Abstract
We previously reported that under the stress condition caused by the addition of 2-hydroxyethyl disulfide, a thiol-specific oxidant, to growing cultures of Escherichia coli BL21(DE3), a population of stress-responsive proteins [peptidyl-prolyl cis–trans isomerase B (PpiB), bacterioferritin (Bfr), putative HTH-type transcriptional regulator yjdC (YjdC), dihydrofolate reductase (FolA), chemotaxis protein cheZ (CheZ), and glutathione synthetase (GshB)] were significantly upregulated when compared with the nonstress condition. When those stress-responsive proteins were used as fusion partners for the expression of human granulocyte colony-stimulating factor (hG-CSF), the solubility of hG-CSF was dramatically enhanced in E. coli cytoplasm, whereas almost all of the directly expressed hG-CSF were aggregated to inclusion bodies. In addition, the spectra of circular dichroism measured with the purified hG-CSF were identical to that of standard hG-CSF, implying that the synthesized hG-CSF has native conformation. These results indicate that the bacterial stress-responsive proteins could be potent fusion expression partners for aggregation-prone heterologous proteins in E. coli cytoplasm.
Keywords: G-CSF, Escherichia coli BL21(DE3), E. coli stress-responsive proteins, fusion partner
Introduction
Human granulocyte colony-stimulating factor (hG-CSF) is a hematopoietic growth factor that plays an important role in hematopoietic cell proliferation, differentiation of hemopoietic precursor cells, and activation of mature neutrophilic granulocytes (Metcalf, 1985; Nomura et al., 1986). In addition, hG-CSF has been widely used for treating neutropenia caused by cancer chemotherapy. The mature hG-CSF is an 18.7-kDa glycoprotein that predominantly consists of 174 amino acids with two intramolecular disulfide bonds, although another minor form comprised of 177 amino acids as a result of alternative splicing of mRNA has also been described (Nagata et al., 1986; Lu et al., 1992; Hill et al., 1993). Furthermore, native hG-CSF has only one single O-glycosylation site at Thr133, which is not essential for biological activity (Kubota et al., 1990). Therefore, nonglycosylated recombinant hG-CSF has the same specific biological activity as natural glycosylated hG-CSF (Souza et al., 1986; Oh-eda et al., 1990). Because hG-CSF is synthesized in Escherichia coli as inclusion bodies, solubilization and renaturation steps are necessary to obtain the native conformation (Misawa & Kumagai, 1999), which means that highly complicated downstream processes with low-recovery yield are required.
Escherichia coli is one of the most widely used organisms for the commercial production of therapeutic and industrial recombinant proteins (Baneyx & Mujacic, 2004) for several reasons, including low manufacturing costs, rapid product accumulation, and well-established tools for genetic manipulation (Eiteman & Altman, 2006). Despite these advantages, overproduced heterologous proteins in E. coli often form nonproductive inclusion bodies, and fusion expression using solubility enhancer proteins as fusion partner has emerged as an efficient production strategy to overcome the inclusion body formation (Baneyx & Mujacic, 2004; Sørensen & Mortensen, 2005). Currently, Shistosoma japonicum glutathione S-transferase (Smith & Johnson, 1988), E. coli maltose-binding protein (Bach et al., 2001), E. coli N utilization substance A (De Marco et al., 2004) and E. coli thioredoxin (LaVallie et al., 1993) are the most extensively examined fusion partners (Davis et al., 1999; Nallamsetty & Waugh, 2006), but they are not free from proprietary protection upon commercial application. Moreover, the traditional solubility-enhancing fusion partner proteins are not equally effective in promoting the folding of fusion partners (Dyson et al., 2004; Park et al., 2008). In this study, we report a novel and efficient strategy for the synthesis of soluble and correctly folded hG-CSF using various E. coli stress-responsive proteins that we previously found through a proteome-wide analysis of E. coli proteins under 2-hydroxyethyl disulfide (2-HEDS)-induced stress condition (Han et al., 2008). Using the six stress-responsive proteins of E. coli, we successfully produced hG-CSF with correct conformation as well as demonstrated using circular dichroism (CD).
Materials and methods
Bacterial strain and plasmid vectors
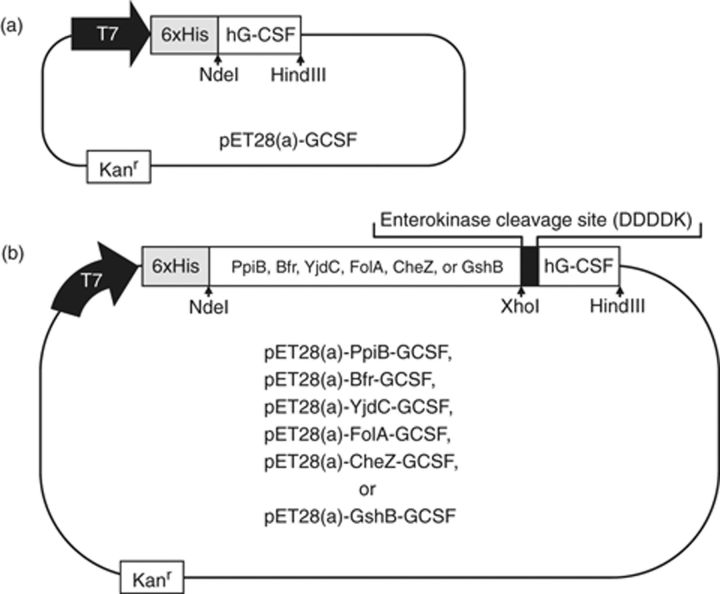
Escherichia coli strain BL21(DE3) [F−ompT hsdSB(rB−mB−)] was used for the hG-CSF synthesis. After PCR amplification using appropriate primers, hG-CSF gene and its various fusion mutants (YjdC∷hG-CSF, GshB∷hG-CSF, FolA∷hG-CSF, Bfr∷hG-CSF, CheZ∷hG-CSF, PpiB∷hG-CSF) were inserted into the NdeI–HindIII site of plasmid pET28a (Novagen) to construct the fusion expression vector (Fig. 1). After complete DNA sequencing of all gel-purified plasmid vectors, the E. coli BL21(DE3) was transformed with the plasmid expression vectors, and kanamycin-resistant transformants were subsequently selected using Luria–Bertani (LB) agar plates supplemented with kanamycin (50 mg L−1).
1.
Plasmid vectors used for the direct (a) and fusion (b) expression of hG-CSF. As shown in (b), the plasmid vectors for the fusion expression of hG-CSF contain hexa-histidine tag (His)6 and the specific sequence for enterokinase cleavage (DDDDK).
Recombinant E. coli culture, gene expression, and recombinant hG-CSF purification
For shake flask experiments, 250-mL Erlenmeyer flasks containing 50 mL LB media with 50 mg L−1 kanamycin were incubated at 37 °C and 200 r.p.m. When the culture turbidity (OD600 nm) reached 0.5, gene expression was induced by the addition of isopropyl-β-d-thiogalactoside (1 mM), and after an additional 4 h of cultivation all of the recombinant cells were harvested by centrifugation (MICRO17TR, Hanil Science Industrial, Korea); 16 609 g for 5 min. The cell pellets were then resuspended in 5 mL lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM EDTA) and were disrupted using a Branson Sonifier (Branson Ultrasonics Corp., Danbury, CT). The cell-free supernatant and insoluble protein aggregates were then separated at 16 609 g (MICRO17TR, Hanil Science Industrial) for 10 min. The isolated inclusion bodies, if any, were washed twice with 1% Triton X-100. The cell-free supernatants and the washed inclusion bodies were then subjected to polyacrylamide gel electrophoresis (PAGE) analysis, and Coomassie-stained protein bands were scanned and analyzed by densitometry (Duoscan T1200, Bio-Rad, Hercules, CA).
The purification of recombinant hG-SCF was accomplished using metal affinity chromatography. Briefly, polyhistidine-tagged hG-CSF fusion mutants [(His)6∷fusion partner∷(D4K)∷hG-CSF] were loaded onto a ProBond resin (Ni+2) column. Before sample loading, the resin was washed twice with 10 column volumes of binding buffer (50 mM potassium phosphate, 300 mM KCl, 20 mM imidazole, pH 7.0), which contained 20 mM imidazole to minimize nonspecific binding of untagged protein contaminants. Binding was then conducted in batch mode at 4 °C, after which the resin was washed twice with 5–8 mL Tris-HCl (10 mM Tris, pH 8.0) before the enterokinase digestion step. Enterokinase digestion was carried out in batch mode at 4 °C for 10 h using 5 U of enterokinase (Invitrogen, CA). Next, the proteolytic product was collected and centrifuged (MICRO17TR, Hanil Science Industrial); 16 609 g for 10 min, and the supernatant fraction was then analyzed by sodium dodecyl sulfate (SDS)-PAGE and Western blotting. Removal of the enterokinase was conducted following the manufacturer's protocol (EK-Away™ resin, Cat. No. R180-01, Invitrogen, Germany).
SDS-PAGE and Western blot analysis
hG-CSF digests were resuspended in 0.5 M Tris-HCl, pH 6.8, 10% SDS, 10% glycerol, 0.05% bromophenol blue, 5% 2-mercaptoethanol, and then subjected to reducing 12% SDS-PAGE using Tris-glycine gel. Peptides were transferred onto a nitrocellulose membrane (Schleicher & Schuell BioScience GmbH, Germany) for 2 h at 4 °C in 1 × transfer buffer (glycine 2.9 g L−1, Tris base 5.8 g L−1, SDS 0.37 g L−1, MEOH 200 mL, distilled water 800 mL). Nonspecific antibody-binding sites on the membrane were blocked by incubating for 1 h at room temperature with blocking buffer [1% nonfat skim milk in phosphate-buffered saline (PBS)]. After washing three times with PBS, the membrane was incubated for an additional 1 h at room temperature with 1 : 1000 diluted hG-CSF primary antibody (mouse anti-human G-CSF monoclonal antibody; Clone 3D1, Santa Cruz Biotechnology Inc., CA). The membrane was washed again as described above, after which it was incubated with 1 : 1000 diluted goat anti-mouse immunoglobulin G-conjugated horseradish peroxidase (HRP) secondary antibody (Santa Cruz Biotechnology Inc.). After being washed, the membrane was developed using an HRP conjugate substrate kit (Sigma-Aldrich, MO).
CD
The CD spectra of commercial standard hG-CSF [Grasin Prefilled-Syringe (Filgrastim), Kirin, Japan] and the purified hG-CSF (74 mg L−1) were measured using a JASCO J-710 spectropolarimeter (Korea Basic Science Center, Ochang, Korea) at room temperature.
Results and discussion
Direct expression of hG-CSF gene in the cytoplasm of E. coli
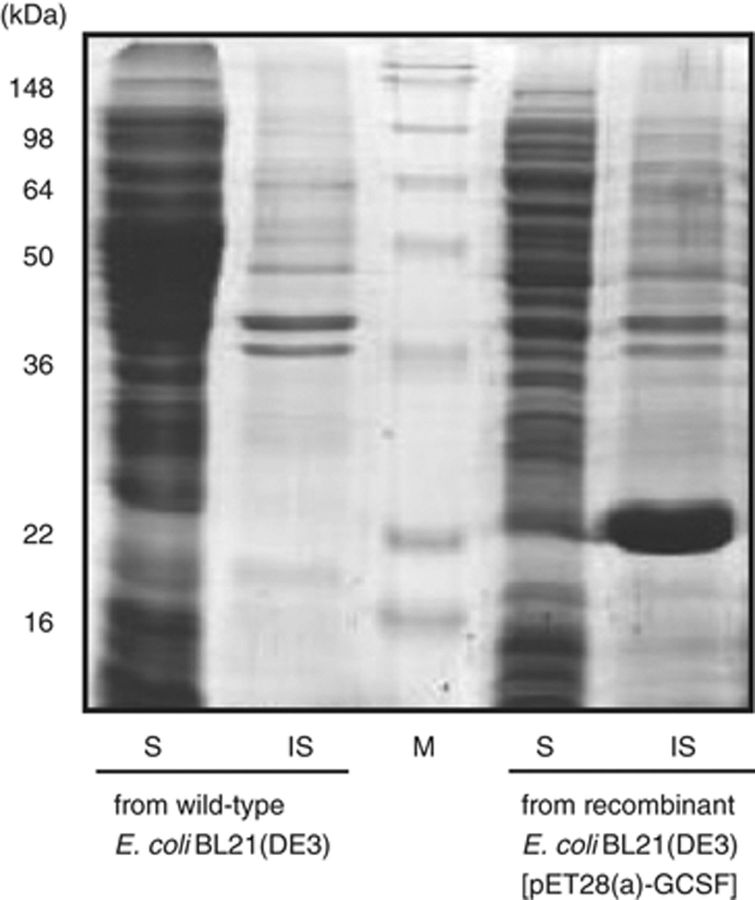
For the direct expression of mature hG-CSF gene in E. coli cytoplasm, the plasmid vector, pT7-GCSF was constructed by inserting the mature hG-CSF gene into the NdeI–HindIII site of the pT7-7 vector. However, as previously reported by Devlin et al. (1988), the native hG-CSF gene is hardly transcribed in E. coli because of the high (G+C) contents at 5′ end of coding strand. Therefore, the 5′-end coding sequences of hG-CSF gene were modified by changing the first five codons (5′ ACC CCC CTG GGC CCT 3′) to the high-frequency codon of E. coli (5′ ACT CCG TTA GGT CCA 3′) (Jeong & Lee, 2001). As shown in Fig. 2, the expression of the modified hG-CSF gene resulted in the synthesis of a large amount (>40% of total E. coli proteins) of recombinant hG-CSF. However, most of the synthesized hG-CSF was present in the form of insoluble inclusion bodies.
2.
Results of SDS-PAGE analysis of directly expressed hG-CSF. S and IS, soluble and insoluble fractions of Escherichia coli cell lysates, respectively.
Strategy to enhance solubility of recombinant hG-CSF using E. coli stress-responsive proteins
The stress induced by overexpression of recombinant proteins resembles environmental stress conditions such as heat shock or amino acid starvation (Sørensen & Mortensen, 2005). Similarly, when proteins are exposed to oxidative stressors such as oxygen radicals and hydrogen peroxide (H2O2), E. coli decreases the protein synthesis rate and enhances the protein degradation pathway (Koo et al., 2004). Previously, VanBogelen et al. (1987) reported that the expression level of molecular chaperones such as DnaK and GroES increased in the presence of H2O2-derived oxidative stress. It has also been reported that addition of 2-HEDS, a thiol-specific oxidant, dramatically reduced the NADPH/NADP+ ratio consistently in response to severe oxidative stress (Halleck et al., 1997; Ayene et al., 2002). In our recent report (Han et al., 2008), we investigated the E. coli proteome under 2-HEDS-induced stress conditions and found stress-responsive proteins that are significantly upregulated by the stressor through the comparative and quantitative analysis of E. coli proteomes under nonstress and stress conditions. Because many proteins were aggregated by 2-HEDS-induced stress, it seems reasonable to assume that such stress-responsive proteins have an intrinsically higher folding efficiency than other proteins that are aggregated in response to the same stress conditions. Among the stress-responsive proteins that we previously found (Han et al., 2008), we were interested in the six stress-responsive proteins [peptidyl-prolyl cis–trans isomerase B (PpiB, 18 kDa), bacterioferritin (Bfr, 19 kDa), putative HTH-type transcriptional regulator yjdC (YjdC, 22 kDa), dihydrofolate reductase (FolA, 18 kDa), chemotaxis protein cheZ (CheZ, 24 kDa), and glutathione synthetase (GshB, 36 kDa)] that have relatively lower molecular mass compared with the other stress-responsive proteins. We found that these relatively small stress-responsive proteins were highly effective in enhancing the solubility of recombinant hG-CSF when used as fusion partner, which will be discussed next.
Fusion expression of hG-CSF using E. coli stress-responsive proteins as fusion partners
The six stress-responsive proteins (PpiB, Bfr, YjdC, FolA, CheZ, and GshB) were used as N-terminus fusion expression partner to synthesize the hybrid proteins, i.e. NH2-[fusion partner]∷[D4K]∷[hG-CSF]-COOH in E. coli cytoplasm. Proteins with high molecular mass are usually not used as fusion partner because the big fusion partners may reduce the actual amount of target protein that is obtained after the fusion partner is removed. PpiB (18 kDa), Bfr (19 kDa), YjdC (22 kDa), FolA (18 kDa), CheZ (24 kDa), and GshB (36 kDa) are relatively small proteins compared with the other proteins above and were also the most effective in enhancing the solubility of hG-CSF. As shown in Fig. 3a and b, the N-terminus fusion of six stress-responsive proteins led to a significant increase in both the expression level and solubility of hG-CSF. Inclusion body formation presumably occurs due to protein overexpression in a heterologous environment in which nascent polypeptide chains are continuously synthesized and then aggregate because of nonspecific hydrophobic interactions among the partially folded intermediates of the recombinant proteins, various host proteins, or a combination of the two within the cytoplasm. In many cases, broad exposure of hydrophobic surfaces on newly synthesized polypeptides results in intermolecular aggregation and misfolding. To prevent such occurrences, molecular chaperones (DnaK/DnaJ/GrpE system, GroEL/GroES, etc.) that recognize and interact with hydrophobic patches of nascent proteins can be used (Maier et al., 2005). Hydrophobic amino acids are usually buried within the core of folded proteins but become exposed in the partially folded polypeptide intermediates (Rüdiger et al., 1997; Suh et al., 1999). Because of this, the solubility-enhancing function of stress-responsive proteins may be similar to that of cis-acting molecular chaperones in the context of fusion proteins (Kapust & Waugh, 1999) or as a ‘chaperone magnet’ (Fox et al., 2001; Ahn et al., 2005) that effectively recruits chaperone binding and prohibits undesired and nonspecific protein–protein interactions.
3.
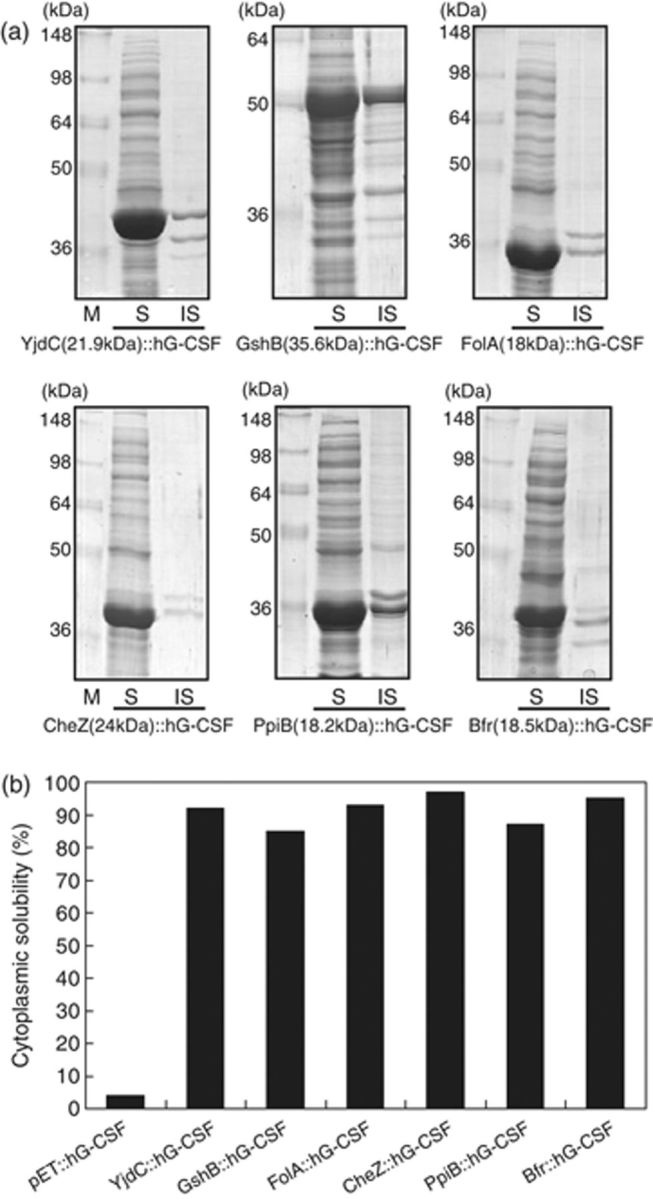
(a) Results of SDS-PAGE analyses of the fusion-expressed hG-CSF. M, molecular markers; S and IS, soluble and insoluble fractions of cell lysates of recombinant Escherichia coli, respectively. (b) Solubility (%) of six recombinant fusion mutants of hG-CSF and directly expressed hG-CSF in E. coli cytoplasm.
Purification and characterization of fusion partner-free hG-CSF
Because the addition of a fusion partner to the native peptide can influence the biological activities and the physical properties of the protein (Esposito & Chatterjee, 2006), the removal of the fusion tags is of crucial importance especially when the recombinant protein is a human therapeutic reagent. In order to purify the fusion partner-free hG-CSF in E. coli, we already inserted the hybrid gene encoding NH2-[fusion tag]∷[D4K]∷[hG-CSF]-COOH into the NdeI–HindIII site of the pET28a(+) plasmid vector. Most of the expressed fusion protein, i.e. NH2-[(His)6]∷[fusion tag]∷[D4K]∷[hG-CSF]-COOH was present in the soluble fraction of the cell lysates, and then the soluble fraction was subjected to metal affinity purification using an Ni2+–NTA column (Wülfing et al., 1994). After loading the soluble fraction containing the fusion proteins onto the Ni2+-NTA column, we subsequently added enterokinase to the column to remove the fusion tags. Because the sequence D4K for enterokinase cleavage is downstream of the fusion tag, the cleaved fusion tags as well as any uncleaved fusion proteins still have the (His)6 tag and hence remains bound to the Ni2+–NTA column, while the released hG-CSF is found in the flow-through from the Ni2+–NTA column. The flow-through containing the released hG-CSF was centrifuged, and the insoluble pellet and supernatant were then analyzed by SDS-PAGE and Western blot (Fig. 4). Figure 4 clearly indicates that the recombinant hG-CSF released from the fusion protein was still present in the form of soluble protein.
4.
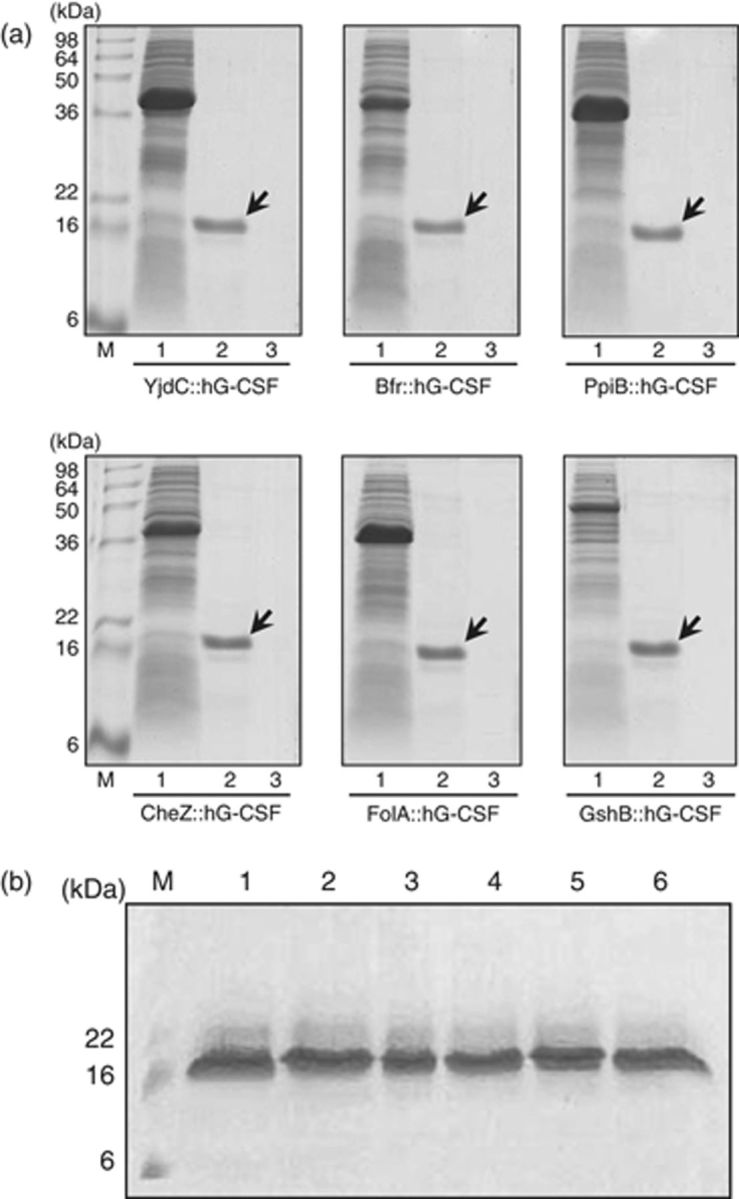
(a) Results of SDS-PAGE analyses showing the solubility of recombinant hG-CSF purified after the removal of fusion partner by enterokinase treatment. M, molecular markers; 1, soluble fraction of recombinant cell lysates; 2, soluble fraction of purified hG-CSF; 3, insoluble fraction of purified hG-CSF; each arrow indicates the purified hG-CSF. (b) Results of Western blot analyses of purified hG-CSF that were run from YjdC∷hG-CSF (lane 1), GshB∷hG-CSF (lane 2), FolA∷hG-CSF (lane 3), CheZ∷hG-CSF (lane 4), PpiB∷hG-CSF (lane 5), and Bfr∷hG-CSF (lane 6).
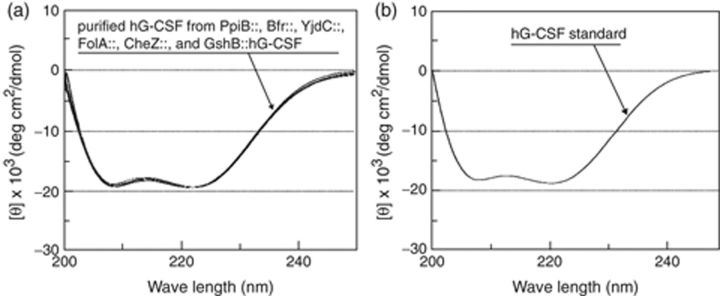
With the purified hG-CSF released from all the six fusion proteins, the spectra of CD were analyzed and compared with the CD spectrum of commercial standard hG-CSF [Grasin Prefilled-Syringe (Filgrastim), Kirin]. CD is very useful for the analysis of secondary structures of polypeptides and proteins (Purdie et al., 1989). Recent studies also verified the proper folding of recombinant hG-CSF using CD spectra (Bae et al., 1998, 1999; Jeong & Lee, 2001). In the other recent studies using CD spectrum, properly folded conformations of recombinant human and viral proteins were structurally analyzed (Leopoldino et al., 2006; Brucz et al., 2007; Tolun et al., 2007; Vamvakas et al., 2007). The CD spectra of the purified fusion-free hG-CSF (Fig. 5a) were identical to the spectrum of commercial hG-CSF standard (Fig. 5b), thereby indicating that the recombinant hG-CSF has a conformation with the correct secondary structure. It is also assumed that the recombinant hG-CSF possesses biological activities similar to native hG-CSF based on the proper conformation of recombinant hG-CSF, even though the biological activity was not directly determined (Bae et al., 1998, 1999; Jeong & Lee, 2001).
5.
CD spectra measured with the purified recombinant hG-CSF that were run from PpiB∷, Bfr∷, YjdC∷, FolA∷, CheZ∷, and GshB∷hG-CSF (a) and CD spectrum of commercial standard of hG-CSF (b).
Conclusion
We used the upregulated stress-responsive proteins as N-terminus fusion expression partners upon the synthesis of hG-CSF, and then the hybrid proteins designated as NH2-[fusion partner]∷[D4K]∷[hG-CSF]-COOH were synthesized in E. coli cytoplasm. The synthesized hG-CSF was aggregated and formed inclusion bodies when directly expressed without the N-terminus fusion tag. However, the solubility of hG-CSF in E. coli cytoplasm dramatically increased when the stress-responsive proteins were used as fusion partners, indicating that the fusion expression partners were highly effective solubility enhancers. In addition, even after the fusion partner was removed by enzymatic cleavage, the released, i.e. fusion-free recombinant hG-CSF was present in the form of soluble protein. The CD spectra of the affinity-purified hG-CSF was investigated, and the results showed that the recombinant hG-CSF had a conformation with the correct secondary structure. Consequently, the six stress-responsive E. coli proteins were highly effective cis-acting solubility and folding enhancers for the production of hG-CSF and could be used for the bacterial synthesis of other aggregation-prone heterologous proteins in the form of soluble and active proteins.
Acknowledgements
We thank Professor Hang Chul Shin at Soongsil University for kindly providing the gene clones of hG-CSF. This study was supported by the National Research Laboratory Project of the Ministry of Science and Technology (grant no. ROA-2007-000-20084-0) and by the Korea Health 21 R&D Project of the Ministry of Health & Welfare of the Republic of Korea (grant no. A050750). This work was also supported by the Second Brain Korea 21 Project. Additional support from the Korea Science and Engineering Foundation (grant no. R01-2005-000-10355-0) and the Korea Research Foundation (grant no. KRF-2004-041-D00180) are also appreciated.
Footnotes
Editor: Peter Lund
References
- Ahn JY, Choi H, Kim YH, Han KY, Park JS, Han SS, Lee J. (2005) Heterologous gene expression using self-assembled supra-molecules with high affinity for HSP70 chaperone. Nucleic Acids Res 33: 3751–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayene IS, Stamato TD, Mauldin SK, Biaglow JE, Tuttle SW, Jenkins SF, Koch CJ. (2002) Mutation in the glucose-6-phosphate dehydrogenase gene leads to inactivation of Ku DNA end binding during oxidative stress. J Biol Chem 277: 9929–9935. [DOI] [PubMed] [Google Scholar]
- Bach H, Mazor Y, Shaky S, Shoham-Lev A, Berdichevsky Y, Gutnick DL, Benhar I. (2001) Escherichia coli maltose-binding protein as a molecular chaperone for recombinant intracellular cytoplasmic single-chain antibodies. J Mol Biol 312: 79–93. [DOI] [PubMed] [Google Scholar]
- Bae CS, Yang DS, Chang KR, Seong BL, Lee J. (1998) Enhanced secretion of human granulocyte colony-stimulating factor directed by a novel hybrid fusion peptide from recombinant Saccharomyces cerevisiae at high cell concentration. Biotechnol Bioeng 57: 600–609. [DOI] [PubMed] [Google Scholar]
- Bae CS, Yang DS, Lee J, Park YH. (1999) Improved process for production of recombinant yeast-derived monomeric human G-CSF. Appl Microbiol Biot 52: 338–344. [DOI] [PubMed] [Google Scholar]
- Baneyx F, Mujacic M. (2004) Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 22: 1399–1408. [DOI] [PubMed] [Google Scholar]
- Brucz K, Miknis ZJ, Schultz LW, Umland TC. (2007) Expression, purification and characterization of recombinant severe acute respiratory syndrome coronavirus non-structural protein 1. Protein Expres Purif 52: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GD, Elisee C, Newham DM, Harrison RG. (1999) New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol Bioeng 65: 382–388. [PubMed] [Google Scholar]
- De Marco V, Stier G, Blandin S, De Marco A. (2004) The solubility and stability of recombinant proteins are increased by their fusion to NusA. Biochem Bioph Res Co 322: 766–771. [DOI] [PubMed] [Google Scholar]
- Devlin PE, Drummond RJ, Toy P, Mark DF, Watt KW, Devlin JJ. (1988) Alteration of amino-terminal codons of human granulocyte-colony-stimulating factor increases expression levels and allows efficient processing by methionine aminopeptidase in Escherichia coli. Gene 65: 13–22. [DOI] [PubMed] [Google Scholar]
- Dyson MR, Shadbolt SP, Vincent KJ, Perera RL, McCafferty J. (2004) Production of soluble mammalian proteins in Escherichia coli: identification of protein features that correlate with successful expression. BMC Biotechnol 4: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiteman MA, Altman E. (2006) Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol 24: 530–536. [DOI] [PubMed] [Google Scholar]
- Esposito D, Chatterjee DK. (2006) Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotech 17: 353–358. [DOI] [PubMed] [Google Scholar]
- Fox JD, Kapust RB, Waugh DS. (2001) Single amino acid substitutions on the surface of Escherichia coli maltose-binding protein can have a profound impact on the solubility of fusion proteins. Protein Sci 10: 622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halleck MM, Liu H, North J, Stevens JL. (1997) Reduction of trans-4,5-dihydroxy-1,2-dithiane by cellular oxidoreductases activates gadd153/chop and grp78 transcription and induces cellular tolerance in kidney epithelial cells. J Biol Chem 272: 21760–21766. [DOI] [PubMed] [Google Scholar]
- Han KY, Park JS, Seo HS, Ahn KY, Lee J. (2008) Multiple stressor-induced proteome responses of Escherichia coli BL21(DE3). J Proteome Res 7: 1891–1903. [DOI] [PubMed] [Google Scholar]
- Hill CP, Osslund TD, Eisenberg D. (1993) The structure of granulocyte-colony-stimulating factor and its relationship to other growth factors. P Natl Acad Sci USA 90: 5167–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KJ, Lee SY. (2001) Secretory production of human granulocyte colony-stimulating factor in Escherichia coli. Protein Expres Purif 23: 311–318. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Waugh DS. (1999) Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci 8: 1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BM, Yoon MJ, Lee CR, Nam TW, Choe YJ, Jaffe H, Peterkofsky A, Seok YJ. (2004) A novel fermentation/respiration switch protein regulated by enzyme IIAGlc in Escherichia coli. J Biol Chem 279: 31613–31621. [DOI] [PubMed] [Google Scholar]
- Kubota N, Orita T, Hattori K, Oh-eda M, Ochi N, Yamazaki T. (1990) Structural characterization of natural and recombinant human granulocyte colony-stimulating factors. J Biochem 107: 486–492. [DOI] [PubMed] [Google Scholar]
- LaVallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM. (1993) A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Nat Biotechnol 11: 187–193. [DOI] [PubMed] [Google Scholar]
- Leopoldino AM, Canduri F, Cabral H. et al (2006) Expression, purification, and circular dichroism analysis of human CDK9. Protein Expres Purif 47: 614–620. [DOI] [PubMed] [Google Scholar]
- Lu HS, Clogston CL, Narhi LO, Merewether LA, Pearl WR, Boone TC. (1992) Folding and oxidation of recombinant human granulocyte colony stimulating factor produced in Escherichia coli. J Biol Chem 267: 8770–8777. [PubMed] [Google Scholar]
- Maier T, Ferbitz L, Deuerling E, Ban N. (2005) A cradle for new proteins: trigger factor at the ribosome. Curr Opin Struc Biol 15: 204–212. [DOI] [PubMed] [Google Scholar]
- Metcalf D. (1985) The granulocyte-macrophage colony-stimulating factors. Science 229: 16–22. [DOI] [PubMed] [Google Scholar]
- Misawa S, Kumagai I. (1999) Refolding of therapeutic proteins produced in Escherichia coli as inclusion bodies. Biopolymers 51: 297–307. [DOI] [PubMed] [Google Scholar]
- Nagata S, Tsuchiya M, Asano S, Yamamoto O, Hirata Y, Kubota N, Oheda M, Nomura H, Yamazaki T. (1986) The chromosomal gene structure and two mRNAs for human granulocyte colony-stimulating factor. EMBO J 5: 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamsetty S, Waugh DS. (2006) Solubility-enhancing proteins MBP and NusA play a passive role in the folding of their fusion partners. Protein Expres Purif 45: 175–182. [DOI] [PubMed] [Google Scholar]
- Nomura H, Imazeki I, Oheda M, Kubota N, Tamura M, Ono M, Ueyama Y, Asano S. (1986) Purification and characterization of human granulocyte colony-stimulating factor (G-CSF). EMBO J 5: 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh-eda M, Hasegawa M, Hattori K, Kuboniwa H, Kojima T, Orita T, Tomonou K, Yamazaki T, Ochi N. (1990) O-linked sugar chain of human granulocyte colony-stimulating factor protects it against polymerization and denaturation allowing it to retain its biological activity. J Biol Chem 265: 11432–11435. [PubMed] [Google Scholar]
- Park JS, Han KY, Lee JH, Song JA, Ahn KY, Seo HS, Sim SJ, Kim SW, Lee J. (2008) Solubility enhancement of aggregation-prone heterologous proteins by fusion expression using stress-responsive Escherichia coli protein, RpoS. BMC Biotechnol 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdie N, Swallows KA, Murphy LH, Purdie RB. (1989) Analytical applications of circular dichroism. J Pharmaceut Biomed 7: 1519–1526. [DOI] [PubMed] [Google Scholar]
- Rüdiger S, Buchberger A, Bukau B. (1997) Interaction of Hsp70 chaperones with substrates. Nat Struct Biol 4: 342–349. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. (1988) Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67: 31–40. [DOI] [PubMed] [Google Scholar]
- Sørensen HP, Mortensen KK. (2005) Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol 115: 113–128. [DOI] [PubMed] [Google Scholar]
- Souza LM, Boone TC, Gabrilove J. et al (1986) Recombinant human granulocyte colony-stimulating factor: effects on normal and leukemic myeloid cells. Science 232: 61–65. [DOI] [PubMed] [Google Scholar]
- Suh WC, Lu CZ, Gross CA. (1999) Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J Biol Chem 274: 30534–30539. [DOI] [PubMed] [Google Scholar]
- Tolun AA, Dickerson IM, Malhotra A. (2007) Overexpression and purification of human calcitonin gene-related peptide-receptor component protein in Escherichia coli. Protein Expres Purif 52: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamvakas SS, Leondiadis L, Pairas G, Manessi-Zoupa E, Spyroulias GA, Cordopatis P. (2007) Expression, purification, and physicochemical characterization of the N-terminal active site of human angiotensin-I converting enzyme. J Pept Sci 13: 31–36. [DOI] [PubMed] [Google Scholar]
- VanBogelen RA, Kelley PM, Neidhardt FC. (1987) Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J Bacteriol 169: 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wülfing C, Lombardero J, Plückthun A. (1994) An Escherichia coli protein consisting of a domain homologous to FK506-binding proteins (FKBP) and a new metal binding motif. J Biol Chem 269: 2895–2901. [PubMed] [Google Scholar]