Abstract
Identifying the epitope to which an antibody binds is central for many immunological applications such as drug design and vaccine development. The Pepitope server is a web-based tool that aims at predicting discontinuous epitopes based on a set of peptides that were affinity-selected against a monoclonal antibody of interest. The server implements three different algorithms for epitope mapping: PepSurf, Mapitope, and a combination of the two. The rationale behind these algorithms is that the set of peptides mimics the genuine epitope in terms of physicochemical properties and spatial organization. When the three-dimensional (3D) structure of the antigen is known, the information in these peptides can be used to computationally infer the corresponding epitope. A user-friendly web interface and a graphical tool that allows viewing the predicted epitopes were developed. Pepitope can also be applied for inferring other types of protein–protein interactions beyond the immunological context, and as a general tool for aligning linear sequences to a 3D structure.
Availability: http://pepitope.tau.ac.il/
Contact: talp@post.tau.ac.il
1 INTRODUCTION
Mapping the epitope of an antibody on its corresponding antigen is fundamental to our understanding of the mechanisms of molecular recognition and provides the basis for rational vaccine and drug design (Irving et al., 2001). Solving the three-dimensional (3D) structure of an antibody–antigen complex is the most accurate way to determine an epitope. When such a complex is not available (or cannot be obtained), a customary approach is to map the location of the epitope using a phage display library (Barbas et al., 2001). In this technology, peptides that bind the antibody of interest with high affinity are selected from a large pool of random peptides. These affinity-selected peptides are assumed to mimic the genuine epitope in terms of their physicochemical properties and spatial organization. Hence, a resemblance of a selected peptide to a linear region of the antigen is often indicative of the genuine epitope location. However, epitopes are often conformational, being comprised of a number of discontinuous segments of the antigen brought together via folding. Thus, an affinity-selected peptide may have similarity only to non-linear fragments of the antigen. Advanced computational methods are hence needed to correctly infer the epitope from the selected peptides.
Here we present the Pepitope server, which implements three algorithms for epitope mapping: PepSurf, Mapitope, and a combination of the two. Thus allowing users to run and compare two epitope prediction algorithms that rely on two different methodological approaches via a single web platform. These algorithms were proved successful in predicting discontinuous epitopes based on affinity-selected peptides (Bublil et al., 2006, 2007; Enshell-Seijffers et al., 2003; Mayrose et al., 2007; Tarnovitski et al., 2006). Furthermore, the combined algorithmic option executes both PepSurf and Mapitope and combines their results into a single epitope prediction. Pepitope enables the alignment of short, hypothetical or synthesized, sequences to a 3D structure and is thus not confined to mapping random peptides derived from phage-display experiments. Pepitope also provides a powerful visualization tool to examine the predicted epitopes and each of the peptide-to-structure alignments. It allows non-expert users to predict epitopes with no previous algorithmic knowledge, yet incorporates advanced options for expert users to fine-tune their predictions.
Several algorithms and web servers for epitope mapping were previously developed (Castrignano et al., 2007; Halperin et al., 2003; Moreau et al., 2006; Schreiber et al., 2005). The main differences between Pepitope and the other servers are described at the ‘OVERVIEW’ web section. Although some of the features of Pepitope were previously suggested, it is the only tool that uniquely combines all of the following features: (1) It takes into account both identities and similarities, e.g. using the BLOSUM matrix, when aligning linear peptide sequences to the antigen 3D structure. (2) It allows any number of gaps in the alignment and distinguishes between terminal gaps and non-terminal ones. (3) It clusters the resulting matches on the protein surface, thus providing an epitope prediction, which is based on the entire set of peptides collectively. (4) It optionally accounts for the number of times a peptide was selected in the phage-display experiment. A short description of the Pepitope methodology is provided herein, and a more detailed description is available under the ‘OVERVIEW’, ‘GALLERY’ and ‘QUICK HELP’ web sections. The ease of using Pepitope is exemplified by predicting the human monoclonal antibody (mAb) Bo2C11 epitope for coagulation factor VIII (FVIII).
2 METHODS
The PepSurf algorithm (Mayrose et al., 2007) maps each of the affinity-selected peptides onto the surface of the antigen. This is done by efficiently searching virtually all possible 3D paths (i.e. a sequence of neighboring residues on the surface of the antigen structure) for those that exhibit high similarity to the peptide sequences. Thus, the best alignment of each peptide to antigen residues brought to proximity by folding is obtained. The resulting most significant alignments are then clustered and the epitope location is inferred. The Mapitope algorithm (Bublil et al., 2007) is based on an alternative computational approach in which epitope determinants shared by the entire set of peptides are detected. Specifically, each peptide is first deconvoluted to amino acid pairs. Mapitope then identifies pairs of residues that are significantly overrepresented in the panel of peptides. It then searches for a cluster of these enriched pairs on the antigen 3D structure to obtain predicted epitope regions.
The minimal input requirement for both algorithms is a set of affinity-selected peptide sequences and a PDB identifier of the antigen. Using these two inputs the following steps are carried out: (1) the exposed residues of the given structure are determined using the Surface Racer program (Tsodikov et al., 2002). (2) A graph representing neighboring surface residues is created. (3) The epitope prediction algorithm (PepSurf, Mapitope, or both) is executed. (4) The predicted epitopes are projected and visualized on the 3D structure via a specifically designed 3D structural viewer.
2.1 Pepitope options
Similar to classical sequence alignment algorithms, PepSurf relies on a specified amino acid similarity matrix and a gap penalty. The user can choose the gap penalty as well as a number of substitution matrices. The difference between PepSurf calculations using different matrices tends to be small but not negligible (see the ‘GALLERY’ web section). PepSurf's alignment algorithm guarantees finding the best alignment between each peptide and the PDB structure only up to a certain probability, which can also be set by the user. For Mapitope, a detailed description of its option parameters is available under the ‘OVERVIEW’ and ‘QUICK HELP’ web sections.
2.2 Pepitope output
A graphic visualization of the results using the FirstGlance in Jmol interface (http://firstglance.jmol.org) is available by clicking ‘View Pepitope Results with FirstGlance in Jmol’. For the PepSurf algorithm, the corresponding alignment of each peptide to the 3D structure is also presented using the 3D viewer. The predicted clusters can be viewed in a text format under the ‘Predicted Clusters’ link. The link ‘Resulting alignments for each peptide’ leads to a textual listing of the top-scoring alignments for each peptide. Pepitope also supplies a RasMol command script for viewing the results locally with the RasMol program (Sayle and Milner-White, 1995).
3 EXAMPLE: COAGULATION FACTOR VIII EPITOPE
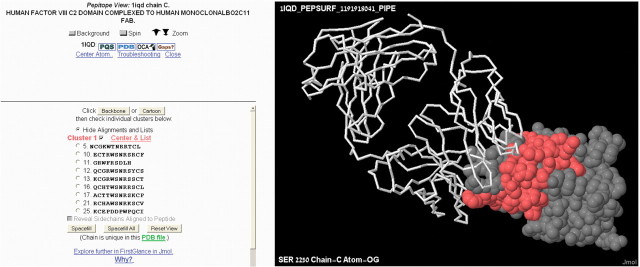
We illustrate the mapping of the human mAb Bo2C11 epitope on the 3D surface structure of its antigen, FVIII. Using the Contact Map Analysis server (Sobolev et al., 2005), 19 surface residues of FVIII were identified as comprising the ‘genuine epitope’ and were used as the benchmark epitope when determining the prediction accuracy. Bo2C11 was used to screen a combinatorial phage-display library resulting in a set of 27 affinity-selected peptides (Villard et al., 2003). Figure 1 illustrates the main Pepitope output. Pepitope predicts an epitope that significantly overlaps the benchmark epitope. The predicted epitope is comprised of 30 residues, of which 12 are part of the genuine epitope. The 18 remaining predicted residues, although false-positive predictions, are located in close proximity to the genuine epitope and may also contribute to the mAb binding.
Fig. 1.
The epitope prediction obtained by the PepSurf algorithm for the Bo2C11−FVIII complex (PDB id 1IQD). The FVIII antigen is shown as a space-filled model with the predicted epitope in red. The Bo2C11 antibody Fab fragment is shown as a backbone model.
ACKNOWLEDGEMENTS
This study was supported by the Wolfson Foundation (T.P.), ISF grants (T.P., J.M.G., E.R.), BSF grant (J.M.G.), the Israeli Ministry of Science and Technology (T.P., R.S.), Alon fellowship (R.S.), Tauber Fund (E.R.), Safra bioinformatics program (N.D.R., O.P.) and by the US National Science Foundation Division of Undergraduate Education (E.M.).
Conflict of Interest: none declared.
REFERENCES
- Barbas CF, et al. Phage Display: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Bublil EM, et al. Computational prediction of the cross-reactive neutralizing epitope corresponding to the monoclonal antibody b12 specific for HIV-1 gp120. FASEB J. 2006;20:1762–1774. doi: 10.1096/fj.05-5509rev. [DOI] [PubMed] [Google Scholar]
- Bublil EM, et al. Stepwise prediction of conformational discontinuous B-cell epitopes using the Mapitope algorithm. Proteins. 2007;68:294–304. doi: 10.1002/prot.21387. [DOI] [PubMed] [Google Scholar]
- Castrignano T, et al. The MEPS server for identifying protein conformational epitopes. BMC Bioinformatics. 2007;8(Suppl. 1):S6. doi: 10.1186/1471-2105-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enshell-Seijffers D, et al. The mapping and reconstitution of a conformational discontinuous B-cell epitope of HIV-1. J. Mol. Biol. 2003;334:87–101. doi: 10.1016/j.jmb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Halperin I, et al. SiteLight: binding-site prediction using phage display libraries. Protein Sci. 2003;12:1344–1359. doi: 10.1110/ps.0237103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving MB, et al. Random-peptide libraries and antigen-fragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr. Opin. Chem. Biol. 2001;5:314–324. doi: 10.1016/S1367-5931(00)00208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I, et al. Epitope mapping using combinatorial phage-display libraries: a graph-based algorithm. Nucleic Acids Res. 2007;35:69–78. doi: 10.1093/nar/gkl975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, et al. Discontinuous epitope prediction based on mimotope analysis. Bioinformatics. 2006;22:1088–1095. doi: 10.1093/bioinformatics/btl012. [DOI] [PubMed] [Google Scholar]
- Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- Schreiber A, et al. 3D-Epitope-Explorer (3DEX): localization of conformational epitopes within three-dimensional structures of proteins. J. Comput. Chem. 2005;26:879–887. doi: 10.1002/jcc.20229. [DOI] [PubMed] [Google Scholar]
- Sobolev V, et al. SPACE: a suite of tools for protein structure prediction and analysis based on complementarity and environment. Nucleic Acids Res. 2005;33:W39–W43. doi: 10.1093/nar/gki398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnovitski N, et al. Mapping a neutralizing epitope on the SARS coronavirus spike protein: computational prediction based on affinity-selected peptides. J. Mol. Biol. 2006;359:190–201. doi: 10.1016/j.jmb.2006.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsodikov OV, et al. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- Villard S, et al. Peptide decoys selected by phage display block in vitro and in vivo activity of a human anti-FVIII inhibitor. Blood. 2003;102:949–952. doi: 10.1182/blood-2002-06-1886. [DOI] [PubMed] [Google Scholar]