Abstract
Type I and type III interferons are important anti-viral cytokines that are massively induced during viral infection. This dynamic process is regulated by many executors and regulators for efficient eradication of invading viruses and protection from harmful, excessive responses. An array of innate sensors recognizes virus-derived nucleic acids to activate their downstream signaling to evoke cytokine responses including interferons. In particular, a cytoplasmic RNA sensor RIG-I (retinoic acid-inducible gene I) is involved in the detection of multiple types of not only RNA viruses but also DNA viruses. Accumulating findings have revealed that activation of nucleic acid sensors and the related signaling mediators is regulated on the basis of post-translational modification such as ubiquitination, phosphorylation and ADP-ribosylation. In addition, long non-coding RNAs (lncRNAs) have been implicated as a new class of regulators in innate signaling. A comprehensive understanding of the regulatory mechanisms of innate sensor activation and its signaling in host–virus interaction will provide a better therapeutic strategy to efficiently control viral infection and maintain immune homeostasis.
Keywords: interferon, pattern-recognition receptors, RIG-I, signal transduction, virus infection
How innate cells detect viral DNA and RNA to trigger interferon production
Introduction
In 1954, Drs Yasuichi Nagano and Yasuhiko Kojima reported the identification of a virus-inhibiting factor during their research to produce a more efficient vaccine for smallpox (1). It is thought that this factor must be ‘interferon’, which was found as a result of a study about interference against viruses by Drs Alick Isaacs and Jean Lindenmann in 1957 (2). Expression of the genes for type I and type III interferons is massively induced in response to viral infection, and the produced interferons evoke anti-viral activities through their binding to specific receptor complexes to induce multiple interferon-stimulated genes. With the remarkable progress in the studies of innate immunity, much more attention was paid to interferons. Molecular sensor-mediated recognition of invading microbes is the first step of host defense against infection to activate the innate immune system. During viral infection, virus-associated molecular patterns—viral nucleic acids (RNA and DNA)—are mainly targeted by pattern-recognition receptors (PRRs), including transmembrane-type sensors, such as Toll-like receptor 3 (TLR3) and TLR9, and cytoplasmic sensors, such as RIG-I (retinoic acid-inducible gene I; also called DDX58) and cGAS (cyclic GMP–AMP synthetase). In most cases, these viral sensors trigger the gene expression of type I and III interferons to induce anti-viral activities.
Accumulating reports revealed that there are multiple mechanisms for the regulation of innate sensor-mediated immune signaling (3, 4). On the other hand, virus proteins are produced to counteract such innate signaling. The balance of power between host and microbe determines the outcome of pathogenicity. In this review, we summarize recent findings on nucleic acid sensor-mediated innate immune signaling and its regulatory mechanisms during virus infection.
Innate sensors for the detection of viral infection
In the innate immune system, viral infection is generally recognized through the detection of virus-derived nucleic acids as pathogen-associated molecular patterns (PAMPs). Recent great progress in studies of innate immunity has revealed a diversity of DNA and RNA sensors involved in host detection of viruses, which leads to the activation of innate immune responses (Fig. 1). In particular, type I and type III interferons are produced to induce anti-viral activities. In this section, we focus particularly on the recent advances in our understanding of the regulation of nucleic acid sensor-mediated innate immune signaling.
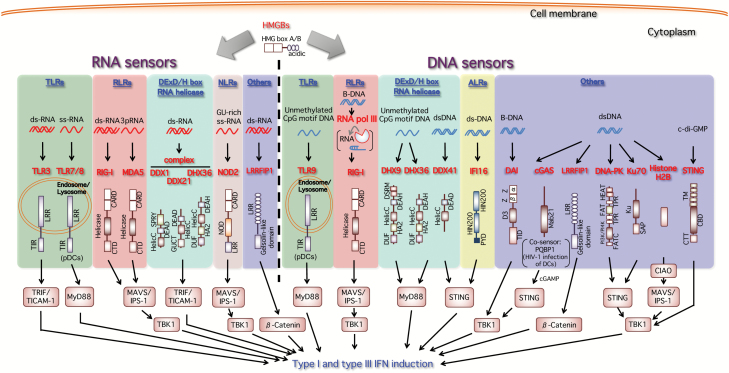
Fig. 1.
Nucleic acid sensors inducing type I and type III interferons. During viral infection, virus-derived nucleic acids (RNA and DNA) are mainly targeted by host nucleic acid sensors triggering the gene expression of type I and III interferons to induce anti-viral activities. As RNA sensors, TLR3 and TLR7/TLR8 sense viral RNAs on the endosomal or lysosomal membrane. RIG-I and MDA5 are ubiquitously expressed and recognize viral RNAs in the cytoplasmic space. Other RNA helicases DDX1, DDX21 and DHX36 form an RNA sensor complex in mDCs. The Nod-like receptor (NLR) NOD2 also detects the RSV-derived ssRNA genome to activate the MAVS/IPS-1 pathway. In the cases of DNA sensing, TLR9 detects DNA containing the unmethylated CpG motif. DAI (DLM-1/ZBP1) interacts with dsDNA and activates TBK1 and IRF-3 as a cytoplasmic DNA sensor. Cytoplasmic AT-rich dsDNA (B-DNA) is transcribed by host RNA polymerase III (RNA pol III) into 5′-triphosphate RNA (3pRNA), which becomes a RIG-I ligand. DHX36 and DHX9 intracellularly sense CpG class A and B, respectively, in mDCs. DNA-stimulated DDX41 and the ALR IFI16 interact with their adaptor protein STING. STING itself also serves as a sensor of CDNs such as bacterium-derived c-di-GMP as well as host-derived cGAMP generated by cGAS in response to microbe- and host-derived cytosolic DNAs. cGAS is a critical cytoplasmic sensor for the detection of both microbial and host DNAs. The cGAS–cGAMP–STING pathway is now recognized as a major intracellular dsDNA sensing pathway. DNA-PK, Ku70 and extrachromosomal histone H2B are also reported to function as cytosolic DNA sensors. As for Ku70, only type III interferon is induced. LRRFIP1 functions possibly as a cytosolic nucleic acid sensor that dually detects both RNA and DNA. HMGBs also bind to both double-stranded and single-stranded immunogenic RNAs and function as universal sensors for nucleic acid-mediated innate immune responses during viral infection.
RNA sensors inducing type I and type III interferon gene expression
RNA sensors, including TLR3, TLR7, TLR8 and RIG-I-like receptors (RLRs) such as RIG-I and MDA5 (melanoma differentiation-associated gene 5), play a central role as viral PRRs to initiate innate immune responses against viral infection. TLR3 and TLR7/TLR8 reside on the endosomal or lysosomal membrane and sense viral RNAs. TLR3 detects double-stranded RNA (dsRNA), whereas TLR7 and TLR8 recognize degradation products of single-stranded RNA (ssRNA) such as mononucleotides [guanine (TLR7) and uridine (TLR8)] and uridine-rich short oligonucleotides (5–7).
TLRs are expressed preferentially in immune cells including macrophages and dendritic cells. TLR3 is known to be also expressed in neuronal tissues, and in certain fibroblasts TLR3 is involved in the detection of DNA virus such as type 1 herpes simplex virus (HSV-1) to activate anti-viral responses (8, 9), although its ligand remains to be defined. TLR7/TLR8 are expressed in plasmacytoid dendritic cells (pDCs) to activate the type I interferon response to RNA virus infection [influenza A virus (IAV), vesicular stomatitis virus (VSV) etc.], in a manner dependent on MyD88 and interferon regulatory factor-7 (IRF-7) (10). Unlike TLR7/TLR8, TLR3 interacts with TRIF/TICAM-1, but not MyD88 as its adaptor protein, and activates not only IRF-7 but also IRF-3 (11).
RIG-I and MDA5 are ubiquitously expressed and recognize viral RNAs in the cytoplasmic space (12, 13). In particular, RIG-I is a key cytoplasmic PRR for the detection of RNA viruses such as influenza virus, hepatitis C virus (HCV) and measles virus (MeV) that are widely known to be pathogenic in humans (14). RNA carrying a 5′-triphosphate modification (3pRNA) is an essential determinant for RIG-I recognition (15). Ligand binding activates the ATPase activity to change the conformation of RIG-I. And then oligomerized RIG-I interacts with the mitochondria-associated adaptor protein, MAVS (also known as VISA, IPS-1 or Cardif), through its CARD domains, which is facilitated by both covalent and non-covalent interactions with K63-linked ubiquitin (Ub) chains. This, in turn, culminates in activation of transcription factors such as IRF-3 and NF-κB, leading to the activation of the downstream gene induction programs such as type I interferon induction (16).
RIG-I was also shown to function as an innate sensor to induce anti-viral responses against hepatitis B virus (HBV) infection in human hepatocytes (17). The 5′-epsilon region of HBV pregenomic RNA (pgRNA), which is known to take a unique hairpin structure, is a target for the RIG-I-mediated sensing. In this case, type III interferons were predominantly induced in vitro and in vivo during infection with HBV genotypes A, B and C, in a RIG-I-dependent manner. In general, RIG-I signaling leads to both type I and type III interferon gene induction, but how this predominant induction of type III interferons during HBV infection occurs remains to be clarified. Since such type III interferon dominancy was also observed during HCV infection and upon treatment with a RIG-I ligand 3pRNA, human hepatocytes seem to have their own intrinsic mechanism to preferentially produce type III interferons. In relation to this, there is an interesting report showing that, in human nasal epithelial cells, type III interferons but not type I interferons are predominantly induced in response to several RNA viruses such as respiratory syncytial virus (RSV), MeV and mumps virus (MuV) (18).
Type III interferon is also reported as the predominant interferon induced by IAV infection in human alveolar type II epithelial cells and in mouse bronchoalveolar lavage (BAL) samples (19–21). In this respect, like these epithelial cells in the mucosal barrier, human hepatocytes might also have some similar characteristics to dominantly produce type III interferons for locally inducing anti-viral defense. The next interesting issue is to address the molecular mechanism for how the predominant expression of type III interferons is regulated.
It was also demonstrated that RIG-I plays a role as an anti-viral factor that directly inhibits HBV replication, in an interferon-independent manner (17). Mechanistically, RIG-I competitively inhibits the binding of HBV P-protein to the 5′-epsilon region of pgRNA, which is essential for the initiation of HBV replication. A similar role of RIG-I as a direct anti-viral restriction factor was also shown in the case of IAV infection (22). Accordingly, RIG-I dually functions as an innate immune sensor inducing interferon expression and as a direct anti-viral restriction factor during infection with certain viruses.
In relation to this, there is another interesting paper, which demonstrated that RIG-I and MDA5 can displace dsRNA-binding viral proteins such as non-structural protein 1 (NS1) of IAV and E3L of vaccinia virus in an ATPase-dependent manner (23). In fact, RIG-I/MDA5 are known to displace NS1/E3L from dsRNA to promote the activity of protein kinase R (PKR), which is suppressed by these viral proteins through the sequestration of dsRNA. In addition to the conventional role of RLRs as an RNA sensor inducing innate anti-viral signaling, these findings provide a novel aspect of RLR-mediated anti-viral activity, whereby RLRs act as direct anti-viral effectors against viral infection by counteracting a dsRNA-binding viral protein-mediated evasion mechanism (see later).
Other DExD/H box RNA helicases—DDX1, DDX21 and DHX36—form a virus sensor complex, together with TRIF/TICAM-1, leading to type I interferon production in myeloid dendritic cells (mDCs). DDX1 binds to poly I:C, whereas DDX21 and DHX36 interact with TRIF/TICAM-1 in the cytoplasm (24). Nucleotide-binding oligomerization domain 2 (NOD2), which was known to respond to bacterial PAMPs, can also bind to synthetic GU-rich ssRNA or the RSV ssRNA genome to activate IRF-3 for the production of IFN-β (a type I interferon), in a MAVS-dependent manner (25).
DNA sensors inducing type I and type III interferon gene expression
TLR9 was first identified as a DNA sensor to detect the unmethylated CpG motif, which is most selectively found in microbial genomes (26). Like other TLR9-subfamily members such as TLR7 and TLR8, TLR9 resides in the endosome. In humans, TLR9 is selectively expressed on pDCs as well as B cells (27). Following TLR9-mediated recognition of the unmethylated CpG motif upon DNA virus infection, pDCs efficiently produce type I interferons in a manner dependent on the MyD88–TRAF6–IRF-7 axis, which is similar to TLR7 and TLR8 signaling (28).
DAI (DLM-1/ZBP1) was the first identified cytoplasmic DNA sensor (29). Its interaction with dsDNA activates TANK-binding kinase 1 (TBK1) and IRF-3 for type I interferon production in mouse fibroblastic L929 cells and an endothelial cell line SVEC4-10 (30, 31). This was supported by subsequent studies, showing that DAI has a role in type I interferon induction in human foreskin fibroblasts upon cytomegalovirus (CMV) infection (32, 33), although DAI does not seem to be essentially involved in DNA sensing in vivo (34). Subsequently, it was reported that the cytoplasmic RNA sensor RIG-I was involved in the induction of the type I interferon response during DNA virus infection. AT-rich dsDNA is transcribed by host RNA polymerase III (RNA pol III) into 5′-triphosphate RNA, which becomes a RIG-I ligand to initiate the RIG-I pathway (35, 36). In this regard, RNA pol III is an intracellular sensor of viral DNA.
Similarly, Epstein–Barr virus (EBV)-encoded RNAs (EBERs) are also transcribed by RNA pol III (37) and sensed by RIG-I (38). RIG-I is also reported to have a role as a viral sensor for the suppression of Kaposi’s sarcoma-associated herpesvirus (KSHV) replication and production. It is speculated that some KSHV-derived non-coding RNAs including viral microRNAs might be ligands for RIG-I (39). In addition, RIG-I is known to be involved in the recognition of HSV-2 infection to induce type I interferon expression, which is dependent on RNase L (40). It is suggested that small self-RNAs generated by RNase L possibly trigger RIG-I activation (41).
Other RNA helicases—DHX36 and DHX9—intracellularly sense CpG class A and B, respectively, leading to MyD88-dependent activation of the type I interferon response. They were shown to be critical for anti-viral immune responses in viral DNA-stimulated human pDCs (42). In addition, DDX41 was reported as an intracellular DNA sensor in mDCs (43). DNA-stimulated DDX41 as well as IFI16, one of the PYHIN family members, interacts with their adaptor protein, stimulator of interferon gene (STING, also known as TMEM173, MITA or ERIS), to activate TBK1 and IRF-3 for type I interferon gene induction (43, 44).
cGAS is now recognized as a critical DNA sensor during viral infection. cGAS senses virus-derived dsDNAs in a sequence-independent manner, which then triggers the catalysis of the formation of a di-nucleotide, cyclic GMP–AMP (cGAMP) (45). Subsequently, cGAMP functions as an endogenous second messenger, and binds to STING protein to induce innate immune responses such as interferon responses (46). In addition, cGAS senses not only pathogen-derived DNA but also aberrantly accumulated self-DNA in the cytosol, and also has important roles in the pathogenesis of autoimmune diseases, inflammation, cellular senescence and cancer (47–49). On the other hand, IFI16 was also identified as an intracellular DNA sensor that mediates the induction of IFN-β (44). IFI16 directly interacts with viral DNAs and activates its downstream signaling via STING (44). However, a recent study showed AIM2-like receptors (ALRs) including IFI16 are dispensable for the DNA-sensing pathway and demonstrated that cGAS is the primary DNA sensor that drives the interferon response to viral DNAs (50). Currently, cGAS–cGAMP–STING pathway is recognized as a major intracellular dsDNA sensing pathway.
STING not only functions as an adaptor protein of innate signaling downstream of IFI16 and DDX41 as mentioned above, but also serves as a sensor of cyclic dinucleotides (CDNs) such as cyclic di-GMP (c-di-GMP), which is known as a bacterial second messenger molecule (51, 52). Thus, the STING- and TBK1-dependent pathway is now known as the major signaling pathway for the induction of cytosolic DNA-induced type I interferons and other inflammatory cytokines and chemokines in microbial infection, inflammation and cancer (53).
In the case of human immunodeficiency virus 1 (HIV-1) infection of DCs, PQBP1 directly binds to reverse-transcribed HIV-1 DNA and interacts with cGAS to initiate an IRF-3-dependent innate response, suggesting a role of PQBP1 as a co-sensor of a HIV-1 infection (54). In contrast, a very recent paper shows that PQBP1 inhibits IFI16/cGAS-induced signaling in response to cytosolic DNA or DNA damage (55).
LRRFIP1, which was originally identified as a binding partner of FLI, which is the human homolog of Drosophila flightless I (Fli-I), one of the gelsolin superfamily members (56), is reported to function possibly as a cytosolic nucleic acid sensor that dually detects both RNA and DNA, inducing another coactivator pathway for IRF-3-mediated interferon production upon infection with VSV and Listeria monocytogenes (57). It is suggested that after binding to dsRNA or dsDNA, LRRFIP1 interacts with β-catenin and induces the activation of β-catenin, which enhances IFN-β expression via its binding to IRF-3. This pathway is MAVS-independent, although its dependency on STING awaits further investigation.
DNA-dependent protein kinase (DNA-PK), which is involved in the DNA repair process upon DNA damage, is known to act as an innate DNA sensor, binding cytoplasmic DNA to trigger a type I interferon response via the activation of the STING–TBK1–IRF-3 pathway (58). Another component involved in DNA repair, Ku70, is known to be a possible DNA sensor inducing the activation of IFN-λ1 (a type III interferon) (59). On the other hand, the extrachromosomal histone H2B is reported to function as a cytosolic sensor to detect dsDNA. This protein interacts with MAVS/IPS-1 through a unique linker, COOH-terminal importin 9-related adaptor organizing histone H2B and MAVS/IPS-1 (CIAO) to activate downstream type I interferon production (60).
High-mobility-group box (HMGB) proteins are known as non-histone chromosomal proteins that participate in many biological processes in the nucleus and also were shown to be released from cells infected with various types of viruses, including dengue virus, HCV and HIV. In addition to their DNA binding activity, HMGBs can also bind to both double-stranded and single-stranded immunogenic RNAs and function as universal sensors for nucleic acid-mediated innate immune responses during viral infection (61). HMGB1 secreted from virus-infected cells triggers anti-viral responses and blocks HCV infection (62). On the contrary, the effect of intracellular HMGB1 in HCV infection seems to be opposite. HMGB1 protein is translocated from the nucleus to the cytoplasm to interact with HCV positive-strand RNA during HCV infection, which then facilitates HCV RNA replication (63). This suggests another role of HMGB1 as a proviral factor to promote HCV infection.
Regulation of RIG-I activation and its innate signaling
During the last decade, there have been accumulating papers showing the identification of both positive and negative regulators that control innate immune signaling at various steps through different mechanisms. These regulatory mechanisms are known to be important to prevent excessive inflammation in order to maintain the balance of innate immune responses. Breakdown of the regulatory system leads to interferogenic and pro-inflammatory syndromes such as Aicardi–Goutieres syndrome (AGS) and systemic lupus erythematosus (SLE) (64–67). In this section, we focus on the regulation of the RIG-I-mediated pathway (Fig. 2). Because of the important role of the RIG-I pathway in anti-viral innate responses to various types of viruses, much attention has recently been paid to studies of the regulatory mechanisms of RIG-I signaling, which is important for both efficient control of viral infection and maintenance of immune homeostasis.
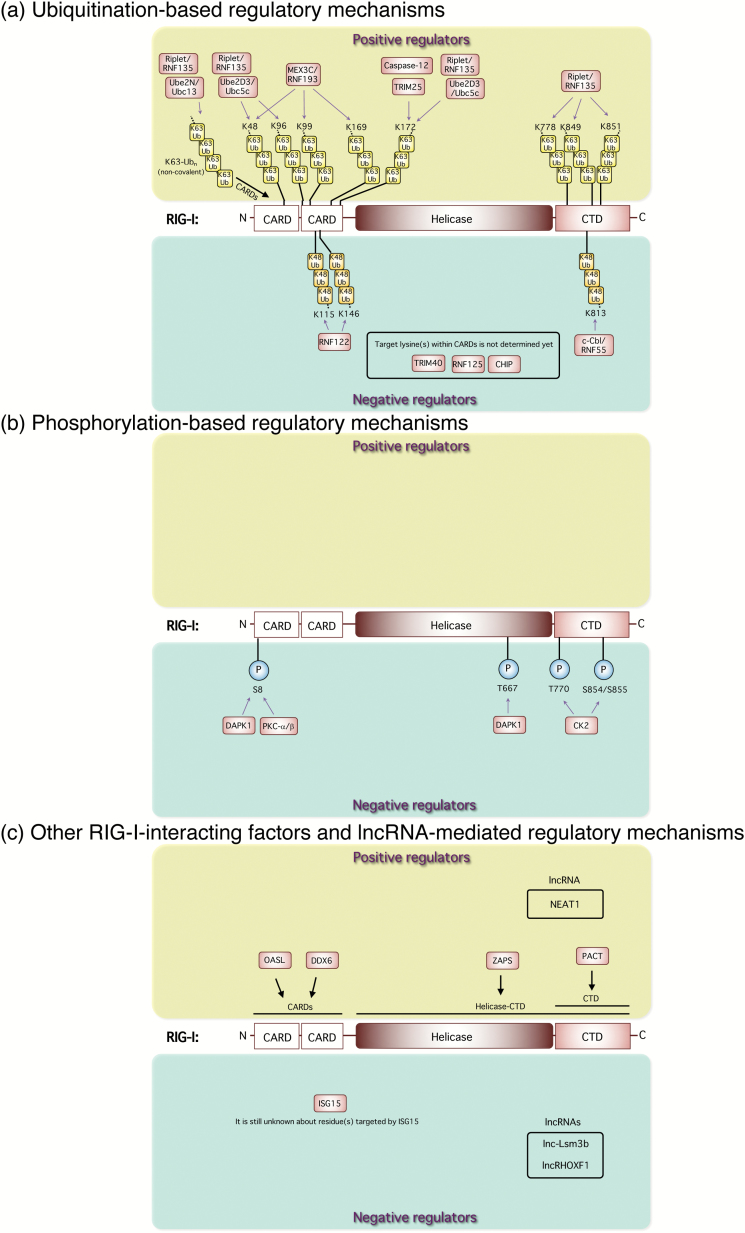
Fig. 2.
Regulatory mechanisms of RIG-I activation and its signaling. (a) Ubiquitination-based regulatory mechanisms. The RING-finger E3 Ub ligase TRIM25 positively mediates the RIG-I pathway by its K63-linked ubiquitination of the N-terminal CARDs of RIG-I. Riplet/RNF135 promotes K63-linked polyubiquitination of the C-terminal region of RIG-I. MEX3C/RNF193 mediates K63-linked polyubiquitination of the CARDs of RIG-I for its activation. Caspase-12 is physically associated with RIG-I and up-regulates TRIM25-mediated ubiquitination of RIG-I, leading to the enhancement of type I interferon responses. Non-covalent binding of K63-Ubn to the RIG-I CARDs induces its tetramer formation for downstream signal activation. Ube2D3/Ubc5c and Ube2N/Ubc13 were also identified as essential Ub-conjugating enzymes (E2) for RIG-I activation. The Ube2D3/Ubc5c–Riplet pair promotes covalent conjugation of polyubiquitin chains to RIG-I, whereas Ube2N/Ubc13 preferentially facilitates production of unanchored polyubiquitin chains. On the contrary, K48-linked ubiquitination of RIG-I by other E3 Ub ligases, such as c-Cbl/RNF55, RNF122, RNF125, CHIP and TRIM40, negatively regulates anti-viral innate responses. (b) Phosphorylation-based regulatory mechanisms. DAPK1 interacts with RIG-I and directly phosphorylates its threonine (T) 667 to impair its RNA ligand binding and abolishes anti-viral signaling. DAPK1 and PKC-α/β phosphorylate the N-terminal serine(S) at position 8 of RIG-I to suppress TRIM25-mediated RIG-I ubiquitination. CK2 is responsible for this regulation, and constitutively phosphorylates T770 and S854 to S855 residues in the RIG-I CTD, which keeps a closed, inactivated conformation. (c) Other RIG-I-interacting factors and lncRNA-mediated regulatory mechanisms. OASL interacts with RIG-I and mimics polyubiquitin through the UBL domains to enhance RIG-I-mediated anti-viral signaling. ISG15-mediated ISGylation of RIG-I reduces the cellular level of active RIG-I and suppresses its downstream anti-viral innate responses. ZAPS, which is a shorter form of PARP-13, functions as a potent stimulator of RIG-I-mediated anti-viral signaling through its interaction with RIG-I to facilitate RIG-I oligomerization and its ATPase activity for the activation of both IRF-3 and NF-κB. PACT directly interacts with the CTD of RIG-I to potentiate the activation of RIG-I. DDX6 interacts with RIG-I and augments type I interferon gene induction.
Ubiquitination-based regulatory mechanisms
RIG-I signaling is critically regulated by ubiquitination of RIG-I itself (Fig. 2a) and its adaptor MAVS. The RING-finger E3 Ub ligase, TRIM25, is an important regulator to positively mediate the RIG-I pathway by its K63-linked ubiquitination of the N-terminal caspase recruitment domains (CARDs) of RIG-I, which is essential for MAVS binding (68). Another E3 Ub-protein ligase, Riplet/RNF135, promotes K63-linked polyubiquitination of the C-terminal region of RIG-I in a TRIM25-independent manner (69). TRIM25 is required for RIG-I oligomerization and interaction with the adaptor MAVS, whereas Riplet/RNF135 is required for the release of RIG-I autorepression of its N-terminal CARDs, which enables RIG-I to interact with TRIM25 (70). Another RNA-binding E3 Ub ligase, MEX3C/RNF193, colocalizes with RIG-I in the stress granules (SGs) of virally infected cells, and mediates K63-linked polyubiquitination of the CARDs of RIG-I for its activation (71).
Some caspases have been shown to be important regulators in anti-viral innate immunity through their effects on innate signaling for type I interferon induction (72). Caspase-12, which is dominantly induced in the central nervous system after West Nile virus (WNV), is physically associated with RIG-I and up-regulates TRIM25-mediated ubiquitination of RIG-I, leading to the enhancement of type I interferon responses (73). Caspase-12-deficient mice were shown to be highly predisposed to WNV encephalitis with increased levels of viral titers and reduced levels of type I interferons.
On the other hand, non-covalent binding of K63-Ubn to the RIG-I CARDs was also shown to induce its tetramer formation, which is critical for downstream signal activation (16, 74). Ube2D3/Ubc5c and Ube2N/Ubc13 were also identified as essential Ub-conjugating enzymes (E2) for RIG-I activation (75). The Ube2D3/Ubc5c–Riplet pair promotes covalent conjugation of polyubiquitin chains to RIG-I, whereas Ube2N/Ubc13 preferentially facilitates production of unanchored polyubiquitin chains. Thus, RIG-I activation is regulated by both covalent and non-covalent interactions with K63-linked Ub chains. Intriguingly, the role of this K63-linked polyubiquitin for RIG-I activation can be substituted with interferon-inducible oligoadenylate synthetases-like (OASL) protein, which lacks 2′–5′ oligoadenylate synthetase activity but contains two tandem Ub-like domains (UBL) in the C terminus. OASL interacts with RIG-I and mimics polyubiquitin through the UBL, thus enhancing RIG-I-mediated anti-viral signaling (76).
On the contrary, K48-linked ubiquitination of RIG-I by other E3 Ub ligases, such as casitas B-lineage lymphoma proto-oncogene (c-Cbl/RNF55), RNF122, RNF125, carboxyl terminus of HSC70-interacting protein (CHIP) and TRIM40 negatively regulates anti-viral innate responses (77–81). In particular, TRIM40 binds to RIG-I and MDA5, and promotes their K27- and K48-linked polyubiquitination, leading to their proteasomal degradation, whereas TRIM40 is down-regulated during RNA virus infection (81). In addition, interferon-induced ISG15, a Ub-like protein, was shown to be conjugated to RIG-I. The ISGylation of RIG-I reduces the cellular level of active RIG-I and suppresses its downstream anti-viral innate responses (82). This is a feedback mechanism mediated by the induced interferons.
Phosphorylation-based regulatory mechanisms
Phosphorylation is also an important post-translational modification for the modulation of RIG-I activity (Fig. 2b). Death-associated protein kinase 1 (DAPK1) is activated by RIG-I signaling and it strongly interacts with RIG-I and directly phosphorylates specific serine/threonine (S/T) residues on RIG-I (83). In particular, phosphorylation of RIG-I threonine T667 impairs its RNA ligand binding and abolishes anti-viral signaling. DAPK1 also phosphorylates the N-terminal serine at position 8 (S8) of RIG-I, which is also reported to undergo phosphorylation by PKC-α/β to suppress TRIM25-mediated RIG-I ubiquitination, thereby negatively regulating RIG-I activity (84). This serine residue is also known to be dephosphorylated by PP1α and PP1γ, which leads to RIG-I activation (85).
RIG-I activity is also regulated by of both phosphorylation and dephosphorylation in the C-terminal domain (CTD). Casein kinase II (CK2) is responsible for this regulation, and constitutively phosphorylates T770 and S854 to S855 residues in the RIG-I CTD, which keeps the interaction between the CTD and the CARDs to form a closed, inactivated conformation (86). On the other hand, these residues are dephosphorylated by an unidentified phosphatase upon RNA virus infection to enable the oligomerization of RIG-I (86). Thus, RIG-I activity is tightly modulated through these modes of protein modification, including ubiquitination, ISGylation and phosphorylation, which may also be reversibly regulated to allow the initiation or termination of RIG-I-mediated signaling in a timely manner. Other post-translational modifications such as methylation, acetylation, SUMOylation and succinylation have also been shown to be implicated in the regulation of innate immune responses (87).
ADP-ribosylation-based regulatory mechanisms
Mono- and poly-ADP-ribosylation is another mode of post-translational protein modification, which is mediated by the poly(ADP-ribose) polymerase (PARP) superfamily (88, 89). Among them, PARP-7, also known as TCDD (2,3,7,8-tetrachlorodibenzo-p-dioxin)-inducible poly(ADP-ribose) polymerase (TIPARP), was shown to be an important mediator to negatively regulate type I interferon production activated by RIG-I and other nucleic acid sensors in the innate immune system (90). Tiparp-deficient cells showed significantly increased levels of IFN-β production following IAV infection, which was accompanied with a marked suppression of viral replication. Further investigation revealed that TIPARP could ADP-ribosylate the TBK1 protein, a pivotal kinase for the interferon pathway, thereby suppressing the TBK1 kinase activity.
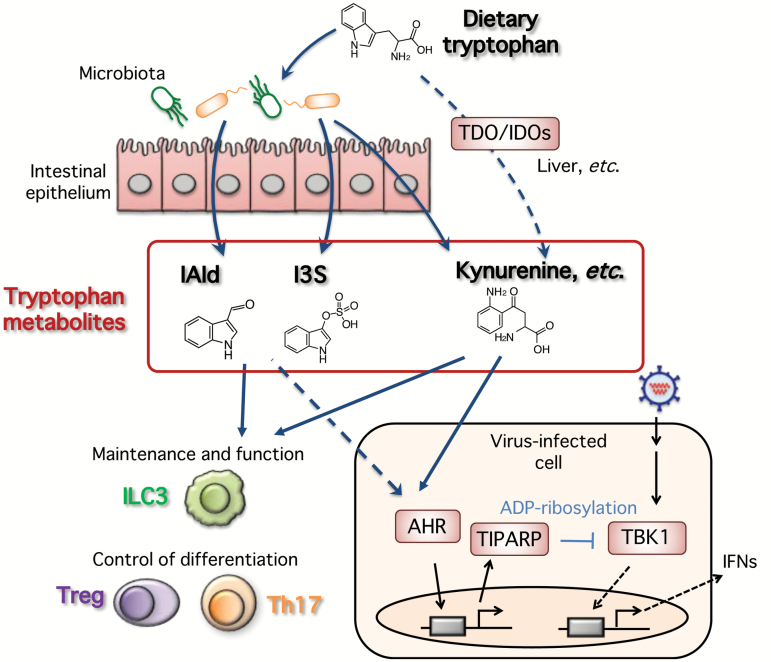
As its name suggests, TIPARP is one of the inducible genes that is expressed following exposure to dioxins, through the aryl hydrocarbon receptor (AHR), which was known to be a receptor for dioxins. Whereas AHR has been mostly studied in terms of its toxic effects on development, cell proliferation and immune responses, but a variety of endogenous metabolites have also been shown to interact with the receptor and are involved in the regulation of multiple biological events (91–93). In this respect, treatment with an AHR antagonist CH-223191 enhanced the interferon response upon viral infection and 3pRNA stimulation, suggesting that constitutive AHR signaling activated by endogenous ligand(s), such as kynurenine, negatively modulates the type I interferon anti-viral response to prevent excessive responses. These findings indicate that this negative regulation is based upon the post-translational ADP-ribosylation of the TBK1 protein mediated by AHR-induced TIPARP (90). An endogenous AHR ligand, kynurenine, is synthesized via oxidation of tryptophan, which is catalyzed by tryptophan 2,3-dioxygenase (TDO) and indoleamine 2,3-dioxygenase 1/2 (IDO1/2). Liver is one of the major places where this enzymatic process takes place by using TDO, which is known to be exclusively expressed at high levels in human liver (94). On the other hand, the liver has large populations of specialized tissue macrophages (namely Kupffer cells) and DCs (95, 96), which have pivotal roles in pathogen recognition and activation of innate immune responses and its subsequent adaptive immune responses. Therefore, it is important to regulate such innate signaling-based responses to maintain immune homeostasis in liver.
In this regard, the AHR–TIPARP axis may contribute to the suppressive regulation of the interferon response, particularly in human hepatocytes. It would be a possible therapeutic intervention to manipulate the activity of the AHR–TIPARP axis to control viral infection and inflammation in liver.
On the other hand, dietary tryptophan is also exploited by the intestinal microbiota. In the gut, some tryptophan metabolites such as kynurenine and indole-3-aldehyde (IAld) are generated by the bacterial catabolism of tryptophan and act as AHR ligands to induce regulatory responses for the maintenance and function of group 3 innate lymphoid cells (ILC3) (97, 98) and the control of Treg and Th17 differentiation (99). Alteration of microbiota may reduce tryptophan metabolites, which leads to higher susceptibility of mice to colitis, as exemplified in the case of Card9 knockout mice (100).
Antibiotic-mediated killing of intestinal microbiota leads to suppression of the synthesis of the AHR ligand indoxyl-3-sulfate (I3S), resulting in the modulation of astrocytes in terms of the pathogenesis of experimental autoimmune encephalomyelitis (EAE), a model for multiple sclerosis (101). Therefore, it could be reasonably speculated that the AHR–TIPARP pathway, activated by microbiota-mediated tryptophan metabolites, may have a role in the regulation of anti-viral innate responses and immune homeostasis not only in the gut but also in other organs (Fig. 3).
Fig. 3.
Proposed model of dietary tryptophan-mediated regulation of anti-viral innate responses and immune homeostasis. An endogenous AHR ligand, kynurenine, is synthesized via oxidation of tryptophan, which is catalyzed by TDO and IDO1/2 in human liver and other organs. Constitutive AHR signaling activated by endogenous ligand(s), such as kynurenine, negatively modulates the type I interferon anti-viral response to prevent excessive responses. On the other hand, in the gut, some tryptophan metabolites such as kynurenine, IAld and I3S are generated by the bacterial catabolism of tryptophan, and act as AHR ligands to induce regulatory responses for the maintenance and function of ILC3 cells and the control of Treg and Th17 differentiation.
Other interacting factors that control RIG-I signaling
RIG-I-interacting positive regulators have been identified (Fig. 2c). ZAPS, which is a shorter form of PARP-13, a member of the PARP superfamily, functions as a potent stimulator of RIG-I-mediated anti-viral signaling through its interaction with RIG-I to facilitate RIG-I oligomerization and its ATPase activity for the activation of both IRF-3 and NF-κB upon 3pRNA stimulation and RNA virus infection in human cells (102). On the other hand, mouse embryonic fibroblasts (MEFs), ZAP is not essential for the RIG-I-dependent production of type I interferons (103).
The dsRNA-binding protein PACT, which was known to function as a partner of Dicer in RNA silencing and to regulate PKR activity, directly interacts with the CTD of RIG-I in a Dicer- or PKR-independent manner. PACT potentiates the activation of the RIG-I-mediated IRF-3 pathway (not NF-κB) by a poly(I:C) molecule of 0.2–1 kb in length, but not 3pRNA (104).
On the other hand, DDX6, which is known to be a member of the DExD/H-box RNA helicases family and a SG and P-body component, interacts with RIG-I and augments type I interferon gene induction (105). Mechanistically, it is speculated that DDX6 can collect viral RNAs and present them in suitable topology to RIG-I to induce its activation.
On the contrary to the above-mentioned negative role of DAPK1 in RIG-I activation, DAPK1 can also function as a positive regulator of RIG-I signaling. DAPK1 interacts with IRF-7 and IRF-3 upon Sendai virus infection, resulting in an increased IFN-β response, which is independent of its kinase activity (106).
DDX25, a DEAD-box RNA helicase, which is up-regulated upon viral infection such as by Dengue virus, negatively modulates the RIG-I signaling pathway and blocks IFN-β production, by interrupting IRF-3 and NF-κB activation (107).
lncRNA-mediated regulation of RIG-I signaling
High-throughput studies have shown that a large number of long non-coding RNAs (lncRNAs) are expressed following viral infection and interferon stimulation. Some of the lncRNAs have been implicated as a new class of regulators in innate signaling (Fig. 2c) (108). Very recently, lnc-Lsm3b, a non-coding alternatively processed isoform of the protein-coding gene Lsm3, was shown to be induced by type I interferons and to inactivate RIG-I function at a late stage of the innate response (109). This study provides a novel lncRNA-mediated negative feedback regulation of innate signaling to prevent detrimental, excessive inflammatory responses, wherein the self-lncRNA lnc-Lsm3b functions as a molecular decoy to occupy the RIG-I-binding sites and sequester RIG-I molecules to restrict the access of ‘non-self’ RNA. Interestingly, lnc-Lsm3b uses GA-rich motifs located at the incomplete pairing strand of the long stem structure to efficiently compete with non-self-viral RNA. lnc-Lsm3b contains a multitude of GA-rich motifs, compared to RIG-I-favored foreign ligands that contain specific sequences, such as polyU/UC motifs or elements in viral genomes. Further structural analyses would reveal its detailed mechanism for the competition and how such RIG-I recognition of self-RNA stops the generation of the downstream signaling. A 5′-triphosphate modification does not seem to be needed for lnc-Lsm3b to bind RIG-I.
Since the critical structural elements for its binding to RIG-I include ‘short’ segments of the RNA duplex structure, it might be speculated that MDA5, another RLR, fails to recognize lnc-Lsm3b. In relation to this, there is a recent paper showing a closely related mechanism for MDA5 to bind a different self-lncRNA, lnc-ITPRIP-1 (110). In this case, its binding confers a conformational change to promote MDA5 oligomerization and its downstream signaling. This lncRNA was studied particularly in terms of its role in anti-viral responses against HCV infection. It is suggested that lnc-ITPRIP-1 may function as a cofactor of MDA5 to mediate the association between MDA5 and HCV RNA.
Another lncRNA, NEAT1, which forms a key structural component of paraspeckles, is robustly up-regulated after Hantaan virus (HTNV) infection to serve as positive feedback for RIG-I signaling (111). Mechanistically, it is likely that the induced NEAT1 attenuated a transcriptional inhibitory regulator, splicing factor proline- and glutamine-rich protein (SFPQ), to enhance the expression of RIG-I and DDX60, thus modulating the innate immune response through a positive feedback mechanism.
lncRHOXF1, which is abundantly expressed in trophectoderm and primitive endoderm cells of human blastocyst-stage embryos, has also been shown to be up-regulated in response to viral infection. Knockdown of this gene leads to up-regulation of IFN-β and interferon-stimulated genes such as IFIH1 (MDA5), DDX58 (RIG-I), MX1 and OAS1. Although it is still unknown whether lncRHOXF1 directly regulates the expression of IFN-β and/or ISGs, it seems to function as a repressor of the viral response during development of human trophoblast cells (112).
Taken together, lncRNAs are also important modules for the regulation of multiple steps of RIG-I-mediated anti-viral innate responses.
Viral evasion from nucleic acid sensor-mediated innate signaling
Viruses have evolved a variety of mechanisms to counteract or escape from host anti-viral systems (Fig. 4). Nucleic acid sensors and downstream signaling molecules are often targeted by these viral proteins (113). Both RIG-I and MDA5 are targets of viral evasion. As shown above, it is reported that IAV NS1 protein interacts with RIG-I in a viral RNA-dependent manner, and inhibits its downstream signaling pathways such as type I interferon induction (114–117). Further analyses demonstrated that NS1 interacts with TRIM25 and leads to inhibition of TRIM25 multimerization and RIG-I CARD ubiquitination, thereby suppressing RIG-I signal transduction (118). A similar observation was made for the effect of NS1 on Riplet, which is also known as another Ub E3 ligase to ubiquitinate RIG-I (119).
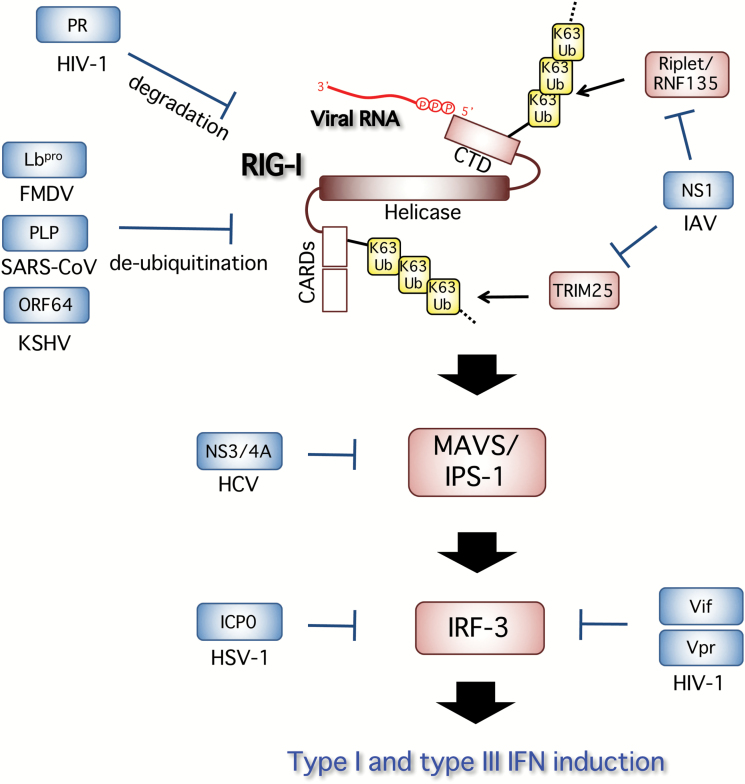
Fig. 4.
Viral evasion from RIG-I activation and its signaling. IAV NS1 interacts with TRIM25, and leads to inhibition of TRIM25 multimerization and RIG-I CARD ubiquitination, thereby suppressing RIG-I signal transduction. NS1 also targets Riplet. Viral proteins that have a deubiquitinase activity, including a PLP from SARS-CoV, FMDV Lbpro (a shorter form of the leader proteinase of FMDV) and a KSHV-derived tegument protein ORF64, directly remove the K63-linked ubiquitination of RIG-I and suppress its activity. HCV-derived NS3/4A protease cleaves MAVS/IPS-1, resulting in a blockade of RLR-signaling. HSV-1 ICP0 inhibits the nuclear accumulation of IRF-3 and enhances the degradation of activated IRF-3. HIV-1 also targets IRF-3 for protein depletion, in which HIV accessory proteins Vif and Vpr mediate the ubiquitination and proteasome-dependent degradation of IRF-3. In addition, HIV-1 PR targets RIG-I through the degradation of RIG-I.
On the other hand, TRIM25 is also reported to bind to influenza virus ribonucleoproteins (vRNPs) to block the onset of RNA chain elongation in an E3 ligase-independent manner (120). NS1 protein is also known to antagonize its binding through a direct interaction with TRIM25. Some viruses directly remove the K63-linked ubiquitination of RIG-I and suppress its pathway by expressing viral proteins that have a deubiquitinase activity, including a papain-like protease (PLP) from severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) (121), FMDV Lbpro, a shorter form of the leader proteinase (Lpro) of foot-and-mouth disease virus (FMDV) (122) and a KSHV-derived tegument protein, ORF64 (123).
Another RLR, MDA5, is targeted by the paramyxovirus V protein, leading to abrogation of its downstream IFN-β induction (124). It is well documented that HCV-derived NS3/4A protease cleaves MAVS/IPS-1, resulting in a blockade of RLR-signaling (125–127). IRF-3 is a transcriptional factor that is activated downstream of most of the nucleic acid sensors, and is critical for the induction of type I and type III interferon genes. HSV-1 inhibits the nuclear accumulation of IRF-3 and enhances the degradation of activated IRF-3, thus interfering with the pathway leading to IFN-β production. Mechanistically, at least ICP0, which is known as an essential component of interferon resistance, was found to be necessary for preventing IRF-3 nuclear accumulation (128).
HIV-1 also targets IRF-3 for protein depletion, in which HIV accessory proteins Vif and Vpr mediate the ubiquitination and proteasome-dependent degradation of IRF-3 (129, 130). In addition, HIV-1 protease (PR) targets RIG-I through the degradation of RIG-I, which is dependent on lysosomes but not proteasomes (131). PR does not directly cleave RIG-I, but redirects RIG-I to the lysosomal compartment for its degradation. Blockade of sensor activation and/or innate signaling modulates the innate responses and attenuates anti-viral defense, which may allow viral persistence. Therefore, it is an important therapeutic strategy to target the host–virus interaction involved in innate immune evasion for the efficient activation of anti-viral innate signaling against viral infection.
Conclusions and future prospects
Innate nucleic acid sensors have been shown to play an important role for the recognition of invading viruses and the subsequent activation of efficient innate and adaptive immune responses, and the regulatory mechanisms for these innate signaling pathways have been extensively studied. In addition, much attention has been paid to the role of these nucleic acid sensors in the detection of self-derived nucleic acids to evoke inflammatory responses. In this respect, several future issues remain to be addressed about what type of self nucleic acids are recognized as ligands for these sensors and which type of cells are key players to evoke inflammatory immune responses.
These innate nucleic acid sensor-derived signaling systems are thus clearly important in host defense against not only viral infection, but also autoimmune inflammatory diseases as well as tumors. In this respect, detailed analyses and understanding of the regulation of these signaling pathways will provide a therapeutic insight and would pave innovative paths towards the development of their clinical application.
Funding
This work was in part supported by grants from the Japan Society for the Promotion of Science (JSPS) [Grant-in-Aid for Challenging Research (Pioneering) (JP17H06265) and Grant-in-Aid for Young Scientists (B) (JP17K15698)], and Japan Agency for Medical Research and Development (AMED) [Program on the Innovative Development and the Application of New Drugs for Hepatitis B (JP18fk0310101)] as well as grants from the Naito Foundation and Asai Germanium Research Institute Co., Ltd.
Conflicts of interest statement: the authors declared no conflicts of interest.
References
- 1. Nagano Y. and Kojima Y. 1954. [Immunizing property of vaccinia virus inactivated by ultraviolets rays]. C. R. Seances Soc. Biol. Fil. 148:1700. [PubMed] [Google Scholar]
- 2. Isaacs A. and Lindenmann J. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B. Biol. Sci. 147:258. [PubMed] [Google Scholar]
- 3. Kumar H., Kawai T. and Akira S. 2011. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30:16. [DOI] [PubMed] [Google Scholar]
- 4. Mcnab F., Mayer-Barber K., Sher A. et al. 2015. Type I interferons in infectious disease. Nat. Rev. Immunol. 15:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alexopoulou L., Holt A. C., Medzhitov R. and Flavell R. A. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732. [DOI] [PubMed] [Google Scholar]
- 6. Heil F.,, Hemmi H.,, Hochrein H., et al. 2004. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 303:1526. [DOI] [PubMed] [Google Scholar]
- 7. Diebold S. S., Kaisho T., Hemmi H., Akira S. and Reis E Sousa C. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303:1529. [DOI] [PubMed] [Google Scholar]
- 8. Zhang S. Y. and Casanova J. L. 2015. Inborn errors underlying herpes simplex encephalitis: from TLR3 to IRF3. J. Exp. Med. 212:1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim H. K.,, Seppänen M.,, Hautala T., et al. 2014. TLR3 deficiency in herpes simplex encephalitis: high allelic heterogeneity and recurrence risk. Neurology 83:1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawai T.,, Sato S.,, Ishii K. J., et al. 2004. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5:1061. [DOI] [PubMed] [Google Scholar]
- 11. Oshiumi H., Matsumoto M., Funami K., Akazawa T. and Seya T. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161. [DOI] [PubMed] [Google Scholar]
- 12. Yoneyama M.,, Kikuchi M.,, Natsukawa T., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730. [DOI] [PubMed] [Google Scholar]
- 13. Yoneyama M.,, Kikuchi M.,, Matsumoto K., et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851. [DOI] [PubMed] [Google Scholar]
- 14. Kell A. M. and Gale M. Jr. 2015. RIG-I in RNA virus recognition. Virology 479-480:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hornung V.,, Ellegast J.,, Kim S., et al. 2006. 5’-Triphosphate RNA is the ligand for RIG-I. Science 314:994. [DOI] [PubMed] [Google Scholar]
- 16. Jiang X.,, Kinch L. N.,, Brautigam C. A., et al. 2012. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36:959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sato S.,, Li K.,, Kameyama T., et al. 2015. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity 42:123. [DOI] [PubMed] [Google Scholar]
- 18. Okabayashi T.,, Kojima T.,, Masaki T., et al. 2011. Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 160:360. [DOI] [PubMed] [Google Scholar]
- 19. Wang J.,, Oberley-Deegan R.,, Wang S., et al. 2009. Differentiated human alveolar type II cells secrete antiviral IL-29 (IFN-lambda 1) in response to influenza A infection. J. Immunol. 182:1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jewell N. A.,, Cline T.,, Mertz S. E., et al. 2010. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J. Virol. 84:11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu W., Zhang W., Duggan E. S., Booth J. L., Zou M. H. and Metcalf J. P. 2015. RIG-I and TLR3 are both required for maximum interferon induction by influenza virus in human lung alveolar epithelial cells. Virology 482:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weber M.,, Sediri H.,, Felgenhauer U., et al. 2015. Influenza virus adaptation PB2-627K modulates nucleocapsid inhibition by the pathogen sensor RIG-I. Cell Host Microbe 17:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yao H.,, Dittmann M.,, Peisley A., et al. 2015. ATP-dependent effector-like functions of RIG-I-like receptors. Mol. Cell 58:541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Z.,, Kim T.,, Bao M., et al. 2011. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity 34:866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sabbah A.,, Chang T. H.,, Harnack R., et al. 2009. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 10:1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hemmi H.,, Takeuchi O.,, Kawai T., et al. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740. [DOI] [PubMed] [Google Scholar]
- 27. Hornung V.,, Rothenfusser S.,, Britsch S., et al. 2002. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 168:4531. [DOI] [PubMed] [Google Scholar]
- 28. Honda K.,, Yanai H.,, Negishi H., et al. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772. [DOI] [PubMed] [Google Scholar]
- 29. Takaoka A. and Taniguchi T. 2008. Cytosolic DNA recognition for triggering innate immune responses. Adv. Drug Deliv. Rev. 60:847. [DOI] [PubMed] [Google Scholar]
- 30. Takaoka A.,, Wang Z.,, Choi M. K., et al. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501. [DOI] [PubMed] [Google Scholar]
- 31. Upton J. W., Kaiser W. J. and Mocarski E. S. 2012. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Defilippis V. R., Alvarado D., Sali T., Rothenburg S. and Früh K. 2010. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J. Virol. 84:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Defilippis V. R., Sali T., Alvarado D., White L., Bresnahan W. and Früh K. J. 2010. Activation of the interferon response by human cytomegalovirus occurs via cytoplasmic double-stranded DNA but not glycoprotein B. J. Virol. 84:8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ishii K. J.,, Kawagoe T.,, Koyama S., et al. 2008. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451:725. [DOI] [PubMed] [Google Scholar]
- 35. Chiu Y. H., Macmillan J. B. and Chen Z. J. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ablasser A., Bauernfeind F., Hartmann G., Latz E., Fitzgerald K. A. and Hornung V. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10:1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Minarovits J., Hu L. F., Marcsek Z., Minarovits-Kormuta S., Klein G. and Ernberg I. 1992. RNA polymerase III-transcribed EBER 1 and 2 transcription units are expressed and hypomethylated in the major Epstein–Barr virus-carrying cell types. J. Gen. Virol. 73(Pt 7):1687. [DOI] [PubMed] [Google Scholar]
- 38. Samanta M., Iwakiri D., Kanda T., Imaizumi T. and Takada K. 2006. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 25:4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. West J. A.,, Wicks M.,, Gregory S. M., et al. 2014. An important role for mitochondrial antiviral signaling protein in the Kaposi’s sarcoma-associated herpesvirus life cycle. J. Virol. 88:5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rasmussen S. B.,, Jensen S. B.,, Nielsen C., et al. 2009. Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene- like receptors, which synergize to induce type I interferon production. J. Gen. Virol. 90(Pt 1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malathi K., Dong B., Gale M. Jr and Silverman R. H. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim T.,, Pazhoor S.,, Bao M., et al. 2010. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl Acad. Sci. USA 107:15181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Z., Yuan B., Bao M., Lu N., Kim T. and Liu Y. J. 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12:959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Unterholzner L.,, Keating S. E.,, Baran M., et al. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11:997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun L., Wu J., Du F., Chen X. and Chen Z. J. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu J.,, Sun L.,, Chen X., et al. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gao D.,, Li T.,, Li X. D., et al. 2015. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl Acad. Sci. USA 112:E5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gray E. E., Treuting P. M., Woodward J. J. and Stetson D. B. 2015. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi-Goutières syndrome. J. Immunol. 195:1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li T. and Chen Z. J. 2018. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215:1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gray E. E., Winship D., Snyder J. M., Child S. J., Geballe A. P. and Stetson D. B. 2016. The AIM2-like receptors are dispensable for the interferon response to intracellular DNA. Immunity 45:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mcwhirter S. M.,, Barbalat R.,, Monroe K. M., et al. 2009. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 206:1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Burdette D. L.,, Monroe K. M.,, Sotelo-Troha K., et al. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barber G. N. 2015. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoh S. M.,, Schneider M.,, Seifried J., et al. 2015. PQBP1 is a proximal sensor of the cGAS-dependent innate response to HIV-1. Cell 161:1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shannon J. L.,, Murphy M. S.,, Kantheti U., et al. 2018. Polyglutamine binding protein 1 (PQBP1) inhibits innate immune responses to cytosolic DNA. Mol. Immunol. 99:182. [DOI] [PubMed] [Google Scholar]
- 56. Liu Y. T. and Yin H. L. 1998. Identification of the binding partners for flightless I, A novel protein bridging the leucine-rich repeat and the gelsolin superfamilies. J. Biol. Chem. 273:7920. [DOI] [PubMed] [Google Scholar]
- 57. Yang P.,, An H.,, Liu X., et al. 2010. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat. Immunol. 11:487. [DOI] [PubMed] [Google Scholar]
- 58. Ferguson B. J., Mansur D. S., Peters N. E., Ren H. and Smith G. L. 2012. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 1:e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang X.,, Brann T. W.,, Zhou M., et al. 2011. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 186:4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kobiyama K.,, Kawashima A.,, Jounai N., et al. 2013. Role of extrachromosomal histone H2B on recognition of DNA viruses and cell damage. Front. Genet. 4:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yanai H.,, Ban T.,, Wang Z., et al. 2009. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462:99. [DOI] [PubMed] [Google Scholar]
- 62. Jung J. H.,, Park J. H.,, Jee M. H., et al. 2011. Hepatitis C virus infection is blocked by HMGB1 released from virus-infected cells. J. Virol. 85:9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yu R.,, Yang D.,, Lei S., et al. 2015. HMGB1 promotes hepatitis C virus replication by interaction with stem-loop 4 in the viral 5’ untranslated region. J. Virol. 90:2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kato H., Oh S. W. and Fujita T. 2017. RIG-I-like receptors and type I interferonopathies. J. Interferon Cytokine Res. 37:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ablasser A., Hertrich C., Waßermann R. and Hornung V. 2013. Nucleic acid driven sterile inflammation. Clin. Immunol. 147:207. [DOI] [PubMed] [Google Scholar]
- 66. Oda H.,, Nakagawa K.,, Abe J., et al. 2014. Aicardi-Goutières syndrome is caused by IFIH1 mutations. Am. J. Hum. Genet. 95:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Van Eyck L.,, De Somer L.,, Pombal D., et al. 2015. Brief report: IFIH1 mutation causes systemic lupus erythematosus with selective IgA deficiency. Arthritis Rheumatol. 67:1592. [DOI] [PubMed] [Google Scholar]
- 68. Gack M. U.,, Shin Y. C.,, Joo C. H., et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916. [DOI] [PubMed] [Google Scholar]
- 69. Oshiumi H., Matsumoto M., Hatakeyama S. and Seya T. 2009. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J. Biol. Chem. 284:807. [DOI] [PubMed] [Google Scholar]
- 70. Oshiumi H., Miyashita M., Matsumoto M. and Seya T. 2013. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 9:e1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kuniyoshi K.,, Takeuchi O.,, Pandey S., et al. 2014. Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I-mediated antiviral innate immunity. Proc. Natl Acad. Sci. USA 111:5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen H., Ning X. and Jiang Z. 2017. Caspases control antiviral innate immunity. Cell. Mol. Immunol. 14:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang P.,, Arjona A.,, Zhang Y., et al. 2010. Caspase-12 controls West Nile virus infection via the viral RNA receptor RIG-I. Nat. Immunol. 11:912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Peisley A., Wu B., Xu H., Chen Z. J. and Hur S. 2014. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shi Y.,, Yuan B.,, Zhu W., et al. 2017. Ube2D3 and Ube2N are essential for RIG-I-mediated MAVS aggregation in antiviral innate immunity. Nat. Commun. 8:15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhu J.,, Zhang Y.,, Ghosh A., et al. 2014. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity 40:936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen W.,, Han C.,, Xie B., et al. 2013. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell 152:467. [DOI] [PubMed] [Google Scholar]
- 78. Arimoto K., Takahashi H., Hishiki T., Konishi H., Fujita T. and Shimotohno K. 2007. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl Acad. Sci. USA 104:7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang W.,, Jiang M.,, Liu S., et al. 2016. RNF122 suppresses antiviral type I interferon production by targeting RIG-I CARDs to mediate RIG-I degradation. Proc. Natl Acad. Sci. USA 113:9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhao K.,, Zhang Q.,, Li X., et al. 2016. Cytoplasmic STAT4 promotes antiviral type I IFN production by blocking CHIP-mediated degradation of RIG-I. J. Immunol. 196:1209. [DOI] [PubMed] [Google Scholar]
- 81. Zhao C.,, Jia M.,, Song H., et al. 2017. The E3 ubiquitin ligase TRIM40 attenuates antiviral immune responses by targeting MDA5 and RIG-I. Cell Rep. 21:1613. [DOI] [PubMed] [Google Scholar]
- 82. Kim M. J., Hwang S. Y., Imaizumi T. and Yoo J. Y. 2008. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 82:1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Willemsen J.,, Wicht O.,, Wolanski J. C., et al. 2017. Phosphorylation-dependent feedback inhibition of RIG-I by DAPK1 identified by kinome-wide siRNA screening. Mol. Cell 65:403–e8.. [DOI] [PubMed] [Google Scholar]
- 84. Maharaj N. P., Wies E., Stoll A. and Gack M. U. 2012. Conventional protein kinase C-α (PKC-α) and PKC-β negatively regulate RIG-I antiviral signal transduction. J. Virol. 86:1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wies E.,, Wang M. K.,, Maharaj N. P., et al. 2013. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 38:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sun Z., Ren H., Liu Y., Teeling J. L. and Gu J. 2011. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J. Virol. 85:1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liu J., Qian C. and Cao X. 2016. Post-translational modification control of innate immunity. Immunity 45:15. [DOI] [PubMed] [Google Scholar]
- 88. Lüscher B., Bütepage M., Eckei L., Krieg S., Verheugd P. and Shilton B. H. 2018. ADP-ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem. Rev. 118:1092. [DOI] [PubMed] [Google Scholar]
- 89. Schreiber V., Dantzer F., Ame J. C. and De Murcia G. 2006. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7:517. [DOI] [PubMed] [Google Scholar]
- 90. Yamada T.,, Horimoto H.,, Kameyama T., et al. 2016. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat. Immunol. 17:687. [DOI] [PubMed] [Google Scholar]
- 91. Nguyen L. P. and Bradfield C. A. 2008. The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 21:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Stockinger B., Di Meglio P., Gialitakis M. and Duarte J. H. 2014. The aryl hydrocarbon receptor: multitasking in the immune system. Annu. Rev. Immunol. 32:403. [DOI] [PubMed] [Google Scholar]
- 93. Opitz C. A.,, Litzenburger U. M.,, Sahm F., et al. 2011. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478:197. [DOI] [PubMed] [Google Scholar]
- 94. Van Baren N. and Van Den Eynde B. J. 2015. Tryptophan-degrading enzymes in tumoral immune resistance. Front. Immunol. 6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Heymann F. and Tacke F. 2016. Immunology in the liver—from homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol. 13:88. [DOI] [PubMed] [Google Scholar]
- 96. Robinson M. W., Harmon C. and O’farrelly C. 2016. Liver immunology and its role in inflammation and homeostasis. Cell. Mol. Immunol. 13:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee J. S.,, Cella M.,, Mcdonald K. G., et al. 2011. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Qiu J.,, Heller J. J.,, Guo X., et al. 2012. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Quintana F. J.,, Basso A. S.,, Iglesias A. H., et al. 2008. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453:65. [DOI] [PubMed] [Google Scholar]
- 100. Lamas B.,, Richard M. L.,, Leducq V., et al. 2016. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22:598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rothhammer V.,, Mascanfroni I. D.,, Bunse L., et al. 2016. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hayakawa S.,, Shiratori S.,, Yamato H., et al. 2011. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat. Immunol. 12:37. [DOI] [PubMed] [Google Scholar]
- 103. Lee H.,, Komano J.,, Saitoh Y., et al. 2013. Zinc-finger antiviral protein mediates retinoic acid inducible gene I-like receptor-independent antiviral response to murine leukemia virus. Proc. Natl Acad. Sci. USA 110:12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kok K. H., Lui P. Y., Ng M. H., Siu K. L., Au S. W. and Jin D. Y. 2011. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe 9:299. [DOI] [PubMed] [Google Scholar]
- 105. Nunez R. D., Budt M., Saenger S. et al. 2018. The RNA helicase DDX6 associates with RIG-I to augment induction of antiviral signaling. Int. J. Mol. Sci. 19:1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhang J., Hu M. M., Shu H. B. and Li S. 2014. Death-associated protein kinase 1 is an IRF3/7-interacting protein that is involved in the cellular antiviral immune response. Cell. Mol. Immunol. 11:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Feng T., Sun T., Li G., Pan W., Wang K. and Dai J. 2017. DEAD-box helicase DDX25 is a negative regulator of type I interferon pathway and facilitates RNA virus infection. Front. Cell. Infect. Microbiol. 7:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Valadkhan S. and Plasek L. M. 2018. Long non-coding RNA-mediated regulation of the interferon response: a new perspective on a familiar theme. Pathog. Immun. 3:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jiang M.,, Zhang S.,, Yang Z., et al. 2018. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 173:906–e13.. [DOI] [PubMed] [Google Scholar]
- 110. Xie Q., Chen S., Tian R. et al. 2018. Long noncoding RNA ITPRIP-1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J. Virol. 92:e00507-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ma H., Han P., Ye W. et al. 2017. The long noncoding RNA NEAT1 exerts anti-hantaviral effects by acting as positive feedback for RIG-I signaling. J. Virol. 91:e02250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Penkala I., Wang J., Syrett C. M., Goetzl L., López C. B. and Anguera M. C. 2016. lncRHOXF1, a long noncoding RNA from the X chromosome that suppresses viral response genes during development of the early human placenta. Mol. Cell. Biol. 36:1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chan Y. K. and Gack M. U. 2016. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 14:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Guo Z.,, Chen L. M.,, Zeng H., et al. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 36:263. [DOI] [PubMed] [Google Scholar]
- 115. Mibayashi M., Martínez-Sobrido L., Loo Y. M., Cárdenas W. B., Gale M. Jr and García-Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Opitz B.,, Rejaibi A.,, Dauber B., et al. 2007. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell. Microbiol. 9:930. [DOI] [PubMed] [Google Scholar]
- 117. Pichlmair A.,, Schulz O.,, Tan C. P., et al. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science 314:997. [DOI] [PubMed] [Google Scholar]
- 118. Gack M. U.,, Albrecht R. A.,, Urano T., et al. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Marc D. 2014. Influenza virus non-structural protein NS1: interferon antagonism and beyond. J. Gen. Virol. 95(Pt 12):2594. [DOI] [PubMed] [Google Scholar]
- 120. Meyerson N. R.,, Zhou L.,, Guo Y. R., et al. 2017. Nuclear TRIM25 specifically targets influenza virus ribonucleoproteins to block the onset of RNA chain elongation. Cell Host Microbe 22:627–e7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Clementz M. A.,, Chen Z.,, Banach B. S., et al. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84:4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wang D.,, Fang L.,, Li P., et al. 2011. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J. Virol. 85:3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Inn K. S.,, Lee S. H.,, Rathbun J. Y., et al. 2011. Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J. Virol. 85:10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Andrejeva J.,, Childs K. S.,, Young D. F., et al. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl Acad. Sci. USA 101:17264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Meylan E.,, Curran J.,, Hofmann K., et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167. [DOI] [PubMed] [Google Scholar]
- 126. Loo Y. M.,, Owen D. M.,, Li K., et al. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl Acad. Sci. USA 103:6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lin R.,, Lacoste J.,, Nakhaei P., et al. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 80:6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Melroe G. T., Deluca N. A. and Knipe D. M. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 78:8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Okumura A., Alce T., Lubyova B., Ezelle H., Strebel K. and Pitha P. M. 2008. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology 373:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Doehle B. P., Hladik F., Mcnevin J. P., Mcelrath M. J. and Gale M. Jr. 2009. Human immunodeficiency virus type 1 mediates global disruption of innate antiviral signaling and immune defenses within infected cells. J. Virol. 83:10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Solis M.,, Nakhaei P.,, Jalalirad M., et al. 2011. RIG-I-mediated antiviral signaling is inhibited in HIV-1 infection by a protease-mediated sequestration of RIG-I. J. Virol. 85:1224. [DOI] [PMC free article] [PubMed] [Google Scholar]