Abstract
The importin-α family of proteins plays an important role in the eukaryotic importin/exportin nuclear transport system. These proteins recognize a nuclear localization signal (NLS) within cargo proteins and import them into the nucleus through nuclear pores, in a process mediated by importin-β. Recent studies have shown that importin-α proteins specifically recognize the NLS of several cellular factors and viral proteins, thus regulating their movement. Dysregulation of importin-α is a common hallmark of many pathologies including, multiple cancers. In this study, we isolated aptamers 76 and 72, which bind specifically and efficiently to KPNA2, a member of a subfamily of importin-α1. Both of these aptamers bind to KPNA2 with an equilibrium dissociation constant (Kd) of 150 nM and discriminate between KPNA2 and other sub-family members of importin-α, such as KPNA1 and KPNA3. These aptamers specifically interfere with the nuclear transport of cargo proteins mediated by KPNA2 but neither with KPNA1 nor KPNA3, which belongs to other subfamily of importins. These results suggest that the selected aptamers (76 and 72) warrant further study to explore not only their application in cancer diagnosis but also their use as a specific reagent to potentially block KPNA2-dependent nuclear transport of macromolecules across the nuclear membrane.
Keywords: Aptamer, Cancer, importin-α, nuclear transport, RNA
In eukaryotic cells, macromolecular trafficking from the cytoplasm to the nucleus and vice versa is tightly regulated by a number of specific transport factors via nuclear pore complexes (NPC). The entire system is referred to as the importin/exportin transport system, which enables the transport of macromolecules larger than 40 kDa. Macromolecules, such as transcription factors, histones, DNA and RNA polymerases and other factors, are imported into the nucleus, whereas RNA transcripts and other soluble factors are exported into the cytoplasm through nuclear pores. It has been reported that the nuclear transport system is indispensable for many types of cellular responses, including development, organogenesis and tissue formation (1). To allow shuttling between the nucleus and cytoplasm, a cargo molecule generally possesses either a nuclear localization signal (NLS) or nuclear export signal sequences (NES). Importin and exportin proteins specifically recognize the NLS and NES, respectively. To transport cargo proteins into the nucleus, the NLS in the cargo proteins can interact either with importin-α or importin-β, whereas only exportins recognize the NES region in the cargo protein, and they export the resulting complex from the nucleus by interacting with another protein complex (RanGTP).
Since the discovery of human importin-α nearly 20 years ago, a total of seven isoforms and over 75 isoform-specific cargo proteins to be imported have been identified, including cellular and viral proteins (2). For example, to import SV40 large T-antigen into the nucleus, the NLS of SV40 T-antigen binds to importin-α in the presence of importin-β, and the resulting importin-α/importin-β/SV40-T-antigen complex shuttles through NPC with the aid of small RanGTPases. Similarly, other identified proteins either cellular or viral have also been shown to import into the nucleus. Importin-α has acquired diversified functions during evolution, to meet the increasing complexity necessary in higher organisms that require a wide range of functions during development and differentiation (3). Human importin-α isoforms are divided into three major sub-classes according to the homology of amino acid sequence; the α1 subclass, which contains importin-α1 and importin-α8; the α2 subclass, which contains importin-α3 and importin-α4; and the α3 subclass, which contains importin-α5, importin-α6 and importin-α7. All of the importin-α isoforms share a common domain structure consisting of a flexible N-terminal importin-β-binding (IBB) domain (1-70 amino acids) and a C-terminal helical core consisting of tandem armadillo (ARM) repeats (Supplementary Fig. 1). The IBB domain of importin-α appears to have a dual role by acting as a motif for binding to importin-β and as an autoinhibitor of its NLS binding pocket (4).
Despite the common structural features observed among the importin-α isoforms, a number of cellular and viral cargo proteins display remarkable specificity toward specific importin-α isoforms for their transport into the nucleus. For example, cellular proteins such as JNK1 (5), NBS1 (6) and TBP-2 (7) as well as the viral protein SARS-CoV ORF6 (8) are exclusively transported by KPNA2 (importin-α1). One possible explanation for this specificity could be due to the tissue-specific expression of the varying importin-α isoforms. A clear case of changes in the expression pattern of one particular importin-α isoform has been observed during the differentiation of mouse embryonic stem cells into neurons, which modulate the shift of expression from KPNA2 to KPNA1 during development (9). Similarly, malfunction of the cellular transport machinery is commonly observed in multiple cancers; in particular, KPNA2, compared to other isoforms, is a major translocator of cancer-associated proteins. In recent years, both cancer cells and tissues have been shown to express elevated transcriptional levels (5–10-fold) of KPNA2 compared to normal cells and tissues (10, 11). Thus, KPNA2 has emerged as a potential biomarker for multiple cancer forms, including melanoma, cervical cancer, esophageal cancer, lung cancer, ovarian cancer, prostate cancer, brain cancer, liver cancer and bladder cancer (12). In addition, it is also important to develop selective inhibitors that block specific isoforms without affecting the bulk of NLS cargo proteins shuttling through the NPC. To this end, peptides have been designed to target specific isoforms of importin-α based on the NLS domains of cellular proteins (13, 14).
Considering the role of KPNA2, it has been an important aim to develop ligands that bind efficiently to KPNA2, because such ligands may have potential biomedical applications. In view of this, the selection of specific aptamers against KPNA2 is an attractive strategy, because they are known to bind specifically and efficiently to their target ligands, similarly to an antibody binding to its target antigen. Moreover, such selected aptamers have been shown to have a higher capacity for molecular discrimination compared to antibodies. In the past, we were able to select efficient aptamers that distinguish between closely related influenza subtypes (15–17) and recognize herpes simplex viruses (18). In this study, we have explored the aptamer selection strategy to isolate aptamers that bind efficiently to KPNA2 and distinguish it from other isoforms of the importin-α family of proteins. We have carried out a total of eight selection cycles using a completely random RNA pool in the presence of KPNA2. The KPNA2 specific aptamers were enriched in the eighth selected pool and, upon sequencing, we identified two major sequences belonging to clone 76 (22%) and clone 72 (4%). These aptamers efficiently bind to KPNA2 and discriminate between KPNA2 and other isoforms such as KPNA1 and KPNA3. Both the 76 and 72 aptamers bind to either the human or mouse-derived KPNA2 protein with equal affinity (Kd ≈ 150 nM). These aptamers were also tested for their ability to interfere with KPNA2 function during the nuclear transport of cargo proteins, and both of these aptamers (76 and 72) were able to interfere with the KPNA2-dependent nuclear transport of cargo protein but not either with KPNA1-dependent or KPNA3-dependent nuclear transport. These results suggest that the selected aptamers (76 and 72) warrant further studies to explore their potential application not only in molecular cancer diagnosis but also as a specific reagent to interfere with KPNA2-depenedent nuclear transport of macromolecules across the nuclear membrane.
Matrial and Methods
Materials
KPNA2 proteins from either mouse or human were expressed in E. coli and purified as previously described (9, 19). Similarly, mouse KPNA1 was expressed in E. coli and purified also as previously described. Human KPNA2 was received as a gift from Dr. Miyamoto (National Institute of Biomedical Innovation). The purity of the mouse KPNA1, mouse KPNA2 and human KPNA2 proteins was confirmed by SDS-PAGE (12.5%) (Supplementary Fig. 2). The mouse KPNA3 protein was prepared and confirmed the purity as mentioned above (data not shown).
RNA pool
The RNA library was designed to contain a central domain of randomized sequences flanked by invariable 5′ and 3′ sequences. The ssDNA library AGTAATACGACTCACTATAGGATCCTGAGCTACTGAC-N60-CACCACT ACTGACCATACAC, with 60-nucleotide contiguous random sequences, was synthesized and flanked by defined sequences, including the T7 promoter (in italics). The 5′ and 3′ defined sequences were 5′- AGTAATACGACTCACTATAGGATCCTGAGCTACTGAC-3′ and 5′-GTGTATGGTCAGTAGTGGTG-3′, respectively. Polymerase chain reaction (PCR) was performed with the random DNA library (1 × 1014 molecules) with 0.25 μM of each of the 5′- and 3′-primers. To preserve the abundance of the original library, the PCR was limited to eight cycles to avoid amplifying a skewed population from the random DNA library. The DNA library was converted into an RNA library by in vitro transcription using a T7 Ampliscribe kit (Epicentre Technologies, USA). This RNA library was used for selection.
In vitro selection
Selection was performed using 16 μg (∼1014 RNA molecules) of the original RNA library in RNA binding buffer (10 mM HEPES, pH 7.4; 150 mM NaCl) at a 7:1 molar ratio of RNA to protein. During the later selection cycles, the concentration of recombinant mouse KPNA2 was decreased to select high affinity RNA molecules. The RNA was briefly heated to 95 °C and then cooled to room temperature to form stable structures before the selection steps. In the selection cycles, tRNA (total tRNA from E. coli, Roche, Switzerland) was used as a non-specific competitor. After the RNA was incubated with mouse KPNA2 at room temperature for 10 min, the protein–RNA complexes were filtered through a pre-wetted nitrocellulose acetate filter (HAWP filter, 0.45 µm, 13.0-mm, Merck Millipore, USA) fitted in a ‘pop-top’ filter holder (Nucleopore) and washed with 1 ml of RNA binding buffer. The RNAs that were retained on the filter were eluted and recovered as previously described (15, 16). The recovered RNAs were reverse-transcribed in 20 µl of a reaction mixture containing 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 10 mM MgCl2, 0.5 mM spermidine, 10 mM DTT, 0.4 mM dNTPs, 0.4 µM primer and 25 U AMV reverse transcriptase (Wako, Japan). The nucleotides and enzymes were added after a denaturation and annealing step (2 min at 95 °C, followed by incubation at room temperature for 10 min). Reverse transcription was performed for 45 min at 37 °C. The resulting cDNA was amplified by PCR and used as a template to obtain RNA for the next round of selection. For amplification of the cDNA by PCR, a 20 µl aliquot of the reverse transcription reaction (cDNA reaction mixture) was diluted in 80 µl of PCR reaction mixture [PrimeSTAR Max Premix (Takara, Japan) with 1 µM of each primer]. The reaction mixture was amplified using the following conditions: 95 °C for 20 s, 54 °C for 10 s and 72 °C for 20 s. This program was repeated for as many cycles as needed to produce a DNA band of the correct size. The PCR product was precipitated in ethanol and used for transcription. In vitro transcription was performed at 37 °C for 3 h with a T7 Ampliscribe kit (Epicentre Technologies). After treatment with DNase I, the reaction mixture was fractionated on an 8% denaturing polyacrylamide gel. The RNA was extracted from the gel, quantified and used for the next cycle of selection and amplification. We manipulated each selection cycle to ensure specificity and high affinity of the KPNA2-bound molecules. For example, we modified the ratio of RNA:KPNA2, competitor concentrations (tRNA) and buffer volumes. To remove the filter binders, pre-filtration of the RNA pools were performed for each selection round. A total of eight selection cycles were carried out and the RNA pool from cycles 1, 3, 5 and 8 were evaluated for binding to the mouse KPNA2 protein.
Binding analysis of aptamers
To obtain individual aptamers, the amplified PCR products from cycle 8 were directly ligated into the pCRII vector (Invitrogen) according to the manufacturer’s protocol. DNA was isolated from individual clones by the alkaline-lysis method and sequenced with a BigDye Terminator Sequencing kit (Life Technologies) on a DNA sequencer (Model 373A, Life Technologies). The secondary structures of the aptamers were predicted using the MFold program (20). To evaluate the mouse KPNA2 binding to the RNA pools from selection cycles 1, 3, 5 and 8 as well as the individual aptamers, internally labeled RNA was prepared using 0.5 mCi/ml [α-32P]ATP and binding experiments were performed using a filter binding assay similar to those previously reported (21). The binding and in vitro transcription conditions were similar to those used for selection, except for the RNA and mouse KPNA2 protein concentrations. For the evaluation of binding of RNA pools (from 1, 3, 5 and 8 selection cycles) to the mouse KPNA2 protein, we used 20 nM of labeled RNA and different concentration (490, 980 or 1960 nM) of mouse KPNA2. The RNA–KPNA2 complexes retained on the filters were washed with 1 ml binding buffer and air dried and the radioactivity was quantified with an image analyzer (BAS2000, Fuji Film, Japan). To ensure that the binding was specific, we performed binding assays in the presence of a 10-fold molar excess of tRNA as a nonspecific competitor. Similarly, these RNA pools were also evaluated under similar conditions for their binding ability to another importin family subclass mouse KPNA1. Individual clones (aptamers 76, 72 and 21) binding ability to the mouse KPNA2 was evaluated using 25 nM of labeled RNA using [α-32P]ATP in the presence of 10-fold molar excess of tRNA and mouse KPNA2 protein (246, 492 and 984 nM). Next, to judge the importance of sequence of the above aptamers for binding, we carried out another filter binding assay using 25 nM of unselected pool RNA (containing random RNA sequences), in the presence of 10-fold tRNA and excess amount of mouse KPNA2 protein (490, 980 and 1960 nM). In order to estimate the equilibrium dissociation constants, we performed a filter binding assay using labeled aptamer RNA for either clone 72 or clone 76 (25 nM) in the presence of various concentrations (37, 74, 148, 296 and 592 nm) of mouse KPNA2. Additionally, aptamers 76 and 72 binding to the human KPNA2 was also evaluated by filter binding assay at different concentrations (37, 74, 148, 296 and 592 nM). The equilibrium dissociation constants were determined for all complexes (aptamer 76-mouse KPNA2, aptamer 76-human KPNA2, aptamer 72-mouse KPNA2, aptamer 72-human KPNA2) using a nonlinear regression algorithm in GraphPad Prism 6.0 (GraphPad Software).
In vitro nuclear transport assay in the presence and absence of aptamers
To evaluate the aptamers’ (76 and 72) ability to interfere with human KPNA2 functions, we performed in vitro nuclear transport assays similar to those previously described (22). Initially, HeLa cells were cultured on an eight-well slide slip for two days in DMEM containing 10% FBS. The cells were then permeabilized with digitonin and then the cytosolic components were washed out with transport buffer (20 mM HEPES–KOH pH 7.3, 110 mM CH3COOK, 5 mM CH3COONa, 2 mM (CH3COO)2Mg, 0.5 mM EGTA, 2 mM DTT, 1 μg/ml each of aprotinin, pepstatin and leupeptin). Nuclear transport was established with 400 nM GST-SV40TAgNLS-GFP, 600 nM importin-β, 400 nM human KPNA2, 4 μM RanGDP, 350 nM NTF2, 0.5 mM GTP and an ATP regeneration system, at 37 °C for 8 min. To perform the same assay in the presence of the RNA aptamers (either 76 or 72) or pool RNA, we denatured the RNA in binding buffer (10 mM HEPES, pH 7.4; 150 mM NaCl) and equilibrated for 10 min at room temperature. The renatured aptamers (at 5.0 μM final concentration) were pre-incubated with human KPNA2 at room temperature for 5 min prior to execution of the transport assay as indicated in the figure legends. Cells were fixed in 3.7% formaldehyde in PBS and observed by fluorescence microscopy. The quantitative analysis of transport efficiency was performed using Image J software (23) as follows: the mean value of the fluorescent signal inside the nucleus was measured and the signal intensity was calculated by normalizing with the mean value of the control cells. A total of 60 cells were randomly selected for each sample and the mean value with standard deviation was reported. To compare the specificity of the aptamers interference of nuclear transport mediated by different sub-class of importins, we performed the above assay by replacing the KPNA2 either with KPNA1 or KPNA3 proteins.
Results
Selection of RNA ligands
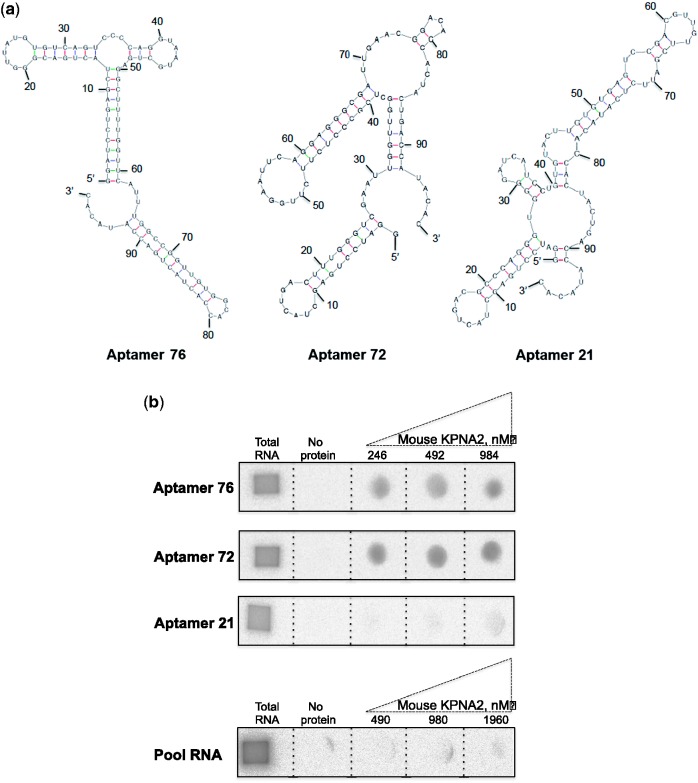
To select an RNA aptamer against KPNA2, random RNA pool over a 60-nucleotide stretch flanked by a fixed 37-nucleotide primer region at the 5′ end and 20-nucleotide primer region at the 3′ end was used. This randomized pool represents approximately 1014 different species under our experimental conditions. For the initial selection, the N-60 RNA pool (3.0 μM) and mouse KPNA2 (0.43 μM) were mixed and incubated for 10 min in the presence of buffering agents and the free RNA was separated from the mouse KPNA2–RNA complexes by filtration. The mouse KPNA2 bound RNAs retained on the filter were recovered in a hot 7 M urea solution and subsequently amplified by reverse transcription, PCR and in vitro transcription. To enrich specific aptamers during the selection process, in each selection cycle, competition between the individual sequences was allowed. For this, a high RNA pool to target protein ratio was used in the initial selection cycles. In the subsequent selection cycles (2–8), the ratio between the mouse KPNA2 protein and RNAs (pool RNA and tRNA) was manipulated to increase the selection stringency (Table 1). To ensure selection of specific aptamers during the selection cycles, a non-specific competitor, tRNA, was used at different molar ratios. After eight rounds of selection cycles, RNA pools (1, 3, 5 and 8) were selected for the evaluation of enrichment of mouse KPNA2-binding aptamers and their specificity. The specificity and binding of RNA pools to the mouse KPNA2 was analyzed by filter binding assay at different concentrations (490, 980 and 1960 nM). The binding results show that the pool RNAs binding increased progressively with selection cycles and also the specificity to KPNA2, compared to the KPNA1 (Table 1). No significant retention of the selected RNA pool occurred on the filter in the absence of mouse KPNA2, indicating that the non-specific filter binders were not co-isolated during the selection process. Approximately 17% of the 8th RNA pool bound to mouse KPNA2 (Table 1). Since the 8th selection cycle RNA pool bound specifically to the mouse KPNA2, we proceeded to the cloning and sequencing steps to evaluate the sequences in the pool. Individual aptamers from the eight cycles were classified based on the sequence similarity. We observed that aptamer 76 represented 22% of the population, whereas aptamer 72 and aptamer 21 represented 4% and 2%, respectively, in the selected pool of cycle 8. The secondary structures of these aptamers were predicted according to the MFold program (20, Fig. 1a). Next, all three aptamers were evaluated with the filter assays for their binding ability to mouse KPNA2 (at concentrations of 246, 492 and 984 nM), in the presence of a 10-fold molar excess of tRNA to avoid non-specific interactions with RNA. These filter binding assays showed that only aptamers 76 and 72 bound to mouse KPNA2, but aptamer 21 showed either minimal or insignificant binding to mouse KPNA1 (Fig. 1b). In order to clearly evaluate that the sequence of 76 and 72 would be important, we performed another binding assay with unselected pool RNA (containing random sequences) and found that the unselected RNA pool had no binding ability even in the excess amount of mouse KPNA2 protein (1960 nM, Fig. 1b). These binding analyses clearly suggest that the sequences of aptamers 76 and 72 are important for their affinity and specificity to the mouse KPNA2 and thus, all further studies were focused using these two aptamers.
Table 1.
Concentration of RNA and protein used and the ability of the RNA pool to bind to the KPNA2 and KPNA1 in different selection cycles
| Selection cycle | Pool RNA (μM) | Competitor RNA (μM) | Mouse KPNA2 (μM) | % of binding to mouse KPNA2 |
% of binding to mouse KPNA1 |
||||
|---|---|---|---|---|---|---|---|---|---|
| 0.49 | 0.98 | 1.96 μM | 0.49 | 0.98 | 1.96 μM | ||||
| 1 | 3 | None | 0.43 | 0 | 0 | 0 | 0 | 1 | 0 |
| 2 | 0.31 | 2 | 0.43 | ||||||
| 3 | 0.31 | 5 | 0.22 | 1 | 2 | 4 | 0 | 0 | 0 |
| 4 | 0.15 | 5 | 0.22 | ||||||
| 5 | 0.31 | 10 | 0.11 | 3 | 7 | 21 | 0 | 1 | 2 |
| 6 | 0.31 | 10 | 0.11 | ||||||
| 7 | 0.15 | 10 | 0.11 | ||||||
| 8 | 0.15 | 10 | 0.11 | 6 | 12 | 27 | 0 | 1 | 1 |
Fig. 1.
In vitro selected aptamers and their ability to bind mouse KPNA2. (a) The predicted secondary structures of aptamers (76, 72 and 21) presented were drawn using mfold program. (b) Filter binding assays were performed with labeled aptamers (20 nM) in the presence of native mouse KPNA2 (246, 492 and 984 nM) and a 10-fold excess of tRNA as a non-specific competitor. Filter binding assay was also carried out using 25 nM of unselected pool RNA in the presence of 10-fold excess of tRNA and an excess amount of mouse KPNA2 protein (490, 980 and 1960 nM).
Binding efficiency of the selected aptamers
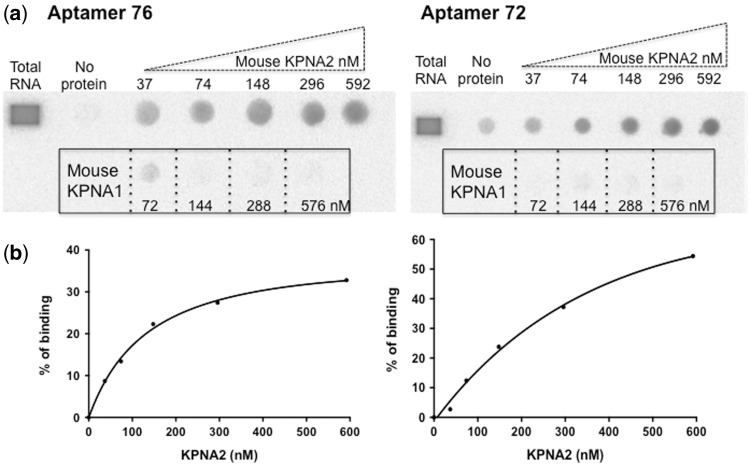
To determine the dissociation constants (Kd) of the selected aptamers (72 and 76), we performed filter binding assays using aptamers labeled with [α-32P]ATP with increasing concentrations of either mouse KPNA2 (37, 74, 148, 296 and 592 nM) or mouse KPNA1 (72, 144, 288 and 576 nM). Both the 76 and 72 aptamers bound to mouse KPNA2 with equal affinity; the calculated dissociation constant (Kd) was approximately 150 nM for both aptamers (Fig. 2a and b). Both aptamers did not show any binding to mouse KPNA1, even at the highest concentration (576 nM, Fig. 2a). Mouse and human KPNA2 are highly conserved, with an amino acid similarity of 94.5% (Supplementary Fig. 3); thus, we hypothesized that these two aptamers would bind with equal efficiency to both mouse and human KPNA2. To test this, we repeated this experiment with human-derived KPNA2 and found that these two aptamers bound with a similar affinity to that of mouse-derived KPNA2 (data not shown). To evaluate the aptamers’ ability to interfere with KPNA2 activity, we used a previously developed in vitro nuclear transport assay using human-derived KPNA2 protein.
Fig. 2.
Evaluation of the binding efficiency of the selected aptamers to mouse KPNA2 and KPNA1. (a) Filter binding assay showing the labeled aptamers binding to mouse KPNA2 at various concentrations. The framed region shows that the aptamers’ binding ability to mouse KPNA1, at the indicated concentrations. (b) The amount of aptamer–KPNA2 complex retained on the filter was quantified at different concentrations of mouse KPNA2, and the dissociation constant (Kd) was determined using GraphPad Prism 6.0 nonlinear regression algorithm (GraphPad Software).
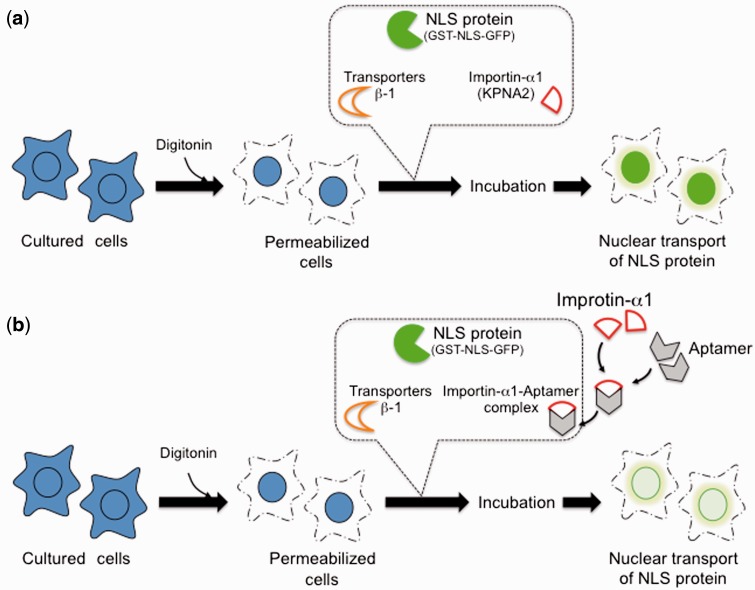
In vitro assay to evaluate the aptamers’ ability to interfere with nuclear transport
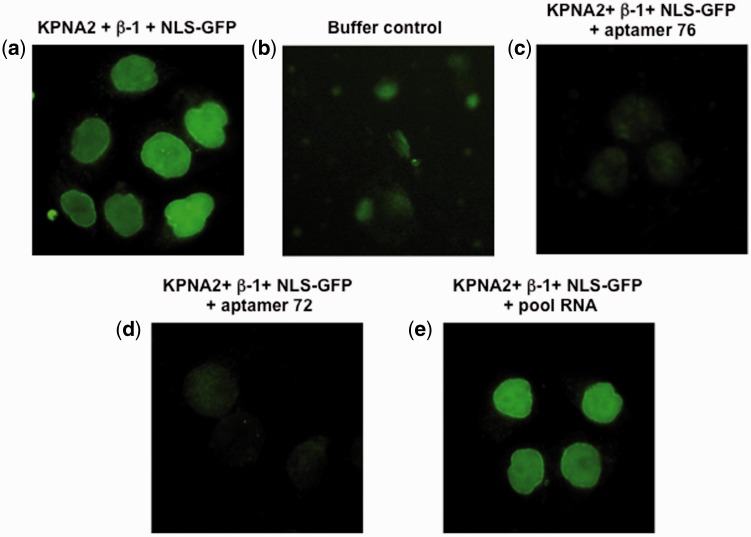
An overview of in vitro nuclear transport is depicted in Fig. 3a, in which cultured cells are initially permeabilized, followed by the addition of a ternary complex (consisting of human-derived KPNA2, importin-β1 and GST-NLS-GFP cargo protein) and observation of GFP positive cells upon transport of the complex into the nucleus. We reasoned that if aptamer binding to human KPNA2 interferes with the binding of either importin-β1 or GST-NLS-GFP (or both proteins), then there would be a lack of GFP accumulation in the cell nucleus (Fig. 3b). In a typical in vitro transport assay, cells with green fluorescence were observed when permeabilized cells were incubated for approximately 10 min with the ternary complex (Fig. 4a) and cells with no fluorescence were observed in the absence of the ternary complex (Fig. 4b). As predicted, in the presence of either the 76 or 72 aptamer (5 μM final concentration), the human KPNA2-dependent transport of cargo protein (GST-NLS-GFP) was significantly decreased, as determined by the decrease in the fluorescence of GFP positive cells (Fig. 4c and d). However, the presence of random RNA sequences (pool RNA) (Fig. 4e) did not affect the transport of the cargo protein, a result similar to that observed in the control (Fig. 4a).
Fig. 3.
Schematic representation of the experiments performed to analyze the ability of the aptamers to interfere with nuclear transport of cargo protein by KPNA2. (a) Permeabilized cells were incubated at 37 °C for 8 min in the presence of ternary transport complex [KPNA2, transporters-β1, and NLS protein (GST-NLS-GFP)]. After incubation, the cells were fixed and observed under microscope for the presence of GFP in the nucleus. (b) A predicted scenario if the selected aptamers were to interfere with KPNA2, KPNA1, KPNA3 or all three.
Fig. 4.
In vitro nuclear transport assay using human KPNA2. (a) Permeabilized cells were incubated at 37 °C for 8 min in the presence of the ternary transport complex (human KPNA2-importin-β1-NLS-GFP) and observed under microscope. (b) A buffer control, in the absence of any transport complex (human KPNA2-importin-β1-NLS-GFP). (c) In the presence of aptamer 76 (5 μM) and the ternary transport complex (human KPNA2-importin-β1-NLS-GFP). (d) In the presence of aptamer 72 (5 μM), and the ternary transport complex (human KPNA2-importin-β1-NLS-GFP). (e) In the presence of pool RNA (5 μM) and the ternary transport complex (human KPNA2-importin-β1-NLS-GFP). For details, see the Materials and Methods. Cells were fixed after 8 min incubation and images were collected.
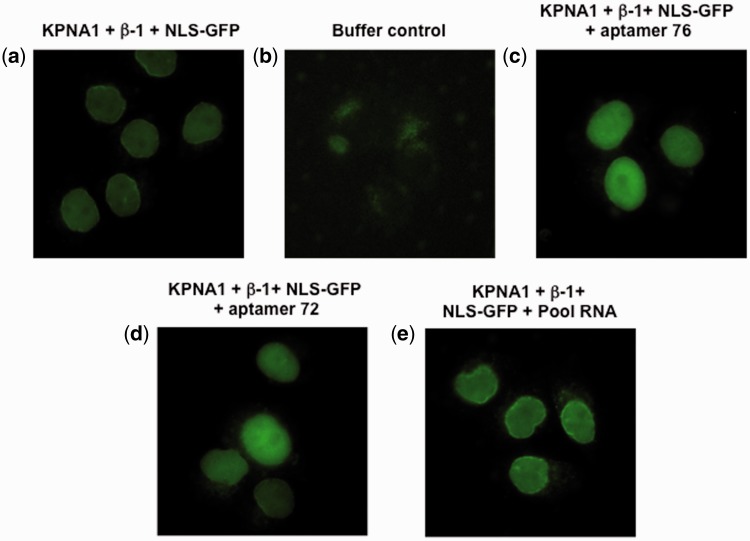
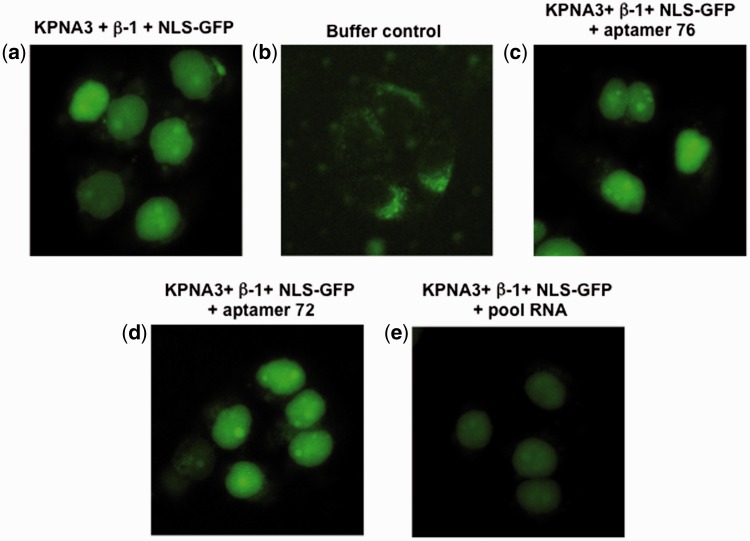
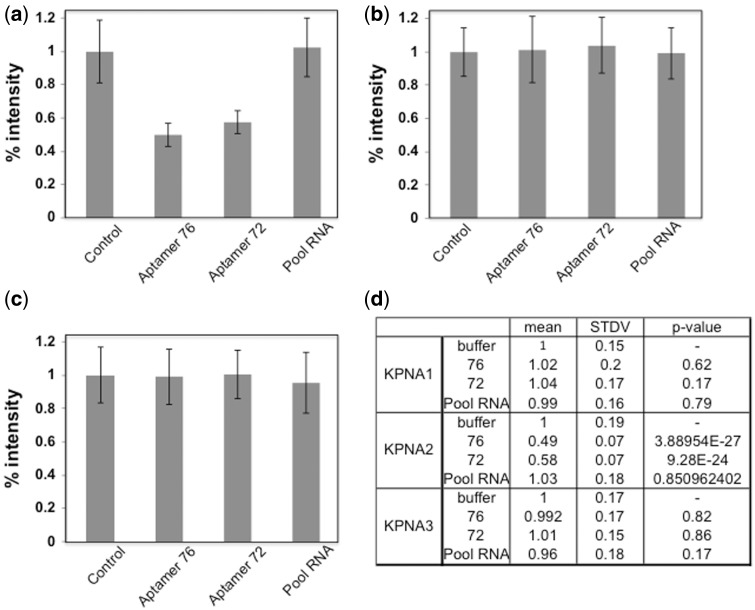
The GST-NLS-GFP cargo protein is also transported into the nucleus by KPNA1 or KPNA3 in the presence of importin-β1. To test whether the aptamers (76 and 72) would be able to interfere with this type of nuclear transport, we performed a similar experiment to the one described above, except that we replaced KPNA2 either with KPNA1 or KPNA3. Similarly, cells with green fluorescence were observed when permeabilized cells were incubated with a ternary complex composed of all three components (KPNA1, importin-β1 and GST-NLS-GFP cargo) (Fig. 5a) and no fluorescent cells were observed in the absence of the ternary complex (Fig. 5b). Interestingly, the population of the GFP positive cells was not significantly affected by the presence of either the aptamers (76 and 72) or pool RNA (Fig. 5c–e). Further, we have carried the nuclear transport assay with KPNA3 in the presence or absence of aptamers (76 and 72), or pool RNA. The permeabilized cells incubated with all three components (KPNA3, importin-β1 and GST-NLS-GFP cargo) show green fluorescence cells (Fig. 6a) and no fluorescent cells were observed in the absence of the ternary complex (Fig. 6b). As observed in the case of KPNA1, the presence of aptamers, or pool RNA was also not affected the population of GFP positive cells (Fig. 6c–e). To quantify this observed reduction of GFP fluorescence, we used Image J (NIH program) to measure the fluorescence resulting from any one of KPNA1-, KPNA2-, and KPNA3–mediated nuclear transport. The GFP fluorescence intensity of a total of 60 cells was measured for all of the samples via random selection of cells. These analyses showed that in the presence of the aptamers (76 and 72), the GFP fluorescence intensity decreased by approximately 40% when KPNA2 was used for the nuclear transport (Fig. 7a). However, these two aptamers (76 and 72) and pool RNA had no effect on either KPNA1-dependent (Fig. 7b) or KPNA3-dependent (Fig. 7c) nuclear transport of cargo protein. The significant of quantitated data was statistically evaluated as shown in Fig. 7d.
Fig. 5.
In vitro nuclear transport assay using KPNA1. (a) Permeabilized cells were incubated at 37 °C for 8 min in the presence of the ternary transport complex (KPNA1-importin-β1-NLS-GFP) and observed under the microscope. (b) A buffer control, in the absence of any transport complex (KPNA1-importin-β1-NLS-GFP). (c) In the presence of aptamer 76 (5 μM), and the ternary transport complex (KPNA1-importin-β1-NLS-GFP). (d) In the presence of aptamer 72 (5 μM), and the ternary transport complex (KPNA1-importin-β1-NLS-GFP). (e) In the presence of pool RNA (5 μM) and the ternary transport complex (KPNA1-importin-β1-NLS-GFP). For details, see the Materials and Methods. Cells were fixed after 8 min incubation and images were collected.
Fig. 6.
In vitro nuclear transport assay using KPNA3. (a) Permeabilized cells were incubated at 37 °C for 8 min in the presence of the ternary transport complex (KPNA3-importin-β1-NLS-GFP) and observed under the microscope. (b) A buffer control, in the absence of any transport complex (KPNA3-importin-β1-NLS-GFP). (c) In the presence of aptamer 76 (5 μM), and the ternary transport complex (KPNA3-importin-β1-NLS-GFP). (d) In the presence of aptamer 72 (5 μM), and the ternary transport complex (KPNA3-importin-β1-NLS-GFP). (e) In the presence of pool RNA (5 μM) and the ternary transport complex (KPNA3-importin-β1-NLS-GFP). For details, see the Materials and Methods. Cells were fixed after 8 min incubation and images were collected.
Fig. 7.
Quantification of GFP fluorescence intensity of nuclear transport cells mediated by KPNA2, KPNA1, and KPNA3 proteins in the presence or absence of aptamer 76, 72 and pool RNA. The GFP fluorescence intensity of 60 cells is quantified for each sample as described in Figs. 4, 5 and 6. (a) The human KPNA2-dependent nuclear transport system. (b) The KPNA1-dependent nuclear transport system. (c) The KPNA3-dependent nuclear transport system. (d) The resulting statistical values are described.
Discussion
The nuclear exportin/importin transport system is a critical process for all eukaryotic cells, affecting gene expression, signaling pathways, the cell cycle and pathogenesis. One of the key components of this transport system is importin α, which initiates the process by recognizing the NLS within the cargo and importing macro proteins into the nucleus through the nuclear pores, as mediated by importin-β. Currently, seven importin-α proteins are grouped into three subfamilies. Despite their similarity, these proteins display remarkable specificity toward their cognate cargo proteins, which may decide the fate of the cell during differentiation, as observed in embryonic stem cells (9, 24). The divergent roles of importin-α in the normal and in diseased cells scenarios have attracted much attention toward the development of ligands that specifically bind to a specific subfamily. In this study, we explored an in vitro selection strategy to identify aptamers from a completely random RNA pool that could bind specifically and efficiently as well as discriminate among subfamily members of importin-α. We executed a total of 8 selection cycles, which resulted in an enrichment of two aptamers, 76 and 72, as major and minor representation in the pool, respectively. These two aptamers bind efficiently (Kd ≈ 150 nM) to KPNA2-derived from either mouse or human. Moreover, both clones failed to bind to KPNA1 (Fig. 2b). The secondary structure predictions suggest that these two aptamers are not related to each other.
Because these two aptamers bound to the human KPNA2 specifically, an in vitro nuclear transport assay was used to evaluate whether the selected aptamers interfered with human KPNA2-mediated nuclear transport. Our analyses showed that ability of human KPNA2 to transport the cargo protein (GST-NLS-GFP) decreased by about 40% in the presence of either aptamer 76 or aptamer 72 (Fig. 6a). The efficiency of interference for these two clones was observed to be the same, correlating well with their affinity towards human KPNA2. However, under similar conditions, random RNA sequences (pool RNA) showed no effects on nuclear transport activity mediated either by KPNA1 or KPNA2, or KPNA3, suggesting that the negatively charged surface of the aptamers had no major roles in binding to KPNA proteins. Nevertheless, structural analyses of the aptamer-KPNA2complex may reveal the specific binding site and shed light on their interference with KPNA2-mediated nuclear transport. Interestingly, both aptamers failed to interfere neither the KPNA1-mediated nor KPNA3-mediated nuclear transport of cargo protein. Taken together these results suggest that the selected aptamers 76 and 72 are able to discriminate efficiently and block specifically the human KPNA2-mediated nuclear transport (that belongs to the sub-family of importin–α1) from KPNA1 (that belongs to the subfamily of importin–α2) or KPNA3 (that belongs to the subfamily of importin–α3) (Figs 4–6).
In short, this study identified two aptamers (76 and 72) that bound with a high affinity to mouse or human KPNA2 and were able to distinguish it from other sub-family members, such as KPNA1 and KPNA3. Importantly, both aptamers efficiently and specifically interfered with KPNA2-mediated nuclear transport. We believe that these selected aptamers will have useful applications as new reagents for cancer diagnosis because many cancer cells and tissues are known to overexpress KPNA2 (10–12). In addition, these aptamers may aid in modulating the specific nuclear transport system mediated by KPNA2; if these aptamers were delivered into the cytoplasm by fusion with internalizing aptamers (25, 26), they would probably be internalized by receptor-mediated endocytosis.
Supplementary Data
Supplementary Data are available at JB Online.
Supplementary Material
Acknowledgements
The authors would like to thank Drs. Yoshihiro Yoneda, Hiroki Kaneko and Hiroshi Mizuno for their help during the course of this study. PKRK obtained the results in the paper were Table 1, figures 1–2 and supplementary Figs S1–S2 and NY was Figs 3–7. PKRK and NY planned and performed the experiments, analyzed the data and wrote the article.
Funding
This study was supported by MEXT KAKENHI Grant Number 25116008.
Conflict of Interest
None declared.
References
- 1. Yasuhara N., Oka M., Yoneda Y. (2009) The role of nuclear transport system in cell differentiation. Sem. Cell Dev. Biol. 20, 590–599 [DOI] [PubMed] [Google Scholar]
- 2. Pumroy R.A., Cingolani G. (2015) Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem. J. 466, 13–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mason D.A., Stage D.E., Goldfarb D.S. (2009) Evolution of the metazoan-specific importin alpha gene family. J. Mol. Evol. 68, 351–365 [DOI] [PubMed] [Google Scholar]
- 4. Kobe B. (1999) Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 6, 388–397 [DOI] [PubMed] [Google Scholar]
- 5. Misheva M., Kaur G., Ngoei K.R., Yeap Y.Y., Ng I.H., Wagstaff K.M., Ng D.C., Jans D.A., Bogoyevitch M.A. (2014) Intracellular mobility and nuclear trafficking of the stress-activated kinase JNK1 are impeded by hyperosmotic stress. Biochim. Biophys. Acta 1843, 253–264 [DOI] [PubMed] [Google Scholar]
- 6. Tseng S.F., Chang C.Y., Wu K.J., Teng S.C. (2005) Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J. Biol. Chem. 280, 39594–39600 [DOI] [PubMed] [Google Scholar]
- 7. Nishinaka Y., Masutani H., Oka S., Matsuo Y., Yamaguchi Y., Nishio K., Ishii Y., Yodoi J. (2004) Importin alpha 1 (Rch1) mediates nuclear translocation of thioredoxin-binding protein-2/vitamin D(3)-up-regulated protein1. J. Biol. Chem. 279, 37559–37565 [DOI] [PubMed] [Google Scholar]
- 8. Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. (2007) Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the endoplasmic reticulum/Golgi membrane. J. Virol. 81, 9812–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yasuhara N., Shibazaki N., Tanaka S., Nagai M., Kamikawam Y., Oe S., Asally M., Kamachi Y., Kondoh H., Yoneda Y. (2007) Triggering neural differentiation of ES cells by subtype switching of importin-α. Nat. Cell Biol. 9, 72–79 [DOI] [PubMed] [Google Scholar]
- 10. Van der Watt P.J., Maske C.P., Hendricks D.T., Parker M.I., Denny L., Govender D., Birrer M.J., Leaner V.D. (2009) The karyopherin proteins, Crm1 and karyopherin beta1, are overexpressed in cervical cancer and rare critical for cancer cell survival and proliferation. Int. J. Cancer 124, 1829–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng M., Tang L., Huang L., Ding H., Liao W.T., Zeng M.S., Wang H.Y. (2010) Overexpression of karyopherin-2 in epithelial ovarian cancer and correlation with poor prognosis. Obstet. Gynecol. 116, 884–891 [DOI] [PubMed] [Google Scholar]
- 12. Christiansen A., Dyrskjot L. (2013) The functional role of the novel biomarker karyopherin a-2 (KPNA2) in cancer. Cancer Lett. 331, 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin Y.Z., Yao S.Y., Veach R.A., Torgerson T.R., Hawiger J. (1995) Inhibition of nuclear translocation of transcription factor NF-kappa B by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J. Biol. Chem. 270, 14255–14258 [DOI] [PubMed] [Google Scholar]
- 14. Zienkiewicz J., Armitage A., Hawiger J. (2013) Targeting nuclear import shuttles, importins/karyopherins alpha by a peptide mimicking the NFkappaB1/p50 nuclear localization sequence. J. Am. Heart Assoc. 2, e000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gopinath S.C., Misono T.S., Kawasaki K., Mizuno T., Imai M., Odagiri T., Kumar P.K.R. (2006) An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J. Gen. Virol. 87, 479–487 [DOI] [PubMed] [Google Scholar]
- 16. Gopinath S.C., Kumar P.K.R. (2013) Aptamer that binds to the haemagglutinin of the recent pandemic influenza virus H1N1 and efficiently inhibit agglutination. Acta Biomater 9, 8932-8841 [DOI] [PubMed] [Google Scholar]
- 17. Suenaga E., Kumar P.K.R. (2014) An aptamer that binds efficiently to the haemagglutinin of highly pathogenic avian influenza viruses (H5N1 and H7N7) and inhibits haemagglutinin-glycan interactions. Acta Biomater 10, 1314–1323 [DOI] [PubMed] [Google Scholar]
- 18. Gopinath S.C., Hayashi K., Kumar P.K.R. (2012) Aptamer that binds to the gD protein of herpes simplex virus 1 and efficiently inhibits viral entry. J. Virol. 86, 6732–6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Imamoto N., Shimamotom T., Takao T., Tachibana T., Kose S., Matsubae M., Sekimoto T., Shimonishi Y., Yoneda Y. (1995) In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. Embo J. 14, 3617–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acid Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar P.K.R., Machida K., Urvil P.T., Kikuchi P.N., Vishnuvardhan D., Shimotohno K., Taira K., Nishikawa S. (1997). Isolation of RNA aptamers specific to the NS3 protein of hepatitis C virus from a completely random RNA. Virology 237, 270–282 [DOI] [PubMed] [Google Scholar]
- 22. Adam S.A., Marr R.S., Gerace L. (1990) Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111, 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Meth. 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yasuhara N., Yamagishi R., Arai Y., Mehmmod R., Kimoto C., Fujita T., Touma K., Kaneko A., Kamikawa Y., Moriyama T., Yanagida T., Kaneko H., Yoneda Y. (2013). Importin alpha subtypes determine differential transcription factor localization in embryonic stem cells maintenance. Dev. Cell 26, 123–135 [DOI] [PubMed] [Google Scholar]
- 25. Zhou J., Rossi J.J. (2009). The therapeutic potential of cell-internalizing aptamers. 2009. The therapeutic potential of cell-internalizing aptamers. Curr. Top. Med. Chem 9, 1144–1157 [DOI] [PubMed] [Google Scholar]
- 26. Zhou J., Satheesan S., Li H., Weinberg M.S., Morris K.V., Burnett J.C., Rossi J.J. (2015) Cell-specific RNA aptamer against the CCR5 specifically targets HIV-1 susceptible cells and inhibits HIV-1 infectivity. Chem. Biol. 22, 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.