Abstract
The FilmArray respiratory virus panel detects 15 viral agents in respiratory specimens using polymerase chain reaction. We performed FilmArray respiratory viral testing in a core laboratory at a regional children’s hospital that provides service 24 hours a day 7 days a week. The average and median turnaround time were 1.6 and 1.4 hours, respectively, in contrast to 7 and 6.5 hours documented 1 year previously at an on-site reference laboratory using a direct fluorescence assay (DFA) that detected 8 viral agents. During the study period, rhinovirus was detected in 20% and coronavirus in 6% of samples using FilmArray; these viruses would not have been detected with DFA. We followed 97 patients with influenza A or influenza B who received care at the emergency department (ED). Overall, 79 patients (81%) were given oseltamivir in a timely manner defined as receiving the drug in the ED, a prescription in the ED, or a prescription within 3 hours of ED discharge. Our results demonstrate that molecular technology can be successfully deployed in a nonspecialty, high-volume, multidisciplinary core laboratory.
Keywords: FilmArray, Respiratory virus, Polymerase chain reaction, PCR
Acute respiratory infection is a leading cause of outpatient visits and hospitalization in young children, especially in winter and spring.1 Most of the acute respiratory infections are caused by viral agents, with primary or secondary bacterial infections occurring less frequently. Without definitive diagnosis, patients with viral infection are more likely to receive unnecessary antibacterial agents.2 Therefore, laboratory tests providing accurate, timely determination of the infectious agents associated with viral respiratory disease are important. A broad array of tests is available to detect viral respiratory agents. Rapid antigen tests are available for individual respiratory viruses such as influenza A, influenza B, and respiratory syncytial virus (RSV). However, these tests have low sensitivity and specificity.3–6 Several molecular tests have been developed to detect viral RNA or DNA using the polymerase chain reaction (PCR) method.7–9 These tests show high sensitivity and specificity, but most of these molecular tests are technically challenging and time consuming and require experienced, specialized medical technologists. They are usually performed in large medical centers in highly specialized molecular or virology laboratories with limited hours of operation.
FilmArray (Idaho Technologies, Salt Lake City, UT) is a small desktop closed single-piece flow real-time PCR system. This end-to-end molecular system includes automation of nucleic acid extraction, an initial reverse transcription and multiplex PCR, followed by singleplex second-stage PCR reactions for detection of specific viral agents in a single-use cartridge. The respiratory virus panel performed on FilmArray is able to detect 15 viral agents from respiratory specimens.10 The test requires 5 minutes hands-on time and 65 minutes of instrumentation time. Comparison studies between FilmArray and other molecular tests for respiratory viral agents showed comparable results.11–15 As part of our preimplementation planning, we surveyed 10 other clinical laboratories in the United States. Only 1 laboratory performed the test in the general laboratory, with the rest performing the test in the microbiology, virology, or molecular laboratories. We reasoned that a general medical technologist with proper training would be able to perform the test.
To provide 24 hours per day, 7 days per week (24/7) service to our emergency department (ED) and urgent care center, we decided to perform the test in the core laboratory. Our core laboratory is a rapid response facility staffed by approximately 35 full-time equivalent (FTE) employees providing analysis encompassing automated chemistry, hematology, coagulation, urinalysis, blood gas analysis, and selected therapeutic drug monitoring. In addition, individual manual tests such as pregnancy tests, cardiac markers, drug abuse screening, sickle cell screening, occult blood, and heterophil antibodies are also performed in the core laboratory. We do not currently have a 24/7 microbiology laboratory, and during the night shift the core technologists perform culture setup and Gram staining. Our core specimen processing and testing are designed based on lean, single-piece flow principles without batching.16 Because the core laboratory was already involved in such work flow, the institution of FilmArray should be relatively simple.
In this study we describe the implementation of FilmArray in the core laboratory to provide rapid 24/7 service for respiratory virus diagnosis.
Materials and Methods
Patients and Samples
Pediatric patients up to 21 years of age who were cared for in our hospital or urgent care clinics and who underwent respiratory viral PCR testing between December 14, 2011, and April 19, 2012, were included in the study. For FilmArray assay, the midturbinate nasal swab was collected using a nylon flocked swab (Copan Diagnostics, Murrieta, CA) that was immediately placed in universal transport media (UTM) (Copan Diagnostics). Nursing staff collected all samples and were trained in the technique before the December initiation of the FilmArray assay. Specimens in UTM were tested as soon as they were received in the laboratory. Before the institution of the FilmArray panel, direct fluorescence assay (DFA) testing was used for respiratory specimens, with the nasal wash the preferred specimen type. More than 95% of the specimens submitted for DFA by the ED the previous year were nasal washes.
Virus Detection by FilmArray Technology
Viruses present in respiratory samples were tested using the FilmArray respiratory panel, which detects 15 viral agents including adenovirus, coronavirus HKU1, coronavirus NL63, human metapneumovirus, rhinovirus/enterovirus, influenza A, influenza A H1, influenza A H1 2009, influenza A H3, influenza B, parainfluenza 1, 2, 3, 4, and RSV. Samples were loaded into the testing cartridge under a ventilation hood. After hydrating the pouch with 1.0 mL of hydration solution, 300 μL of respiratory sample was diluted in 0.5 mL of sample buffer, of which 300 μL was injected into the sample pouch, which was then loaded onto the instrument. After entering the sample identification, the instrument was started and the result was available in approximately 1 hour. The testing pouch contains all the reagents for nucleic acid extraction, reverse transcription, first-step multiplex PCR amplification, and second-step real-time PCR amplification with single specific primer pair. For each viral agent, the second-step PCR was performed in triplicate. The software automatically analyzes the melting curve of the second-step PCR to report the results as positive or negative.
Laboratory Staff Training
This was the first molecular test implemented in the core laboratory, so microbiology staff assisted in the initial test validation and personnel training. Multiple continuing education sessions were given to core laboratory staff on all shifts regarding technical procedures as well as the principles underlying the technology. All core staff competency evaluations were fully completed before the test was made available clinically.
Direct Fluorescence Assay
DFA was performed on nasal wash samples as described.17 The procedure detected 8 viral agents including adenovirus, RSV, human metapneumovirus, influenza A, influenza B, and parainfluenza 1, 2, 3. However, it could not differentiate among parainfluenza 1, 2, and 3.
Collection of Turnaround Time and Viral Epidemiology Data
Turnaround time (TAT) data for FilmArray and DFA were collected using the PathNet laboratory clinical information system (Cerner, Kansas City, KS). The TAT was the interval from when the sample was logged into the laboratory information system (inside the laboratory) to when the result was verified. The information on administration of antiviral therapy was obtained from chart review in the clinical information system. The positive results of viral agents were tabulated daily.
The study was approved by the institutional review board of Seattle Children’s Hospital, Seattle, WA.
Results
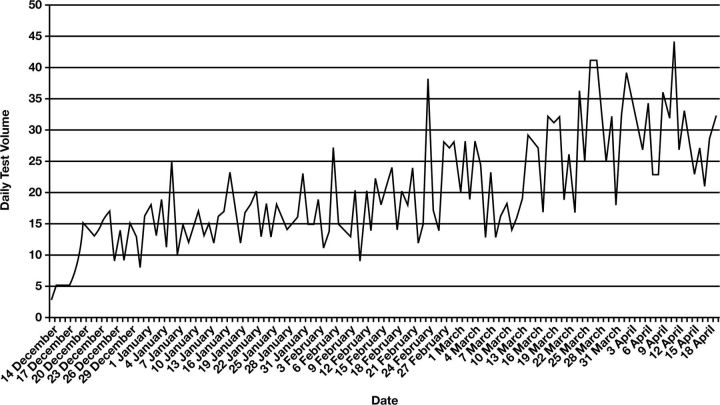
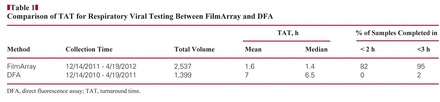
Respiratory virus testing with FilmArray was initiated on December 14, 2011. During the 4-month period ending April 19, 2012, a total of 2,537 specimens were tested. Seventy-one percent of the samples were from the ED or urgent care. The daily testing volume varied from 3 to 44 Figure 1. The average daily volume for the first 2 months and second 2 months were 15 and 25, respectively. Because the FilmArray instrument is designed for single-specimen throughput and requires 65 minutes of instrument time, we purchased 3 instruments that could run simultaneously 24 hours a day. The average and median TAT was 1.6 and 1.4 hours, respectively, with 82% completed in less than 2 hours and 95% in less than 3 hours Table 1. Before this molecular test was available, we sent respiratory viral testing to an on-site reference laboratory for DFA. DFA testing involved multiple handoff steps in our laboratory and specimen transport to the reference laboratory. DFA specimens were run in batches 3 or 4 times per day depending on the volume during respiratory virus season. The testing time was approximately 3 hours. During the same period 1 year previously, 1,399 DFAs for respiratory viruses were performed. The average and median TAT for DFA was 7 and 6.5 hours, respectively, with 2% completed in less than 3 hours (Table 1). The test volume of respiratory virus during the study period was almost doubled compared with the same period last year. During the study period, the instrument failed 4 times. The rate of failed testing, which included quality control failures, insufficient vacuum in the pouch, and pouch leaking, was 1.3%. The TAT was significantly longer when instruments or reagent pouches failed.
Figure 1.
Daily volume of FilmArray respiratory viral testing was tallied during the period from December 14, 2011, to April 19, 2012.
Table 1.
Comparison of TAT for Respiratory Viral Testing Between FilmArray and DFA
During the study period, 63% of all samples tested positive for viral agents. Rhinovirus/enterovirus and RSV were detected in 20% and 18% of total samples, respectively. The other viral agents detected were influenza B (10%), human metapneumovirus (7%), influenza A (6%–2% H12009, 4% H3), coronavirus (HKU1 and NL63) (6%), adenovirus (2%), and parainfluenza virus type 1–4 (2%). DFA does not detect rhinovirus and coronavirus. Thus, in an additional 660 (26%) of 2,537 specimens, FilmArray detected viruses that would not have been detected with DFA.
Current guidelines for treating patients with influenza A or B infection with oseltamivir indicate that the medication should be given within 48 hours of symptom onset to be most effective. In general, children with respiratory symptoms may not seek or be brought to medical attention immediately. A delay in laboratory testing may further prolong the interval between symptom onset and the administration of medication. For treatment benefits, we specifically followed patients (n = 97) who tested positive for influenza A or influenza B admitted to the ED during March 2012. The mean and median length of stay in the ED during the study period was 3.1 and 2.8 hours, respectively. In these 97 patients, respiratory viral results provided by FilmArray were available for 44 patients (45%) before they were discharged from the ED. In 50 patients (52%), positive results were called within 3 hours after discharge. The remaining 3 patients (3%) were notified of the results in more than 3 hours. Of the 97 patients, 22 patients (23%) were treated with oseltamivir in the ED, and 30 patients (31%) were given the prescription while they were in the ED. The prescription was called in to another 27 patients (28%) within 3 hours after discharge from the ED. The remaining 18 (19%) patients received no prescription for oseltamivir. Overall, 79 patients (81%) with influenza virus were given oseltamivir in a timely manner—defined as receiving the drug in the ED, a prescription in the ED, or a prescription within 3 hours of ED discharge. Among the 18 patients who did not receive prescriptions, 6 reported having symptoms for more than 48 hours before the ED visit, 4 patients showed improvement of symptoms according to parents, 4 had a follow-up appointment, 2 received physicians’ decisions not to treat, and no information was available for 2 patients.
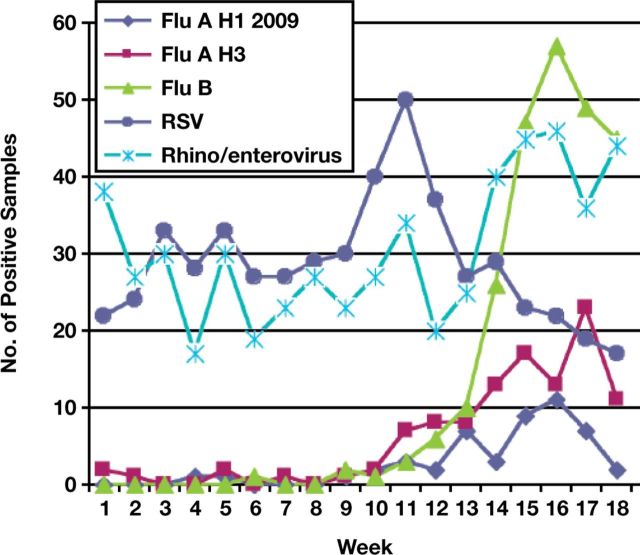
The daily tally of all positive results during the respiratory virus season clearly demonstrated the epidemiology of the multiple respiratory viral agents present in symptomatic children receiving care at our institution Figure 2 and contributed to both hospital-wide as well as community-wide public health information. Unusually, influenza A and B did not emerge until later in the season at the end of February and peaked in April. Before the emergence of influenza viruses, RSV was the major infectious agent, with a peak noted in mid-February. We also found that 1 patient who received live attenuated influenza vaccine less than 7 days before testing was positive for both influenza A and B; in retrospect, we would not recommend viral testing in patients recently immunized with live vaccine.
Figure 2.
Weekly respiratory samples detected as positive for influenza A H1 2009, influenza H3, influenza B, respiratory syncytial virus (RSV), or rhinovirus/enterovirus.
Discussion
Molecular testing for respiratory viruses has become widely available and rapidly deployed in diagnostic laboratories during the past few years. Therefore, the critical operational decision involves which system to choose to meet patient care needs while integrating smoothly into a laboratory’s infrastructure. In general, molecular tests for respiratory viruses have to be conducted in batches in specialized molecular, microbiology, or virology laboratories that rarely offer testing during evening and night shifts. The TAT for most molecular tests is also relatively long, ranging from 6 to 8 hours.
FilmArray is a recent US Food and Drug Administration (FDA)–approved PCR method for detecting 15 respiratory viral agents. Recently, Idaho Technology received FDA approval for 5 additional respiratory viral and bacterial agents including coronavirus 229E, coronavirus OC43, Bordetella pertussis, Chlamydophila pneumoniae, and Mycoplasma pneumoniae. It is simple and fully automated (sample in, result out), with a 65-minute TAT and a single-piece throughput platform. We implemented FilmArray in our core laboratory and ran the test 24/7 during this respiratory viral season. We started with 2 FilmArray instruments. As the volume of tests increased, a third instrument was added and then a fourth one was made available to us as a backup. The redundancy of our instrumentation allowed the laboratory to continue to perform testing if 1 or more instruments were not in operation.
Implementing FilmArray in the core laboratory significantly reduced the TAT of respiratory virus test reporting. In addition, the FilmArray respiratory viral panel includes more viral agents that were not included in previous testing using DFA. Although current treatment for respiratory viral infection is limited to influenza A and B, detection of other viral agents is valuable because clinical suspicion of viral respiratory tract infections can be confirmed, additional workup and therapy can be avoided, and clinicians and parents can be reassured.
In addition to early treatment of patients with influenza virus, early detection of the infectious agent is important to place the patients into appropriate isolation cohorts. Because of its rapid TAT, FilmArray testing for respiratory viruses was also used to place patients into cohorts effectively. In fact, 20% of the respiratory viral tests we performed were primarily for the purpose of patient isolation and forming cohorts. In 2 cases in which patients were admitted for urgent surgery, the rapid respiratory viral testing quickly ruled out influenza infection so that the surgery did not have to be performed under strict isolation procedures using masks and negative pressure in the operating room.
The purchase price of a FilmArray respiratory viral PCR panel was $109 per reagent pouch after discount (list price, $129). Adding 1% to 2% of pouch failure rate and additional quality control testing, the cost was approximately $115 per test. The test took approximately 5 minutes to perform, which was about the same amount of time required for packaging a sample to be sent to a reference laboratory. We did not add any FTE in the core laboratory with the addition of the test. The cost of sending a sample to the reference laboratory for respiratory viral testing by DFA was $98.26. The actual price of FilmArray was only slightly higher than that previous spent on respiratory viral testing. For the DFA testing performed in previous years, nasal wash was the preferred specimen type. The collection of a nasal wash sample resulted in the generation of aerosols, and therefore procedure rooms had to be closed for 30 minutes before the rooms could be used again. Replacing the nasal wash with a midturbinate swab increased efficiency by eliminating the 30-minute room closures. Because 71% of the respiratory viral testing samples were collected from the ED, we potentially saved 900 hours of ED room for the 2,537 respiratory viral tests performed. The cost savings could not be accurately calculated. However, in 1 report it was found that a 1-hour reduction in ED boarding time would result in $9,693 to $13,298 of additional daily revenue.18 In this report the calculation included patients who left without being seen, which represented 7% of all ED visits. The rate of patients who left without being seen at our institution is 0.6%, so opportunity cost savings in our institution are probably lower.
The major drawbacks of FilmArray are single-sample throughput, the relatively low sensitivity of detecting adenovirus,11 and the inability to separate rhinovirus and enterovirus. Furthermore, we were unable to quantify viral load, which may also be important in some patients. In addition, the high detection rate of rhinovirus cannot be necessarily interpreted as high rates of acute rhinovirus infection because prolonged rates of rhinovirus shedding have been reported. Finally, because there is no interface with our computerized data entry system currently, we required 2 technologists to verify the reported results for clerical error reduction. With this peer review verification reporting process we had only 1 data entry error, which was corrected without affecting patient care. The error rate related to data entry was less than 0.04%.
In summary, the implementation of the FilmArray respiratory panel in our core laboratory significantly decreased the time required to detect and report respiratory viruses. Patients with influenza A and B were treated rapidly and appropriately, which had not been possible using DFA. Detection of other viral agents assisted physicians in the differential diagnosis of respiratory syndromes and isolation of patients admitted to the hospital. Overall, we implemented a molecular-based efficient diagnostic test in our core laboratory for the first time, marking a new era in pediatric clinical laboratory medicine.
Footnotes
This study was funded by Seattle Children’s Hospital. Dr Jerome has served on a scientific advisory board for Idaho Technologies.
Acknowledgments: We are grateful to Doris Cruz, MT, Suchinda Chantarasuksom, MT, Jenny Stapp, MT, Matthew Elamparo, MT, and Yalda Honari for data retrieval.
References
- 1. Chapin K. Multiplex PCR for detection of respiratory viruses: can the laboratory performing a respiratory viral panel (RVP) assay trigger better patient care and clinical outcomes? Clin Biochem. 2011;44:496–497. [DOI] [PubMed] [Google Scholar]
- 2. Byington CL, Castillo H, Gerber K, et al. The effect of rapid respiratory viral diagnostic testing on antibiotic use in a children’s hospital. Arch Pediatr Adolesc Med. 2002;156:1230–1234. [DOI] [PubMed] [Google Scholar]
- 3. Miernyk K, Bulkow L, DeByle C, et al. Performance of a rapid antigen test (Binax NOW® RSV) for diagnosis of respiratory syncytial virus compared with real-time polymerase chain reaction in a pediatric population. J Clin Virol. 2011;50:240–243. [DOI] [PubMed] [Google Scholar]
- 4. Ganzenmueller T, Kluba J, Hilfrich B, et al. Comparison of the performance of direct fluorescent antibody staining, a point-of-care rapid antigen test and virus isolation with that of RT-PCR for the detection of novel 2009 influenza A (H1N1) virus in respiratory specimens. J Med Microbiol. 2010;59:713–717. [DOI] [PubMed] [Google Scholar]
- 5. Takahashi H, Otsuka Y, Patterson BK. Diagnostic tests for influenza and other respiratory viruses: determining performance specifications based on clinical setting. J Infect Chemother. 2010;16:155–161. [DOI] [PubMed] [Google Scholar]
- 6. Hurt AC, Baas C, Deng YM, et al. Performance of influenza rapid point-of-care tests in the detection of swine lineage A(H1N1) influenza viruses. Influenza Other Respir Viruses. 2009;3:171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nolte FS, Marshall DJ, Rasberry C, et al. MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J Clin Microbiol. 2007;45:2779–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henrickson KJ, Hall CB. Diagnostic assays for respiratory syncytial virus disease. Pediatr Infect Dis J. 2007;26(11 suppl):S36–S40. [DOI] [PubMed] [Google Scholar]
- 9. Pabbaraju K, Wong S, Tokaryk KL, et al. Comparison of the Luminex xTAG respiratory viral panel with xTAG respiratory viral panel fast for diagnosis of respiratory virus infections. J Clin Microbiol. 2011;49:1738–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poritz MA, Blaschke AJ, Byington CL, et al. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6:e26047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loeffelholz MJ, Pong DL, Pyles RB, et al. Comparison of the FilmArray Respiratory Panel and Prodesse real-time PCR assays for detection of respiratory pathogens. J Clin Microbiol. 2011;49:4083–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rand KH, Rampersaud H, Houck HJ. Comparison of two multiplex methods for detection of respiratory viruses: FilmArray RP and xTAG RVP. J Clin Microbiol. 2011;49:2449–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Babady NE, Mead P, Stiles J, et al. Comparison of the Luminex xTAG RVP FAST and the Idaho Technology FilmArray RP assays for the detection of respiratory viruses in pediatric patients at a cancer hospital. J Clin Microbiol. 2012;50:2282–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hayden RT, Gu Z, Rodriguez A, et al. Comparison of two broadly multiplexed PCR systems for viral detection in clinical respiratory tract specimens from immunocompromised children. J Clin Virol. 2012;53:308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pierce VM, Elkan M, Leet M, et al. Comparison of the Idaho Technology FilmArray system to real-time PCR for detection of respiratory pathogens in children. J Clin Microbiol. 2012;50:364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rutledge J, Xu M, Simpson J. Application of the Toyota Production System improves core laboratory operations. Am J Clin Pathol. 2010;133:24–31. [DOI] [PubMed] [Google Scholar]
- 17. Kuypers J, Wright N, Ferrenberg J, et al. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J Clin Microbiol. 2006;44:2382–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pines JM, Batt RJ, Hilton JA, et al. The financial consequences of lost demand and reducing boarding in hospital emergency departments. Ann Emerg Med. 2011;58:331–340. [DOI] [PubMed] [Google Scholar]