Abstract
Objectives
Enterovirus 71 (EV71) causes serious diseases in humans. The aim of this study was to examine the effects of aurintricarboxylic acid (ATA) on EV71 replication and to explore the underlying mechanism.
Methods
To measure the activity of ATA in inhibiting the cytopathic effect (CPE) of EV71, a cell-based neutralization (inhibition of virus-induced CPE) assay was performed. The effect of ATA was further confirmed using plaque reduction and viral yield reduction assays. A time of addition assay was performed to identify the mechanisms of ATA's anti-EV71 activity. We examined the effects of ATA on the following key steps involved in virus replication: (i) translation of the internal ribosomal entry site (IRES)-mediated viral polyprotein; (ii) the proteolytic activity of viral proteases 2A and/or 3C; and (iii) the viral 3D RNA-dependent RNA polymerase (RdRp) activity.
Results
In this study, ATA was found to be a potent inhibitor of the replication of EV71. In the antiviral neutralization assay, ATA exhibited inhibitory activity against EV71 (TW/4643/98) and EV71 (TW/2231/98). Plaque assay further demonstrated that ATA inhibited EV71 replication with an EC50 (effective concentration at which 50% of plaques were removed) of 2.9 µM. Studies on the mechanism of action revealed that ATA targets the early stage of the viral life cycle after viral entry. ATA was able to inhibit the RdRp activity of EV71, while neither the IRES-mediated translation of viral polyprotein nor the viral 3C protease activity was affected.
Conclusions
Overall, the findings in this study suggest that ATA is able to effectively inhibit EV71 replication through interfering with the viral 3D polymerase.
Keywords: RNA-dependent RNA polymerase, enteroviruses, polyanionic compounds
Introduction
Enteroviruses are members of the family Picornaviridae, which has ∼70 serotypes including poliovirus (types 1, 2 and 3), coxsackievirus A (A1–22 and A24), coxsackievirus B (B1–6), echovirus (1–21 and 24–33) and enterovirus (68–71, 73–78 and 89–91).1 Among the enterovirus strains, enterovirus 71 (EV71) may cause aseptic meningitis, encephalomyelitis or even acute flaccid paralysis similar to paralytic poliomyelitis that is also caused by poliovirus infection.2 Currently, no specific antiviral agents are available for the treatment of EV71.
Enterovirus consists of a single-stranded, positive-sense RNA of ∼7500 nt.1 Upon infection, the internal ribosome entry site (IRES) element in the 5′-untranslated region (UTR) drives the translation of the viral processor polyprotein. This polyprotein, encoded as NH2-VP4-VP2-VP3-VP1-2A-2B-2C-3A-3B-3C-3D-COOH, is then proteolytically cleaved by viral 2A and 3C proteases to form various structural and non-structural viral proteins.1 Many important stages during the viral life cycle have been considered as potential molecular targets for development of antiviral agents.3,4 Virus-encoded proteases and polymerases have been the most prominent targets of consideration for antiviral drug development. Success in the development of antiviral agents against several viruses, including HIV and hepatitis B virus (HBV), has established that virus-encoded proteases and polymerases are valid drug targets for these viruses.
A group of polyanionic compounds including aurintricarboxylic acid (ATA) has been shown to be effective in inhibition of several viruses in cell culture.5 ATA was reported to inhibit the adsorption, and thus block the replication of HIV by interfering with the interaction between viral envelope glycoprotein (gp120) and the CD4 receptor on the cell surface.6 ATA has also been demonstrated to inhibit the replication of several RNA viruses, such as HIV,7,8 vesicular stomatitis virus,9,10 SARS coronavirus (SARS-CoV)11,12 and influenza virus.13 Very recently, ATA has been reported as a replicase inhibitor of hepatitis C virus (HCV).14
In the present study, we examined the effect of ATA on the inhibition of the replication of EV71 virus. Results indicate that ATA mitigates EV71-induced cytopathic effects (CPEs) in Vero cells. Additionally, ATA inhibited the elongation activity of the viral 3D RNA-dependent RNA polymerase (RdRp).
Materials and methods
Cell culture
Vero cells (African green monkey kidney cells, ATCC accession no. CCL-81) were grown in modified Eagle's medium (MEM; Gibco). Rhabdomyosarcoma cells (RD cells, ATCC accession no. CCL-136) and COS-7 cells (African green monkey kidney cells, ATCC CRL-1650) were grown in Dulbecco's modified Eagle's medium (DMEM). All media were supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/mL of penicillin and streptomycin, and 2 mM l-glutamine.
Virus isolation and propagation
EV71 (TW/4643/98 and TW/2231/98) viruses were provided by the Clinical Virology Laboratory of Chang Gung Memorial Hospital (Linkou, Taiwan). Viruses were amplified by using RD cells. Virus titre was determined by a plaque assay using Vero cells.
Neutralization test
To measure the potential of ATA to inhibit the CPE of the enterovirus strains, 96-well tissue culture plates were seeded with 200 µL of 1 × 105 RD cells/mL in DMEM with 10% FBS. After cells were incubated for 18–24 h at 37°C, virus at 100× the TCID50 (50% tissue culture infectious dose) mixed with different concentrations of ATA was added to the cells. After 1 h of viral adsorption, the infected cells were overlaid with 50 µL of DMEM and 0.5% DMSO and incubated at 37°C for 72 h. At the end of the incubation, the cells were fixed with formaldehyde and stained with 0.1% Crystal Violet as described previously.15 The concentration required for the tested compound to reduce the CPE of the virus by 50% (IC50) was determined.
Plaque assay
The plaque assay was performed in 6-well plates containing 90% confluent Vero monolayer cells. The cells were inoculated with enterovirus at a titre of ∼50–100 pfu per well of cells. After 1 h incubation at 35°C, the infection media were aspirated off and each well was then covered with 3 mL of agar overlay medium containing various concentrations of compounds and incubated for 2 days at 35°C in a CO2 incubator. At the end of the incubation, the cells were fixed with formaldehyde and stained with 0.1% Crystal Violet as described previously.15 The concentration required for ATA to reduce the number of plaques by 50% (EC50) was then determined.
Virus yield assay
Vero cells were infected with EV71 at a multiplicity of infection (moi) of 10 and various concentrations of compounds were added into the cell culture media. After 12 h, the culture media and cell lysates were collected following freeze–thaw cycles and then subjected to virus titration by plaque-forming assay as described previously.15
Time of addition experiment
In the time of addition experiment, Vero cells were infected with enterovirus (100 pfu/well) and incubated in E2 medium and, at specified timepoints before or after infection, ATA was added at 80 µM. At 10 h post-infection (hpi), the cells were rinsed with PBS to remove the drugs and the culture medium overlay without drug was applied to detect the viral infectivity by plaque assay.16
Bi-cistronic expression assay
The pRHF-EV71-5′-UTR, containing the EV71 5′-UTR between Renilla and firefly luciferase genes, was used to evaluate the IRES-dependent translation of EV71.17 The bi-cistronic plasmid was transfected into RD cells (1 × 105 cells/well in a 24-well plate) and the OPTI-MEM medium was replaced with E2 medium with or without ATA treatment 5 h post-transfection. After 48 h, cell lysates were collected and analysed to determined Renilla (RLuc) and firefly (FLuc) luciferase activity using a dual-luciferase reporter assay (Promega Corp., Madison, WI, USA), following the manufacturer's instructions.
2A protease in vivo activity
Plasmids encoding the 2A viral protease (pFLAG-CMV2-2A) or the 2A viral protease with a single point mutation (C110S) [pFLAG-CMV2-2A (C110S)] and a reporter plasmid, pRL-SV40, harbouring Renilla luciferase under the control of the SV40 early enhancer/promoter region, were co-transfected into Vero cells (1 × 105 cells/well in a 24-well plate) and the OPTI-MEM medium was replaced with E2 medium with or without ATA treatment 5 h post-transfection. After 48 h, cell lysates were collected and analysed to determined RLuc activity using a Renilla luciferase assay system (Promega Corp.), following the manufacturer's instructions.
In vivo activity assay of 3C protease
COS-7 cells were seeded in a 24-well plate at 6 × 104 cells per well and incubated at 37°C overnight for transient transfection. COS-7 cells were co-transfected with the following combinations of plasmids: (i) pBAK8-MTEGFP-GAL4-SEAP only; (ii) pBAK8-MTEGFP-M3VP16 only; (iii) pBAK8-MTEGFP-GAL4-SEAP and pBAK8-MTEGFP-M3VP16; (iv) pBAK8-MTEGFP-GAL4-SEAP and pBAK8-MTEGFP-M3-3C-VP16; and (v) pBAK8-MTEGFP-GAL4-SEAP and pBAK8-MTEGFP-M3-3Cmut-VP16. pCMV-GAL as an internal control was co-transfected with each expression vector. At 48 h post-transfection, the secreted alkaline phosphatase (SEAP) activity of the culture medium was measured by using a Phospha-Light assay kit (Tropix, Foster City, CA, USA). The chemiluminescence was detected using a TopCount Microplate Scintillation and Luminescence Counter (Packard, Meriden, CT, USA). AG7088 was a gift from J. M. Fang (National Taiwan University, Taipei, Taiwan).18
Detection of viral RNA in EV71-infected cells
A total of 5 × 105 Vero cells were seeded into 6-well plates to reach confluence and then challenged with virus (moi = 1). Total RNA was extracted from cells using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Following phenol–chloroform extraction and isopropanol precipitation, the RNA pellet was washed, dried and dissolved in 20 µL of RNase-free water. RT–PCR amplifications were carried using a Reverse-iT One Step Kit (Abgene, Epsom, Surrey, UK). Each PCR used the following protocol: 30 min at 42°C; 5 min at 94°C; then 35 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 1 min; and an additional 15 min for elongation in the last cycle. RT–PCR products were examined by electrophoresis on 1% agarose gels by loading 3 µL in each gel well. The sequences of the primers used for RT–PCR were as follows: 5′-GGAGATAGGGTGGCAGATGTGATTG-3′ (genomic position: 2339–2463 of TW/2272/98); and 5′-GAGAGTGGTAATTGCTGTGCG-3′ (genomic position: 3309–3329).19
3D polymerase elongation assay
The determination of the expression of EV71 3D polymerase and the polymerase elongation assay were performed as described previously.15 RNA polymerase assays were performed with 1 U of the 3D polymerase in 50 mM HEPES, pH 7.5, 10 mM 2-mercaptoethanol, 5 mM MgCl2, 60 µM ZnCl2, 500 µM UTP, 0.4 µCi/μL [α-32P]UTP, 1.8 µM dT15/0.15 µM poly(rA)300 (primer/template) and various concentrations of ATA. Reactions were incubated for 10 min at 30°C and stopped by the addition of EDTA to 83 mM. An aliquot of 10 µL of the quenched reaction was spotted onto DE81 filter paper discs (Whatman) and dried at room temperature. The discs were washed three times for 10 min in 5% dibasic sodium phosphate and were rinsed in absolute ethanol. The radioactivity of the sample was quantified in 1.5 mL of scintillation fluid. Data points are the means ± SD of triplicate determinations.
Results and discussion
ATA inhibits EV71 replication
We have previously shown that a number of agents can inhibit the replication of EV71.15,19–26 In this study, we found that EV71 replication can also be inhibited by ATA. In the antiviral neutralization test, Vero cells seeded in 96-well plates were all lysed at 64 h after EV71 infection as shown in the VC (virus control) column in Figure 1(a). Without virus infection, ATA did not cause apparent cell lysis at concentrations up to 1000 µM (row 1, Figure 1a). When EV71-infected cells were treated with ATA at various concentrations, ATA showed effects on the protection of lysis of Vero cells in a dose-dependent manner (rows 2–4, Figure 1a, experiments in triplicate). Subsequently, the effect of ATA was further confirmed using the plaque reduction and viral yield reduction assays. In the plaque reduction assay, Vero cells were infected with EV71 (TW/2231/98) at a titre of ∼50–100 pfu per well of cells in 6-well plates. After virus adsorption for 1 h, ATA was added to Vero cells at various concentrations. ATA dose-dependently reduced the number of pfu (Figure 1b). The EC50 of ATA, as measured using the plaque reduction assay, was 2.9 µM. The CC50 (cytotoxic concentration at which 50% of cell viability was inhibited) of ATA in Vero cells was 211 µM as determined by MTS assay (data not shown).27 Therefore, the selective index (SI = CC50/EC50) of ATA ranged from >3 to >75 based on different assays.28
Figure 1.
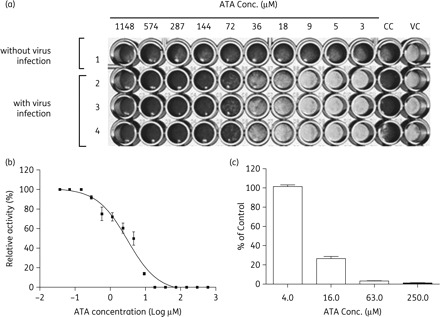
Inhibition of EV71 infection and replication by ATA in Vero cells. (a) ATA inhibits the EV71-induced CPE in cells. In the antiviral neutralization test, Vero cells were all lysed at 64 h after EV71 infection as shown in the virus control (VC) column. The CC column refers to cells without compound treatment and virus infection. ATA was added to EV71-infected cells in 2-fold serial dilutions starting from a concentration of 1148 µM in the left-hand column. (b) ATA inhibits EV71 plaque formation. The anti-EV71 effect of ATA was also assessed by plaque assay. Vero cells were infected with EV71 (TW/2231/98) at a titre of ∼50–100 pfu per well of cells in 6-well plates. After virus adsorption for 1 h, ATA was added to Vero cells at various concentrations. Then the assay was performed as described in the Materials and methods section. (c) Reduction in viral yields from infected cells treated with ATA at different concentrations. Vero cells were infected with an moi of 10 EV71 and treated with ATA at various concentrations. After approximately one virus replication cycle, the culture media and cell lysates were collected for virus titration by plaque assay.
The effect of ATA on the yields of viral progeny in EV71-infected cells was also assessed. In this experiment, Vero cells were infected with EV71 (TW/2231/98) at an moi of 10 accompanied by treatment with ATA at various concentrations. At 12 hpi, viral progeny present in cell culture media and cell lysates were harvested, pooled and then analysed for viral titres using a plaque assay. As shown in Figure 1(c), ATA treatment substantially reduced the titres of EV71 viral progeny produced in infected cells. When cells were treated with ATA at 16 µM, a decrease in viral yield of >60% was evident. The declines in viral yields were 97% and 99% when ATA was applied to treat the virus-infected cells at 64 and 250 µM, respectively. These results showed that ATA could effectively reduce the production of viral progeny in EV71-infected cells. Different EC50 values in different assays were noted, and this may be due to the fact that different infection conditions, such as the moi, were employed in different assays.
ATA acts during the early stage of EV71 replication
To understand the mechanism of ATA's anti-EV71 activity, a time of addition experiment was conducted. ATA (80 µM) was added to Vero cells at different timepoints before (T−1: 1 h before virus infection) or after virus infection (T0, T1, T3, T5, T7 and T9: 0, 1, 3, 5, 7 and 9 h after virus infection, respectively). Then, culture supernatants were removed for determination of the virus yields after approximately one viral replication cycle.16 The yields of viral progeny were normalized based on the vehicle control (same volume of DMSO added at T0). In Figure 2, at T−1 and T1, upon ATA treatment, the viral yields in the EV71-infected cells were reduced by 60%. The antiviral effects of ATA started to decrease when ATA was added after the T3 timepoint. When the EV71-infected cells were treated with 120 µM ATA, a similar phenomenon was observed. Therefore, ATA's anti-EV71 activity was most prominent when it was applied during the early stage of the EV71 replication cycle. Overall, the time of addition experiment indicated that ATA exerts its antiviral effects during the earlier events in the virus replication cycle but not at the viral absorption stage. However, the exact underlying mechanism of inhibition of EV71 by ATA needs further clarification. In this study, we experimentally examined the effects of ATA on the following key steps involved in virus replication: (i) translation of the IRES-mediated viral polyprotein; (ii) the proteolytic activity of viral proteases 2A and/or 3C; and (iii) the viral 3D RdRp elongation activity.
Figure 2.
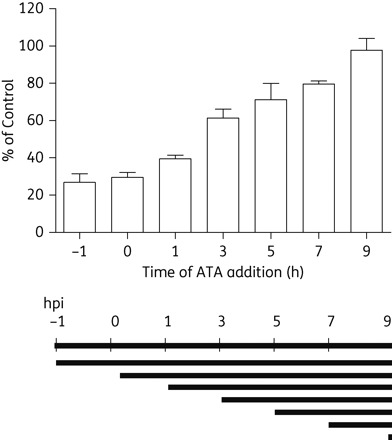
Time of addition examination of the inhibitory effect of ATA. ATA at 80 µM was added to Vero cells at different timepoints as described in detail in the Materials and methods section. The percentages of control values are normalized values according to the no-drug treatment control.
ATA does not affect IRES-driven translation
To examine if ATA interferes with the IRES-mediated translation, a bi-cistronic expression system containing the IRES of EV71, i.e. pRHF-EV71-5′-UTR or pRHF-EV71-5′-UTR-AS (i.e. the antisense EV71 5′-UTR), was employed.17 As shown in the upper panel of Figure 3(a), the IRES-directed translation initiation efficiencies would be reflected by the ratio of RLuc and FLuc levels. The bi-cistronic plasmid was transfected into RD cells and then cell lysates were collected at 48 hpi and analysed to determine the RLuc and FLuc activity. No significant decrease in IRES-mediated translation was observed in the presence of ATA even at concentrations up to 118 µM (Figure 3a). Thus, these results indicated that ATA does not affect the IRES-mediated translation.
Figure 3.
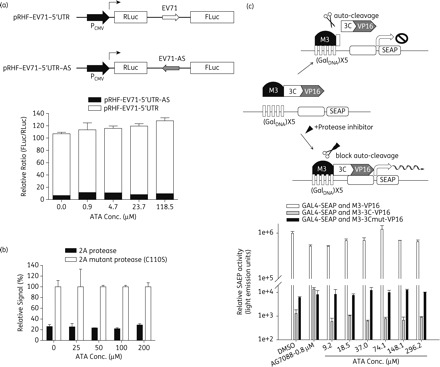
Effects of ATA on viral factors. (a) Effect of ATA on IRES activity of EV71. The diagrams of the constructs of the bi-cistronic reporter plasmids are shown in the upper panel. RD cells were transfected with pRHF-EV71-5′-UTR or pRHF-EV71-5′-UTR-AS (antisense EV71 5′-UTR) in the presence of ATA at different concentrations. (b) Effect of ATA on 2A protease activity of EV71. Vero cells were transfected with pFLAG-CMV2-2A/pFLAG-CMV2-2A (C110S) and reporter plasmid pRL-SV40, which harbours the SV40 early enhancer/promoter region upstream of Renilla luciferase, in the presence of ATA or DMSO. At 48 hpi, the Renilla luciferase (RLuc) and firefly luciferase (FLuc) activities were analysed. (c) Effect of ATA on the 3C protease activity of EV71. The principle of this cell-based 3C protease activity assay is depicted in the upper panel. COS-7 cells were co-transfected with the combinations of plasmids indicated. The cells were incubated in the absence or presence of 9.2, 18.5, 37.0, 74.1, 148.1 or 296.2 µM ATA for 12 h, and culture media were assayed for SEAP activity. Each data point represents the average of three independent experiments.
ATA does not affect 3C and 2A protease activity
We then examined if ATA interferes with the effect of 2A protease in Vero cells using a method described in previous studies.29,30 Cells were co-transfected with a reporter plasmid, pRL-SV40, harbouring Renilla luciferase, and the plasmid pFLAG-CMV2-2A encoding the 2A viral protease or, as a control, plasmid pFLAG-CMV2-2A (C110S) encoding an inactive 2A protease with a single point mutant (C110S). As shown in Figure 3(b), there was an 80% decrease in the Renilla luciferase signal in the cell lysates with 2A protease expression as compared with those with inactive 2A protease. When cells were treated with ATA, at concentrations up to 200 µM, there was no restoration of the reporter signal in Vero cells co-transfected with the reporter plasmid and the 2A expression plasmid (Figure 3b), indicating that ATA does not inhibit the activity of EV71 viral 2A protease.
To examine if ATA inhibits the viral 3C protease, we employed the mammalian cell-based reverse two-hybrid system for functional analysis of 3C viral protease activity of EV71 as described in a previous study.18 As shown in the upper panel of Figure 3(c), the proteolytic activity of EV71 3C protease was monitored in a cell culture system based on the reverse two-hybrid system. When M3 and the VP16 domains were separated by self-cleavage of 3C, the M3-3C-VP16 failed to turn on SEAP expression in the cells. In contrast, the co-expression of GAL4-SEAP and M3-3Cmut-VP16 gave rise to high levels of SEAP enzymatic activity in the culture supernatant of transfected cells while only a background level of SEAP activity was detected in the culture supernatant of cells co-expressing GAL4-SEAP and M3-3C-VP16. The level of SEAP activity in the culture supernatant of cells co-expressing GAL4-SEAP and M3-3C-VP16 was significantly enhanced with the treatment with a known 3C protease inhibitor, AG7088, showing that the assay can be used to monitor the activity of 3C protease inhibitors. No increase of SEAP activity was seen even when transfected cells were treated with ATA at concentrations up to 300 µM (Figure 3c). Thus, ATA does not interfere with viral 3C protease activity as revealed in the cell-based protease activity assay.
ATA reduces EV71 viral RNA synthesis and inhibits EV71 3D RdRp activity
To examine the effect of ATA on the synthesis of EV71 RNA, we treated the virus-infected cells with ATA and then the viral RNA in cells was analysed using RT–PCR. As shown in Figure 4(a), while no RT–PCR product was detected in cells harvested at 7 hpi, the bands corresponding to the RT–PCR product became prominent for the cell lysates harvested at 9, 11 or 13 hpi. In the presence of ATA at 80 or 120 µM, the levels of the RT–PCR products were greatly reduced. The results suggested that ATA can slow down the viral RNA synthesis at early stages after a single round of viral replication in EV71-infected cells.
Figure 4.
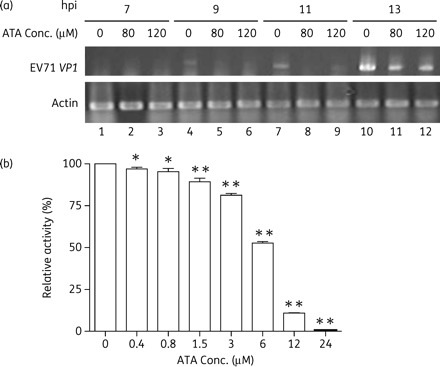
Effect of ATA on EV71 viral RNA synthesis and EV71 3D viral polymerase activity. (a) ATA reduces the level of viral RNA synthesis. Total cellular RNA (0.5 µg) extracted from infected cells was used in each RT–PCR experiment, and 3 µL of reaction solution was added to each well. (b) ATA inhibits the activity of EV71 3D RdRp. RNA polymerase assays were performed using various concentrations of ATA as described in the Materials and methods section. Data are represented as means ± SEM from three independent experiments. The difference among different groups was assessed by one-way analysis of variance followed by post-hoc Tukey's multiple comparison test (*P < 0.05, **P < 0.001 versus the wild-type control group polymerase without inhibitor).
To examine if the reduced viral RNA synthesis upon ATA treatment was due to the ability of ATA to inhibit EV71 3D polymerase (RdRp), the RdRp enzymatic activity was measured as described by detecting the amount of 32P-labelled uridine monophosphate (UMP) incorporated into poly(U) RNA in the presence of a poly(A) template and oligo(dT) primers.15 The rate of UMP incorporation of recombinant EV71 3D RdRp in the presence of 5% DMSO (the vehicle used for dissolving ATA) was employed as a control. In the presence of ATA, UMP incorporation into the RdRp reaction products was clearly inhibited (Figure 4b). When ATA was added to the RdRp reaction at concentrations <0.2 µM, no inhibition was observed. However, ATA began to inhibit RdRp activity at 0.8 µM. At 12.0 µM, ATA reduced the RdRp elongation activity by almost 90% of the control and the IC50 was estimated to be 6.3 µM. Thus, the results suggested that ATA inhibits the 3D RdRp encoded by EV71.
In summary, ATA is a unique compound that has a pleiotropic effect on cellular functions.31–34 The broad-spectrum antiviral activity of ATA is summarized in Table 1.34 Several previous studies have described ATA as an inhibitor of polymerase.11,14 In addition, Yap et al.11 reported the mechanisms of binding of ATA to the RdRp of SARS-CoV and other positive-stranded RNA viruses by a molecular docking method. Recently, Chen et al.14 also reported that ATA was an NS5B replicase inhibitor of HCV. Results in this study demonstrated that ATA is able to effectively inhibit EV71 replication through the inhibition of the elongation activity of the virus-encoded 3D polymerase (RdRp). It is noteworthy that the inhibition of EV71 by ATA may not be due solely to RdRp inhibition, and further investigations are needed to explore if ATA also affects other viral or cellular functions that are important for EV71 replication. Overall, results from this study will prove to be important for the development of more potent agents against EV71.
Table 1.
Effects of ATA on other viruses
| Viral proteins | Virus | EC50a | IC50 | Reference(s) |
|---|---|---|---|---|
| NS5B (replicase) | HCV | 75.0 nM | 150.0 nM | Chen et al. (2009)14 |
| MTase (methyltransferase) | flavivirus | — | >2.3 µM | Milani et al. (2009)35 |
| NA (neuraminidase) | influenza | 9.4 µM | >4.1 µM | Hung et al. (2009)13 |
| H1L (tyrosine phosphatase) | vaccinia | 59.0 µM | 5.3 µM | Myskiw et al. (2007)9 |
| Pol (RdRp, polymerase) | SARS | 473.0 µM | — | Yap et al. (2005)11 and He et al. (2004)36 |
| gp120 (coat protein) | HIV | >10.7 µM | — | Cushman et al. (1991)37 |
aEC50 values were determined using cell-based assays.
Funding
This work was supported by the National Health Research Institutes (NHRI) in Taiwan.
Transparency declarations
None to declare.
References
- 1.Pallansch M, Roos R. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Knipe D, Howley P, editors. Fields Virology. 4th edn. Baltimore: Williams and Wilkins; 2001. pp. 840–93. [Google Scholar]
- 2.Shih SR, Ho MS, Lin KH, et al. Genetic analysis of enterovirus 71 isolated from fatal and non-fatal cases of hand, foot and mouth disease during an epidemic in Taiwan, 1998. Virus Res. 2000;68:127–36. doi: 10.1016/s0168-1702(00)00162-3. [DOI] [PubMed] [Google Scholar]
- 3.Chen TC, Weng KF, Chang SC, et al. Development of antiviral agents for enteroviruses. J Antimicrob Chemother. 2008;62:1169–73. doi: 10.1093/jac/dkn424. [DOI] [PubMed] [Google Scholar]
- 4.De Palma AM, Vliegen I, De Clercq E, et al. Selective inhibitors of picornavirus replication. Med Res Rev. 2008;28:823–84. doi: 10.1002/med.20125. [DOI] [PubMed] [Google Scholar]
- 5.Urbinati C, Chiodelli P, Rusnati M. Polyanionic drugs and viral oncogenesis: a novel approach to control infection, tumor-associated inflammation and angiogenesis. Molecules. 2008;13:2758–85. doi: 10.3390/molecules13112758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Clercq E. New developments in anti-HIV chemotherapy. Biochim Biophys Acta. 2002;1587:258–75. doi: 10.1016/s0925-4439(02)00089-3. [DOI] [PubMed] [Google Scholar]
- 7.Santhosh KC, Paul GC, De Clercq E, et al. Correlation of anti-HIV activity with anion spacing in a series of cosalane analogues with extended polycarboxylate pharmacophores. J Med Chem. 2001;44:703–14. doi: 10.1021/jm000290u. [DOI] [PubMed] [Google Scholar]
- 8.Witvrouw M, Fikkert V, Pluymers W, et al. Polyanionic (i.e., polysulfonate) dendrimers can inhibit the replication of human immunodeficiency virus by interfering with both virus adsorption and later steps (reverse transcriptase/integrase) in the virus replicative cycle. Mol Pharmacol. 2000;58:1100–8. [PubMed] [Google Scholar]
- 9.Myskiw C, Deschambault Y, Jefferies K, et al. Aurintricarboxylic acid inhibits the early stage of vaccinia virus replication by targeting both cellular and viral factors. J Virol. 2007;81:3027–32. doi: 10.1128/JVI.02531-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geber WF, Lefkowitz SS, Hung CY. Effect of ascorbic acid, sodium salicylate, and caffeine on the serum interferon level in response to viral infection. Pharmacology. 1975;13:228–33. doi: 10.1159/000136908. [DOI] [PubMed] [Google Scholar]
- 11.Yap Y, Zhang X, Andonov A, et al. Structural analysis of inhibition mechanisms of aurintricarboxylic acid on SARS-CoV polymerase and other proteins. Comput Biol Chem. 2005;29:212–9. doi: 10.1016/j.compbiolchem.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Clercq E. Potential antivirals and antiviral strategies against SARS coronavirus infections. Expert Rev Anti Infect Ther. 2006;4:291–302. doi: 10.1586/14787210.4.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hung HC, Tseng CP, Yang JM, et al. Aurintricarboxylic acid inhibits influenza virus neuraminidase. Antiviral Res. 2009;81:123–31. doi: 10.1016/j.antiviral.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Bopda-Waffo A, Basu A, et al. Characterization of aurintricarboxylic acid as a potent hepatitis C virus replicase inhibitor. Antivir Chem Chemother. 2009;20:19–36. doi: 10.3851/IMP1286. [DOI] [PubMed] [Google Scholar]
- 15.Chen TC, Chang HY, Lin PF, et al. Novel antiviral agent DTriP-22 targets RNA-dependent RNA polymerase of enterovirus 71. Antimicrob Agents Chemother. 2009;53:2740–7. doi: 10.1128/AAC.00101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verheyden B, Andries K, Rombaut B. Mode of action of 2-furylmercury chloride, an anti-rhinovirus compound. Antiviral Res. 2004;61:189–94. doi: 10.1016/j.antiviral.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Lin JY, Li ML, Shih SR. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009;37:47–59. doi: 10.1093/nar/gkn901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JC, Shih SR, Chang TY, et al. A mammalian cell-based reverse two-hybrid system for functional analysis of 3C viral protease of human enterovirus 71. Anal Biochem. 2008;375:115–23. doi: 10.1016/j.ab.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Shih SR, Tsai KN, Li YS, et al. Inhibition of enterovirus 71-induced apoptosis by allophycocyanin isolated from a blue-green alga Spirulina platensis. J Med Virol. 2003;70:119–25. doi: 10.1002/jmv.10363. [DOI] [PubMed] [Google Scholar]
- 20.Chen TC, Liu SC, Huang PN, et al. Antiviral activity of pyridyl imidazolidinones against enterovirus 71 variants. J Biomed Sci. 2008;15:291–300. doi: 10.1007/s11373-007-9228-5. [DOI] [PubMed] [Google Scholar]
- 21.Chern JH, Chang CS, Tai CL, et al. Synthesis and antipicornavirus activity of (R)- and (S)-1-[5-(4′-chlorobiphenyl-4-yloxy)-3-methylpentyl]-3-pyridin-4-yl-imidazolidin-2-one. Bioorg Med Chem Lett. 2005;15:4206–11. doi: 10.1016/j.bmcl.2005.06.069. [DOI] [PubMed] [Google Scholar]
- 22.Chang CS, Lin YT, Shih SR, et al. Design, synthesis, and antipicornavirus activity of 1-[5-(4-arylphenoxy)alkyl]-3-pyridin-4-ylimidazolidin-2-one derivatives. J Med Chem. 2005;48:3522–35. doi: 10.1021/jm050033v. [DOI] [PubMed] [Google Scholar]
- 23.Chern JH, Lee CC, Chang CS, et al. Synthesis and antienteroviral activity of a series of novel, oxime ether-containing pyridyl imidazolidinones. Bioorg Med Chem Lett. 2004;14:5051–6. doi: 10.1016/j.bmcl.2004.07.084. [DOI] [PubMed] [Google Scholar]
- 24.Chern JH, Shia KS, Hsu TA, et al. Design, synthesis, and structure–activity relationships of pyrazolo[3,4-d]pyrimidines: a novel class of potent enterovirus inhibitors. Bioorg Med Chem Lett. 2004;14:2519–25. doi: 10.1016/j.bmcl.2004.02.092. [DOI] [PubMed] [Google Scholar]
- 25.Shia KS, Li WT, Chang CM, et al. Design, synthesis, and structure–activity relationship of pyridyl imidazolidinones: a novel class of potent and selective human enterovirus 71 inhibitors. J Med Chem. 2002;45:1644–55. doi: 10.1021/jm010536a. [DOI] [PubMed] [Google Scholar]
- 26.Shih SR, Tsai MC, Tseng SN, et al. Mutation in enterovirus 71 capsid protein VP1 confers resistance to the inhibitory effects of pyridyl imidazolidinone. Antimicrob Agents Chemother. 2004;48:3523–9. doi: 10.1128/AAC.48.9.3523-3529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cory AH, Owen TC, Barltrop JA, et al. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991;3:207–12. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
- 28.Pauwels R, Balzarini J, Baba M, et al. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–21. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 29.Castello A, Alvarez E, Carrasco L. Differential cleavage of eIF4GI and eIF4GII in mammalian cells. Effects on translation. J Biol Chem. 2006;281:33206–16. doi: 10.1074/jbc.M604340200. [DOI] [PubMed] [Google Scholar]
- 30.Castello A, Izquierdo JM, Welnowska E, et al. RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J Cell Sci. 2009;122:3799–809. doi: 10.1242/jcs.055988. [DOI] [PubMed] [Google Scholar]
- 31.Andrew DJ, Hay AW, Evans SW. Aurintricarboxylic acid inhibits apoptosis and supports proliferation in a haemopoietic growth-factor dependent myeloid cell line. Immunopharmacology. 1999;41:1–10. doi: 10.1016/s0162-3109(98)00049-6. [DOI] [PubMed] [Google Scholar]
- 32.Benchokroun Y, Couprie J, Larsen AK. Aurintricarboxylic acid, a putative inhibitor of apoptosis, is a potent inhibitor of DNA topoisomerase II in vitro and in Chinese hamster fibrosarcoma cells. Biochem Pharmacol. 1995;49:305–13. doi: 10.1016/0006-2952(94)00465-x. [DOI] [PubMed] [Google Scholar]
- 33.Cho H, Lee DY, Shrestha S, et al. Aurintricarboxylic acid translocates across the plasma membrane, inhibits protein tyrosine phosphatase and prevents apoptosis in PC12 cells. Mol Cells. 2004;18:46–52. [PubMed] [Google Scholar]
- 34.De Clercq E. The next ten stories on antiviral drug discovery (part E): advents, advances, and adventures. Med Res Rev. 2009 doi: 10.1002/med.20179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milani M, Mastrangelo E, Bollati M, et al. Flaviviral methyltransferase/RNA interaction: structural basis for enzyme inhibition. Antiviral Res. 2009;83:28–34. doi: 10.1016/j.antiviral.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He R, Adonov A, Traykova-Adonova M, et al. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem Biophys Res Commun. 2004;320:1199–203. doi: 10.1016/j.bbrc.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cushman M, Wang PL, Chang SH, et al. Preparation and anti-HIV activities of aurintricarboxylic acid fractions and analogues: direct correlation of antiviral potency with molecular weight. J Med Chem. 1991;34:329–37. doi: 10.1021/jm00105a052. [DOI] [PubMed] [Google Scholar]


