Abstract
During the past years, human coronaviruses (HCoVs) have been increasingly identified as pathogens associated with more-severe respiratory tract infection (RTI). Diagnostic tests for HCoVs are not frequently used in the routine setting. It is likely that, as a result, the precise role that HCoVs play in RTIs is greatly underestimated. We describe a rapid, sensitive, and highly specific quantitative real-time reverse-transcriptase polymerase chain reaction (RT-PCR) for the detection of HCoV that can easily be implemented in the routine diagnostic setting. HCoV was detected in 28 (11%) of the 261 clinical specimens obtained from patients presenting with symptoms of RTI ranging from common cold to severe pneumonia. Only 1 (0.4%) of the 243 control specimens obtained from patients without symptoms of RTI showed the presence of HCoV. We conclude that HCoVs can be frequently detected in patients presenting with RTI. Real-time RT-PCR provides a tool for large-scale epidemiological studies to further clarify the role that coronavirus infection plays in RTI in humans.
Coronaviruses are enveloped RNA viruses that can cause disease in humans and animals. The human coronaviruses (HCoVs) were first identified in 1962. They belong to the family Coronaviridae, genus coronavirus, and the 2 human strains, HCoV 229E and OC43, are divided into 2 antigenic groups. HCoVs are recognized as the second-most frequent cause of the common cold syndrome [1]. During the past years, HCoVs have more often been believed to be responsible for severe upper and lower respiratory-tract infection (RTI). They have occasionally been identified as a cause of pneumonia in older adults, infants, and immunocompromised patients [2–5]. Also, in otherwise healthy adults—for example, in military recruits—clusters of infections have been reported as a cause of pneumonia [6]. Moreover, in a recent outbreak of HCoV in Normandy, the clinical manifestation ranged from mild symptoms to pneumonia [7]. Recently, a heightened interest in the coronavirus has been documented, because a previously unknown type that does not resemble the known HCoVs is believed to be responsible for the outbreaks of severe acute respiratory syndrome (SARS) in Hong Kong and Toronto [8–11]. These studies indicate that coronaviruses are increasingly identified as a pathogen causing severe respiratory illnesses and that there is a need for reliable and rapid identification of coronaviruses.
The diagnosis of HCoV infections is hampered, in part, by the difficulty to replicate in-cell cultures, whereas serologic testing is time-consuming and therefore has little clinical significance. As a consequence, efforts have been made to develop more-sensitive molecular detection methods, such as reverse-transcriptase polymerase chain reaction (RT-PCR) and nested RT-PCR [12, 13]. These methods have been shown to be very valuable for the detection of HCoV in different populations of patients, such as children with otitis media, patients with multiple sclerosis, immunocompromised patients with pneumonia, and frail elderly persons with symptoms of RTI [3, 5, 8, 9, 14, 15].
Although they are highly sensitive and specific, the current RT-PCR and nested RT-PCR methods are less suitable for routine laboratory detection because they are prone to contamination and still require time-consuming sample handling and post-PCR analysis. Here, we describe the detection of HCoV in a variety of clinical specimens obtained from patients presenting with RTIs ranging from common cold to severe pneumonia, using a novel, sensitive, and highly specific Taqman-based realtime PCR. In addition, we tested a multiplex-format real-time RT-PCR assay for the detection of HCoVs and the novel coronavirus that has been identified in patients with SARS.
Materials and Methods
Virus stocks and viral culture. HCoV 229E and OC43 were provided by the Laboratory for Virology, National Institute for Public Health and the Environment (RIVM; Bilthoven, The Netherlands), and were propagated on 2 human embryonic lung cell lines (MRC5 and HEL). Cells and supernatants were harvested after 24, 48, and 72 h, respectively, and were frozen at −70°C. After RNA extraction of each stock, series of 10-fold serial dilutions were used to determine, by use of an in-house nested PCR, which propagated stock contained the most virus particles. The stocks, 1 of each strain, that contained the most virus particles were used for further experiments to evaluate the Taqman-based real-time PCR. A panel of various respiratory viruses—including influenza virus A/PR/8/34, influenza virus B/Lee/40, parainfluenza viruses 1–4 (American Type Culture Collection), and reference strains of rhinovirus 1A, rhinovirus 14, rhinovirus 16, echovirus 12, coxsackie virus A9, respiratory synctial virus (RSV) A Long strain, RSV B 9320, and SARS-associated coronavirus—were used to determine the specificity of the Taqman-based real-time PCR.
Clinical specimens. Clinical specimens were obtained at the hospital's virology laboratory and consisted of the following: (1) nasal wash (NW) specimens and combined nose and throat swabs (NTSs) from patients presenting with symptoms of upper or lower RTIs and (2) bronchoalveolar lavage (BAL) specimens and NTSs from adult patients admitted to the hospital with pneumonia. NTSs from healthy volunteers and NTSs obtained at set time points from patients without symptoms of RTI who participated in a prospective 6-month follow-up study to assess the role of respiratory viruses following bone marrow transplantation were used as control specimens. Each specimen was transported in 5 mL of virus transport medium.NWspecimens, NTSs, and BAL specimens were vortexed for 10 s and centrifuged at 2000 g for 15 min. One milliliter of the supernatant was used directly for routine virus culture of other respiratory viruses (influenza viruses, RSV, parainfluenza viruses, picornaviruses, and adenovirus). The remaining material was stored at −70°C until further processing.
Viral RNA isolation and cDNA synthesis. RNA extraction was performed by use of the MagnaPure LC Total Nucleic Acid Kit (Roche Diagnostics), as described elsewhere [16]. The RNA was then either eluted in 100 µL of 40 ng/µL polyA RNA before performing a 1-tube RT-PCR or eluted in 100 µL of elution buffer and directly used for cDNA synthesis. The reverse transcription and cDNA synthesis were both performed as described elsewhere [17], and the products were stored at −70°C until further use.
In-house nested PCR. An in-house nested PCR was performed for HCoV 229E and OC43. First-round amplification primers and nested primers were derived from the literature [12] and targeted the nucleocapsid (N) gene, with 1 minor modification: in contrast to the published sequence, we omitted an excess T on position 13 from the nested antisense primer. A 1- tube RT-PCR followed by a second (nested) amplification was applied, as described elsewhere [16], by use of a PE 9600 Thermocycler (Perkin Elmer). PCR products were visualized on an ethidium bromide-stained agarose gel by use of UV illumination. A 5-µL 100-bp marker was used to control fragment lengths.
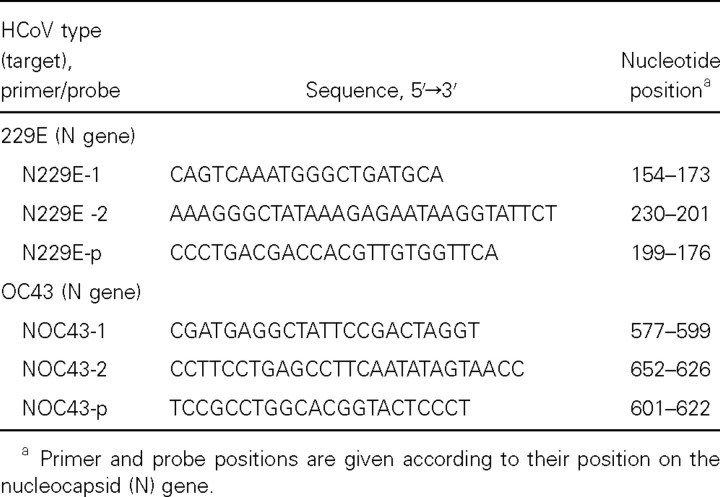
Taqman-based real-time PCR. Type-specific primers and probes for HCoV 229E and OC43 were selected by use of primer express software (PE Applied Biosystems) and were based on the genomic regions of high conservation of the N gene. The forward and reverse primers (N229E-1, N229E-2, NOC43-1, and NOC43-2) and probes (N229E-p and NOC43-p) that were used are shown in table 1. The primers and probes for HCoV 229E and OC43 were tested for possible interactions, to make sure that they could be used in combination. After optimization of the primer and probe concentrations, specimens were assayed in duplicate in a 25-µL reaction mixture containing 5 µL of cDNA, 12.5 µL of 2× Taqman Universal PCR Master Mix (PE Applied Biosystems), 150 nmol/L and 450 nmol/L HCoV 229E and OC43 forward primers, respectively, 150 nmol/L and 450 nmol/L HCoV 229E and OC43 reverse primers, respectively, 50 nmol/L HCoV 229E probe, and 100 nmol/L HCoV OC43 probe. The fluorogenic probes that can be labeled with different fluorogenic dyes were both labeled with a 5′ reporter dye, 6-carboxy-fluorescein, and with a 3′ quencher dye, 6-carboxytetramethyl- rhodamine. Amplification and detection were performed by use of the ABI Prism 7700 sequence-detection system by use of the following conditions: 2 min at 50°C to acquire optimal AmpErase UNG activity and 10 min at 95°C to activate AmpliTaq Gold DNA polymerase, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. The primers and probes for the SARS-associated coronavirus were used, targeting the polymerase gene, as recently described elsewhere [18].The primers and probes for HCoV 229E and OC43 and the SARS-associated coronavirus were tested for possible interactions, to make sure that they could be used in combination in a multiplex assay.
Table 1.
Selected primers and probes for the real-time reverse-transcriptase polymerase chain reaction of human coronavirus (HCoV)-229E and HCoV-OC43.
Virus quantification. To estimate the quantity of the virus load, virus particles were expressed as relative units (RU). Above a threshold cycle of 36, the quantitative value of RNA copies can no longer be considered to be accurate. Therefore, every value above a threshold cycle of 36 and below the detection limit threshold cycle of 45 was assumed to be 2. Every amplification cycle represents a 2-fold increase in viral RNA copies. The virus load was expressed as a 2-fold increase per cycle, relative to a baseline value of 2 copies at a threshold cycle of 36 (RU = 236-threshold cycle).
Results
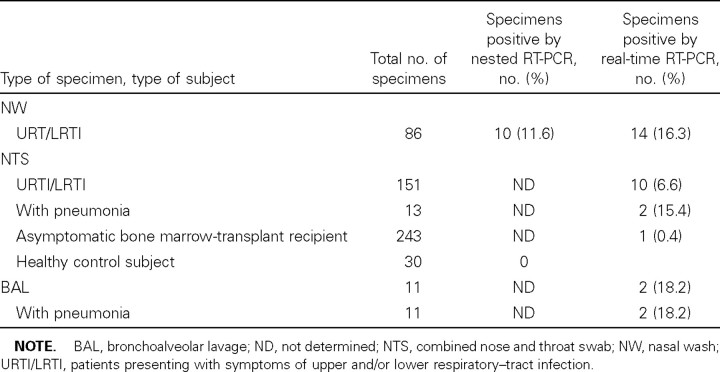
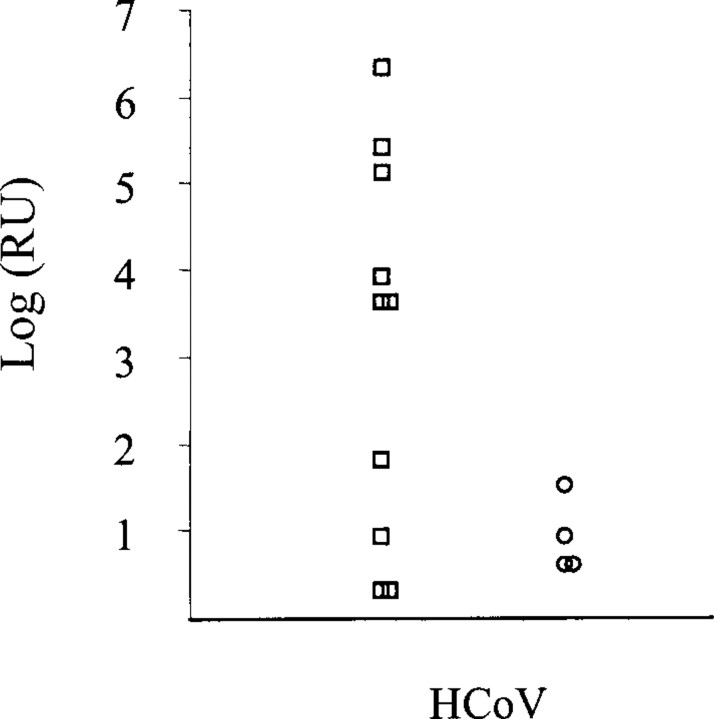
Sensitivity and specificity. Use of limiting-dilution series showed that, for HCoV, the in-house nested PCR and the realtime RT-PCR have similar sensitivity. To compare the sensitivity of the in-house nested PCR with that of the real-time RT-PCR, on clinical specimens, for HcoV, a total of 86 NW specimens obtained from asthmatic and otherwise healthy subjects with symptoms of upper and/or lower RTI were analyzed for HCoV by both the in-house nested PCR and the real-time RT-PCR. As shown in table 2, 14 of 86 specimens were found to be positive by real-time RT-PCR, compared with 10 of 86 specimens tested by the in-house nested PCR. The real-time RTPCR performed better on the specimens containing a low virus load, compared with the nested PCR (figure 1).
Table 2.
Detection of human coronavirus (HCoV) by real-time reverse-transcriptase polymerase chain reaction (RT-PCR) and/or nested RT-PCR, in clinical specimens.
Figure 1.
Virus quantity expressed as relative units (RU) of human coronavirus (HCoV) that could be detected in the clinical specimens (n = 14). ◯, Clinical specimens tested by Taqman-based real-time polymerase chain reaction (PCR) only; □, clinical specimens tested by both the in-house nested PCR and the Taqman-based real-time PCR.
For HCoV, the real-time RT-PCR was highly specific: none of the other respiratory viruses (rhinovirus 1A, rhinovirus 14, and rhinovirus 16, RSV A Long strain, RSV B 9320, parainfluenza viruses 1–4, influenza virus B/Lee/40, influenza virus A/PR/8/34, coxsackievirus A9, echovirus 12, or the SARS-associated coronavirus) were detected by the real-time RT-PCR assay. To evaluate the possibility of clinically false-positive results, NTSs were obtained from 30 asymptomatic subjects during the winter season. HCoV was not detected in any of the NTSs by real-time RT-PCR (table 2).
The real-time RT-PCR for HCoV could successfully be combined with a real-time RT-PCR for SARS-associated coronavirus. Use of limiting-dilution series using a single (primers and probes for the SARS-associated coronavirus only) and multiplex (combination of primers and probes for the HCoV and SARSassociated coronavirus) format showed similar sensitivity in the detection of SARS-associated coronavirus RNA and HCoVs.
Detection in clinical specimens and control specimens. To evaluate the real-time RT-PCR for HCoV, we analyzed a total of 261 clinical specimens obtained at the hospital's virology laboratory: (1) 86 NW specimens and 151 NTSs were obtained from patients presenting with symptoms of upper and/or lower RTI, and (2) 11 BAL specimens and 13 NTSs were obtained from patients admitted to the hospital with pneumonia. Moreover, 243 control NTSs were evaluated from bone marrow-transplant recipients without symptoms of RTI. In total, 28 (11%) of 261 clinical specimens tested positive for HCoV. HCoV was detected in the BAL specimens from 2 (18.2%) of 11 patients. In addition, 2 (15.4%) of 13 NTSs obtained from patients admitted to the hospital with pneumonia tested positive for HCoV (table 2). In contrast, HCoV RNA was detected in only 1 (0.4%) of 243 NTSs that were obtained at set time points from patients without obvious symptoms of RTI (table 2).
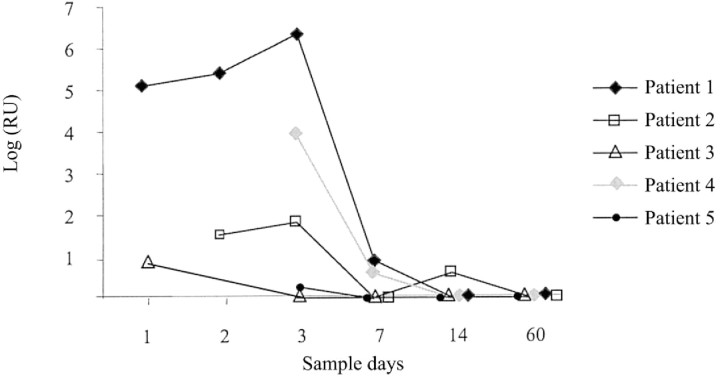
Five patients with symptoms of upper RTI and whose specimens tested positive for coronavirus by real-time RT-PCR were followed during the course of their infections. A total of 24 NW specimens were obtained just after the presentation of common cold symptoms, at several time points up to 60 days. The virus load was expressed as RU (RU = 236-threshold cycle). As shown in figure 2, we were able to detect and quantify HCoV in NW specimens up to 7 days after the initial presentation of common cold symptoms in 3 patients, and up to 14 days in 1 patient. In 1 patient (patient 5), the virus load was below the level of reliable quantitation.
Figure 2.
Longitudinal follow-up of 5 patients with either human coronavirus OC43 or 229E infection. Quantitative analysis was performed by use of the multiplex Taqman-based polymerase chain reaction. The quantity is expressed on the Y-axis as relative units (RU). RU = 236-threshold cycle
Discussion
Our findings have demonstrated that HCoV is frequently detected in clinical specimens obtained at the hospital's virology laboratory from patients presenting with RTI. The novel realtime RT-PCR assay allows rapid and specific detection of HCoVs in patients with various presentations of RTI.
Since there is increasing evidence suggesting that either the known or newly identified HCoVs might be involved in moresevere disease, there is a need for more-rapid and more-reliable diagnostic tools. At present, a great deal of attention has been directed toward patients with SARS. A novel coronavirus that has been identified in the majority of patients is the primary cause of SARS [10, 11, 18–20]. Genetic characterization of this novel coronavirus shows considerable differences between it and HCoVs 229E and OC43 [18]. The real-time RT-PCR described here can detect the novel SARS-associated coronavirus when used in a multiplex format. However, it has yet to be determined whether this is a favorable format, since the clinical presentation of SARS differs from the assumed clinical presentation of HCoV infection. However, for example, advanced age and underlying disease have also been associated with a more severe presentation of HCoV infection [8, 9]. Also, in case reports, HCoV has been associated with pneumonia after autologous bone marrow transplantation, and we recently identified HCoV by use of nested PCR in the BAL specimen of a severely immunocompromised patient with pneumonia [3, 5]. Interestingly, in the present study, we detected HCoV by realtime RT-PCR in the BAL specimens from 2 patients presenting with severe pneumonia and in the NTSs from patients admitted to the hospital with pneumonia, which again suggests that HCoV may be the cause of severe disease in some patients.
By use of molecular detection methods, Nicholson et al. [21] have already shown that 26% of upper RTI in elderly people living at home are due to HCoVs, and the identification of a recent community outbreak of HCoV OC43 in France was facilitated by the use of RT-PCR [7–11]. Although they are valuable in a research setting, these methods are less suitable for routine laboratory detection, because they still require timeconsuming sample handling and post-PCR analysis and are consequently prone to contamination. Besides being rapid, the real-time RT-PCR assay has the advantage of a standardized protocol that can easily be applied to the detection of other respiratory viruses: the RT-PCR can be performed under uniform amplification conditions, thereby using target-specific primerand probe sets.
Another disadvantage is that most studies using RT-PCR for the detection of HCoV lack proper control groups to evaluate the clinical value of a positive result by RT-PCR. To gain insight into the relevance of a positive result by this assay, we followed 5 symptomatic patients during the course of coronavirus infection and also obtained specimens from asymptomatic individuals. The follow-up of the 5 symptomatic patients showed that HCoV RNA could be detected by real-time RT-PCR up to 14 days after infection. Moreover, we tested specimens obtained from patients without obvious signs or symptoms of RTI. None of the specimens obtained from healthy individuals contained HCoV RNA. At 1 time point, just after the bone marrow transplantation, we detected HCoV in an NTS from a bone marrow-transplant recipient without obvious upper and/ or lower RTI. It might be that the patient was suffering from a minor cold and that these symptoms remained unnoticed. From these results we have concluded that an HCoV-positive finding by real-time RT-PCR in a specimen obtained from a symptomatic patient has diagnostic significance.
Diagnostic tests for HCoVs are not frequently used in the routine setting. Serologic testing methods do not allow rapid identification of virus, and, although both HCoV 229E and OC43 can be propagated on specialized cells, the approach lacks sensitivity, is time-consuming, and often requires the expertise of a specialist. In addition, virus isolation is often considered to be redundant and without clinical consequence, since HCoVs are thought to be mainly associated with the common cold syndrome. It is likely that, as a result, the precise role that coronaviruses play in RTIs is greatly underestimated because of the lack of practical diagnostic tools.
We realize that the specimens analyzed in the present study, which were obtained at the hospital's virology laboratory, probably represent specimens from a selected group of patients in which a respiratory virus is considered to be a possible pathogen on clinical grounds. The results, however, indicate that HCoV is frequently detected and that the novel real-time RT-PCR assay provides a tool for large-scale epidemiological studies to further clarify the role that coronavirus infection plays in RTI in humans.
Acknowledgment
We thank H.W. Doerr (Institute of Medical Virology, Johann Wolfgang Goethe University, Frankfurt am Main, Germany) for the gift of severe acute respiratory syndrome-associated coronavirus.
References
- 1. Makela MJ, Puhakka T, Ruuskanen O, et al. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 199836:539–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. El Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin Infect Dis. 200031:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Folz RJ, Elkordy MA. Coronavirus pneumonia following autologous bone marrow transplantation for breast cancer. Chest. 1999115:901–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McIntosh K, Chao RK, Krause HE, Wasil R, Mocega HE, Mufson MA. Coronavirus infection in acute lower respiratory tract disease of infants. J Infect Dis. 1974130:502–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Elden LJ, van Kraaij MG, Nijhuis M, et al. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin Infect Dis. 200234:177–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wenzel RP, Hendley JO, Davies JA, Gwaltney JM., Jr Coronavirus infections in military recruits: three-year study with coronavirus strains OC43 and 229E. Am Rev Respir Dis. 1974109:621–4 [DOI] [PubMed] [Google Scholar]
- 7. Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Infect Dis. 200336:985–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falsey AR, McCann RM, Hall WJ, et al. The “common cold” in frail older persons: impact of rhinovirus and coronavirus in a senior daycare center. J Am Geriatr Soc. 199745:706–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Falsey AR, Walsh EE, Hayden FG. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J Infect Dis. 2002185:1338–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poutanen SM, Low DE, Henry B, et al. Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003348:1995–2005 [DOI] [PubMed] [Google Scholar]
- 11. Tsang KW, Ho PL, Ooi GC, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003348:1948–51 [DOI] [PubMed] [Google Scholar]
- 12. Myint SH, Johnston SL, Sanderson G, Simpson H. Evaluation of nested polymerase chain methods for the detection of human coronaviruses 229E and OC43. Mol Cell Probes. 19948:357–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vabret A, Mouthon F, Mourez T, Gouarin S, Petitjean J, Freymuth F. Direct diagnosis of human respiratory coronaviruses 229E and OC43 by the polymerase chain reaction. J Virol Methods. 200197:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arbour N, Day R, Newcombe J, Talbot PJ. Neuroinvasion by human respiratory coronaviruses. J Virol. 200074:8913–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pitkaranta A, Virolainen A, Jero J, Arruda E, Hayden FG. Detection of rhinovirus, respiratory syncytial virus, and coronavirus infections in acute otitis media by reverse transcriptase polymerase chain reaction. Pediatrics. 1998102:291–5 [DOI] [PubMed] [Google Scholar]
- 16. Nijhuis M, Boucher CA, Schuurman R. Sensitive procedure for the amplification of HIV-1 RNA using a combined reverse-transcription and amplification reaction. Biotechniques. 199519:178–80 182 [PubMed] [Google Scholar]
- 17. van Elden LJ, Nijhuis M, Schipper P, Schuurman R, van Loon AM. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J Clin Microbiol. 200139:196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003348:1967–76 [DOI] [PubMed] [Google Scholar]
- 19. Kuiken T, Fouchier RA, Schutten M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003362:263–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003348:1986–94 [DOI] [PubMed] [Google Scholar]
- 21. Nicholson KG, Kent J, Hammersley V, Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population based study of disease burden. BMJ. 1997315:1060–4 [DOI] [PMC free article] [PubMed] [Google Scholar]