Vazquez J, Pineiro M, Jarillo JA (2019) Arabidopsis YAF9 histone readers modulate flowering time through NuA4‐complex‐dependent H4 and H2A.Z histone acetylation at FLC chromatin. New Phytol 222: 1893–1908 [DOI] [PubMed] [Google Scholar]
- Dai X, Bai Y, Zhao L, Dou X, Liu Y, Wang L, Li Y, Li W, Hui Y, Huang X (2017) H2A.Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in Arabidopsis . Mol Plant 10: 1274–1292 [DOI] [PubMed] [Google Scholar]
- Dann GP, Liszczak GP, Bagert JD, Müller MM, Nguyen UT, Wojcik F, Brown ZZ, Bos J, Panchenko T, Pihl R (2017) ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 548: 607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB (2007) Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Gao Z, Liu S, Li G, Yang Z, Huang H, Xu L (2013) SLIDE, the protein interacting domain of imitation switch remodelers, binds DDT‐domain proteins of different subfamilies in chromatin remodeling complexes. J Integr Plant Biol 55: 928–937 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Ferrari S, Langst G, Straub T, Imhof A, Varga‐Weisz P, Wilm M, Becker PB (2001) Acf1, the largest subunit of CHRAC, regulates ISWI‐induced nucleosome remodelling. EMBO J 20: 3781–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen‐Hughes T (2011) A role for Snf2‐related nucleosome‐spacing enzymes in genome‐wide nucleosome organization. Science 333: 1758–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Zambrano Á, Crevillén P, Franco‐Zorrilla JM, López JA, Moreno‐Romero J, Roszak P, Santos‐González J, Jurado S, Vázquez J, Köhler C (2018) Arabidopsis SWC4 binds DNA and recruits the SWR1 complex to modulate histone H2A.Z deposition at key regulatory genes. Mol Plant 11: 815–832 [DOI] [PubMed] [Google Scholar]
- Hale CJ, Potok ME, Lopez J, Do T, Liu A, Gallego‐Bartolome J, Michaels SD, Jacobsen SE (2016) Identification of multiple proteins coupling transcriptional gene silencing to genome stability in Arabidopsis thaliana . PLoS Genet 12: e1006092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Wu MF, Cui S, Wagner D (2015) Roles and activities of chromatin remodeling ATP ases in plants. Plant J 83: 62–77 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huanca‐Mamani W, Garcia‐Aguilar M, León‐Martínez G, Grossniklaus U, Vielle‐Calzada J‐P (2005) CHR11, a chromatin‐remodeling factor essential for nuclear proliferation during female gametogenesis in Arabidopsis thaliana . Proc Natl Acad Sci 102: 17231–17236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii J, Kondo T, Makino H, Ogura A, Matsuda F, Kondo A (2014) Three gene expression vector sets for concurrently expressing multiple genes in Saccharomyces cerevisiae . FEMS Yeast Res 14: 399–411 [DOI] [PubMed] [Google Scholar]
- Jarillo JA, Piñeiro M (2015) H2A.Z mediates different aspects of chromatin function and modulates flowering responses in Arabidopsis . Plant J 83: 96–109 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Deal RB, McKinney EC, Meagher RB (2005) Silencing the nuclear actin‐related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J 41: 845–858 [DOI] [PubMed] [Google Scholar]
- Kapoor P, Shen X (2014) Mechanisms of nuclear actin in chromatin‐remodeling complexes. Trends Cell Biol 24: 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorasanizadeh S (2004) The nucleosome: from genomic organization to genomic regulation. Cell 116: 259–272 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J (2004) A protein complex containing the conserved Swi2/Snf2‐related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol 2: e131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA (2010) H2A.Z‐containing nucleosomes mediate the thermosensory response in Arabidopsis . Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Langdon WB (2015) Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min 8: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zhang J, Li J, Yang Z, Huang H, Xu L (2012) Imitation Switch chromatin remodeling factors and their interacting RINGLET proteins act together in controlling the plant vegetative phase in Arabidopsis . Plant J 72: 261–270 [DOI] [PubMed] [Google Scholar]
- Li G, Liu S, Wang J, He J, Huang H, Zhang Y, Xu L (2014) ISWI proteins participate in the genome‐wide nucleosome distribution in Arabidopsis . Plant J 78: 706–714 [DOI] [PubMed] [Google Scholar]
- Li D, Liu J, Liu W, Li G, Yang Z, Qin P, Xu L (2017) The ISWI remodeler in plants: protein complexes, biochemical functions, and developmental roles. Chromosoma 126: 365–373 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2013) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ (2012) New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 13: 436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y‐X, Han Y‐F, Zhao Q‐Y, Du J‐L, Dou K, Li L, Chen S, He X‐J (2018) Sumoylation of SUVR2 contributes to its role in transcriptional gene silencing. Sci China Life Sci 61: 235–243 [DOI] [PubMed] [Google Scholar]
- March‐Díaz R, García‐Domínguez M, Florencio FJ, Reyes JC (2007) SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol 143: 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March‐Díaz R, Reyes JC (2009) The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol Plant 2: 565–577 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu W‐H, Sen S, Wu C (2004) ATP‐driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Ranjan A, Stengel F, Wei D, Aebersold R, Wu C, Leschziner AE (2013) Molecular architecture of the ATP‐dependent chromatin‐remodeling complex SWR1. Cell 154: 1220–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie WF, Lei M, Zhang M, Tang K, Huang H, Zhang C, Miki D, Liu P, Yang Y, Wang X et al (2019) Histone acetylation recruits the SWR1 complex to regulate active DNA demethylation in Arabidopsis . Proc Natl Acad Sci 116: 16641–16650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolayeva O, Robinson MD (2014) edgeR for differential RNA‐seq and ChIP‐seq analysis: an application to stem cell biology. Methods Mol Biol 1150: 45–79 [DOI] [PubMed] [Google Scholar]
- Noh Y‐S, Amasino RM (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis . Plant Cell 15: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Cui Y, Bi YM, Rothstein SJ (2006) AtMBD9: a protein with a methyl‐CpG‐binding domain regulates flowering time and shoot branching in Arabidopsis . Plant J 46: 282–296 [DOI] [PubMed] [Google Scholar]
- Potok ME, Wang Y, Xu L, Zhong Z, Liu W, Feng S, Naranbaatar B, Rayatpisheh S, Wang Z, Wohlschlegel JA et al (2019) Arabidopsis SWR1‐associated protein methyl‐CpG‐binding domain 9 is required for histone H2A.Z deposition. Nat Commun 10: 3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD (2005) Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T (2014) deepTools: a flexible platform for exploring deep‐sequencing data. Nucleic Acids Res 42: W187–W191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M, Von Harder M, Cigliano RA, Schlögelhofer P, Scheid OM (2013) The Arabidopsis SWR1 chromatin‐remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 25: 1990–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ (2011) Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145: 692–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijacic P, Holder DH, Bajic M, Deal RB (2019) Methyl‐CpG‐binding domain 9 (MBD9) is required for H2A.Z incorporation into chromatin at a subset of H2A.Z‐enriched regions in the Arabidopsis genome. PLoS Genet 15: e1008326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Muiño JM, Blanvillain R, Busscher M, Busscher‐Lange J, Dinh QP, Liu S, Westphal AH, Boeren S (2012) Characterization of MADS‐domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sura W, Kabza M, Karlowski WM, Bieluszewski T, Kus‐Slowinska M, Pawełoszek Ł, Sadowski J, Ziolkowski PA (2017) Dual role of the histone variant H2A.Z in transcriptional regulation of stress‐response genes. Plant Cell 29: 791–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S (2014) Environmental responses mediated by histone variants. Trends Cell Biol 24: 642–650 [DOI] [PubMed] [Google Scholar]
- Tan LM, Zhang CJ, Hou XM, Shao CR, Lu YJ, Zhou JX, Li YQ, Li L, Chen S, He XJ (2018) The PEAT protein complexes are required for histone deacetylation and heterochromatin silencing. EMBO J 37: e98770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Tan D, Lakshminarasimhan M, Washburn MP, Hong E‐JE, Walz T, Peterson CL (2015) Structural analyses of the chromatin remodelling enzymes INO80‐C and SWR‐C. Nat Commun 6: 7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W‐H, Wu C‐H, Ladurner A, Mizuguchi G, Wei D, Xiao H, Luk E, Ranjan A, Wu C (2009) N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J Biol Chem 284: 6200–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Grullon S, Ge K, Peng W (2014) Spatial clustering for identification of ChIP‐enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol Biol 1150: 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadon AN, Tsukiyama T (2011) SnapShot: chromatin remodeling: ISWI. Cell 144: 453.e1 [DOI] [PubMed] [Google Scholar]
- Yaish MW, Peng M, Rothstein SJ (2009) AtMBD9 modulates Arabidopsis development through the dual epigenetic pathways of DNA methylation and histone acetylation. Plant J 59: 123–135 [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR (2005) Genome‐wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C‐J, Hou X‐M, Tan L‐M, Shao C‐R, Huang H‐W, Li Y‐Q, Li L, Cai T, Chen S, He X‐J (2016) The Arabidopsis acetylated histone‐binding protein BRAT1 forms a complex with BRP1 and prevents transcriptional silencing. Nat Commun 7: 11715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman‐Derr D, Ballinger T, Henikoff S (2008) Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Dataset EV7
Review Process File
Data Availability Statement
Raw RNA‐seq, ChIP‐seq, and MNase‐seq data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE139465; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139465).


