Abstract
Quantitative real-time polymerase chain reaction (qRT-PCR) is a common and robust tool for accurate quantification of mRNA transcripts. To normalize results, a housekeeping gene ([HKG], reference gene or endogenous control gene) is mandatory. Soybean aphid, Aphis glycines Matsumura (Hemiptera: Aphididae), is a significant soybean, Glycine max (L.) Merr., pest, yet gene expression and functional genomics studies are hindered by a lack of stable HKGs. We evaluated seven potential HKGs (SDFS, succinate dehydrogenase flavoprotein subunit; EF1a, elongation factor-1α; HEL, helicase; GAPDH, glyceraldehyde-3 phosphate dehydrogenase; RPS9, ribosomal protein S9; TBP, TATA-box binding protein; and UBQ, ubiquitin-conjugating protein) to determine the most efficient HKGs that have stable expression among tissues, developmental stages, and aphids fed on susceptible and host plant–resistant soybean. HKG stability was determined using GeNorm and NormFinder. Results from three different experimental conditions revealed high stability of TBP compared with the other HKGs profiled across the samples assayed. RPS9 showed stable expression among aphids on susceptible and resistant plants, whereas EF1a showed stable expression in tissues and developmental stages. Therefore, we recommend the TBP as a suitable HKG for efficient normalization among treatments, tissues, and developmental stages of A. glycines. In addition, RPS9 may be used for host-plant resistance experiments and EF1a could be considered for testing differential expression across tissues or developmental stages. These results will enable a more accurate and reliable normalization of qRT-PCR data in A. glycines.
Keywords: housekeeping gene, quantitative real-time polymerase chain reaction, normalization, Aphis glycines
Quantitative real-time PCR (qRT-PCR) has transformed genetic research (gene expression analysis and molecular diagnostics) because of its rapidity, sensitivity, and reproducibility in quantifying mRNA transcripts (Wong and Medrano 2005, Espy et al. 2006, Bustin 2010). The ability of detecting transcripts expressed at low levels by qRT-PCR has made it a standard protocol for gene expression analysis, thus replacing the conventional mRNA quantification methods (e.g., Northern blot analysis, competitive RT-PCR, RNase protection assay, and microarrays) (Vandesompele et al. 2002, Klie and Debener 2011).
Given the sensitivity and reproducibility of qRT-PCR technique, a suitable housekeeping gene (HKG) is a prerequisite for accurate quantification of mRNA transcripts at a given condition (Vandesompele et al. 2002). Also referred as reference genes or internal control genes, HKGs are thought to be expressed constitutively across different physiological conditions because they are involved in basic functions of a cell (Butte et al. 2001). Because of their inherent property of stable expression across different treatments, various tissues, and developmental stages, HKGs are widely accepted as internal controls in mRNA quantification studies (Bustin 2010). However, several normalization studies using qRT-PCR have demonstrated the variability of HKG expression (Vandesompele et al. 2002, Klie and Debener 2011), supporting the hypothesis that there is no universal reference gene for all biological systems (Gutierrez et al. 2008). Although a few HKGs have been reported in insects, a species-specific HKG is recommended for precise mRNA quantification (Scharlaken et al. 2008, de Boer et al. 2009, Hiel et al. 2009, Hornakova et al. 2010, Jiang et al. 2010, Lord et al. 2010, Mamidala et al. 2011, Rajarapu et al. 2011).
The soybean aphid, Aphis glycines Matsumura (Hemiptera: Aphididae), a native of Asia, has emerged as a serious invasive pest in North America and has caused huge losses to soybean, Glycine max (L.) Merr., production (Heimpel et al. 2010, Ragsdale et al. 2011, Tilmon et al. 2011). The traditional measures to control A. glycines such as the use of chemicals and resistant plants have been hindered because of the high cost and evolution of virulent biotypes, respectively (Kim et al. 2008, Song and Swinton 2009, Hill et al. 2010, Tilmon et al. 2011). Development of novel management strategies necessitates the exploration of the physiology of A. glycines; studies have been severely lacking because of the recent North American invasion (first found in year 2000). Our long-term research goal is focused on understanding the genetic basis of biotype evolution and to explore novel targets based on molecular physiology. However, validation of any gene expression data requires identification and validation of stable HKGs. Here, we have evaluated the stability of seven commonly used reference genes (SDFS, succinate dehydrogenase flavoprotein subunit; EF1a, elongation factor-1α; HEL, helicase; GAPDH, glyceraldehyde-3 phosphate dehydrogenase; RPS9, ribosomal protein S9; TBP, TATA-box binding protein; and UBQ, ubiquitin-conjugating protein). These HKGs were tested across various A. glycines samples, including aphids fed on resistant and susceptible plants; adult tissues; and developmental stages. Recommendations were suggested as to their utility for A. glycines gene expression studies.
Materials and Methods
cDNA Library Construction and Sequencing.
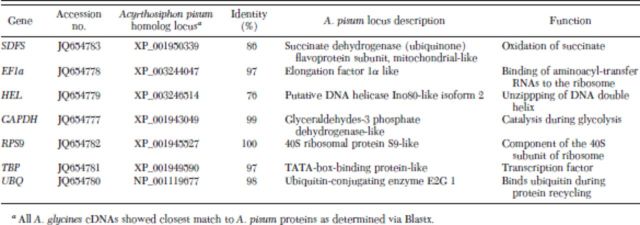
The cDNA library from A. glycines adults was constructed using cDNA Synthesis System kit (Roche Applied Science, Indianapolis, IN) and was sequenced on 454 GS Titanium platform. More details on library construction, sequencing, and data analysis are provided previously (Bai et al. 2010). The cDNA library of A. glycines contained 19,293 high-quality transcripts in total. Homology searches for transcript sequences were performed using Blast2GO software [E-value cut-off 10−3] (Conesa et al. 2005, Conesa and Götz 2008; Götz et al. 2008, 2011). Based on information available on commonly used reference genes in the literature, we selected cDNA contigs of various candidate reference genes for A. glycines (Table 1). The identity of putative cDNAs was further confirmed by Blastx search in GenBank (National Center for Biotechnology Information, Bethesda, MD). The cDNA sequences of candidate reference genes were deposited in GenBank (for accession numbers, see Table 1).
Table 1.
Description of candidate reference genes for qRT-PCR studies in A. glycines
Insect Culture.
For all qRT-PCR experiments, A. glycines insects were obtained from a laboratory colony, referred as biotype 1 (B1) that originated from insects collected from Urbana, IL (40° 06′ N, 88° 12′ W) in 2000 (Hill et al. 2004). At Ohio Agricultural Research and Development Center (Wooster, OH), a laboratory population of these insects is maintained on susceptible soybean seedlings [SD01-76R (2)] in a rearing room at 23–25°C and a photoperiod of 15:9 (L:D) h.
qRT-PCR Analysis in A. glycines Fed With Resistant and Susceptible Plants.
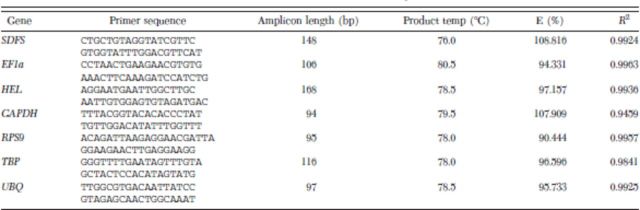
Freshly hatched first-instar nymphs of A. glycines (60–70 individuals) were allowed to feed on resistant [LD-05 16060, contains Rag1] and susceptible soybean [SD01-76R (2)] plants (Tinsley et al. 2012). After 12 h of feeding, insects were collected and placed in −80°C. There were three biological replications for each treatment. The collected insect samples were processed for total RNA extraction by using TRI reagent (Molecular Research Center Inc, Cincinnati, OH), following the protocol provided by the manufacturer. RNA samples were treated with TURBO DNase (Applied Biosystems/Ambion, Austin, TX) to remove any DNA contamination. Using iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA), first strand cDNA was prepared with 500 ng of RNA. qRT-PCR reactions were performed with iQ SYBR Green super mix on a CFX-96 thermocycler system (Bio-Rad Laboratories) (Bansal et al. 2011). Specific primers for each candidate reference gene were designed using Beacon Designer version 7.0 (Premier Biosoft, Palo Alto, CA) (Table 2). Each reaction was performed with 1 μl of cDNA, 0.5 μM of each primer, and 12.5 μl of iQ SYBR Green super mix in a 25-μl total volume. Each reaction was done in duplicate in a 96-well optical-grade PCR plates, sealed with optical sealing tape (Bio-Rad Laboratories). The PCR amplifications were done with the following cycling conditions: one cycle at 95°C (3 min), followed by 35 cycles of denaturation at 95°C (30 s), annealing and extension at 55°C for 45 s. Finally, melt curve analyses were done by slowly heating the PCR mixtures from 55 to 95°C (1°C per cycle of 10 s) with simultaneous measurements of the SYBR Green signal intensities. PCR amplification efficiencies and correlation coefficients (R2) for each primer pair were calculated as described in Real-Time PCR Applications Guide (catalog no. 170-9799, Bio-Rad Laboratories).
Table 2.
Primer sequences and amplicon characteristics of candidate reference genes for qRT-PCR studies in A. glycines
qRT-PCR Analysis in A. glycines Tissues and Developmental Stages.
To obtain selected tissue samples (gut, fat body, integument, and embryo developing inside adults), A. glycines adults (5 d old) were dissected out in phosphate-buffered saline, pH 8.0, under a dissecting microscope. During dissection, other tissues of A. glycines such as salivary glands and bacteriocytes were discarded. To determine the expression of candidate reference genes in different developmental stages, all the four nymphal and adult samples were collected from insects feeding on susceptible soybean [SD01-76R (2)] plants. Both tissue and developmental stages' samples were processed for total RNA extraction, DNase treatment, first strand cDNA synthesis, and qRT-PCR as described in the previous section. There were two biological and two technical replications for each. The first strand cDNA was prepared with 150 and 500 ng of RNA (DNA free) from tissue and developmental stages samples, respectively.
Stability Analysis of Candidate Reference Genes.
Two software algorithms, i.e., GeNorm (Vandesompele et al. 2002) and Normfinder (Andersen et al. 2004) were used to determine the stability of candidate reference genes. The raw expression values of each gene, calculated by equation 2(−ΔCt) were used as input data for both GeNorm and Normfinder. GeNorm calculated the M-score, and the lower the value for M is indicative of a more stable expression or low variation (Vandesompele et al. 2002). This value is calculated by a geometric averaging of all the reference genes used in the study and mean pairwise variation of a reference gene from other reference genes. It is important to note that the HKGs showing higher M value (M > 1.5) are not considered for normalization studies. NormFinder also determines the expression stability but by taking account of intra- and intergroup variations for candidate reference genes (Andersen et al. 2004). NormFinder provides the stability value for each gene, a direct measure for the estimated expression variation enabling evaluation of the systematic error introduced when using the gene for normalization (Andersen et al. 2004).
Results and Discussion
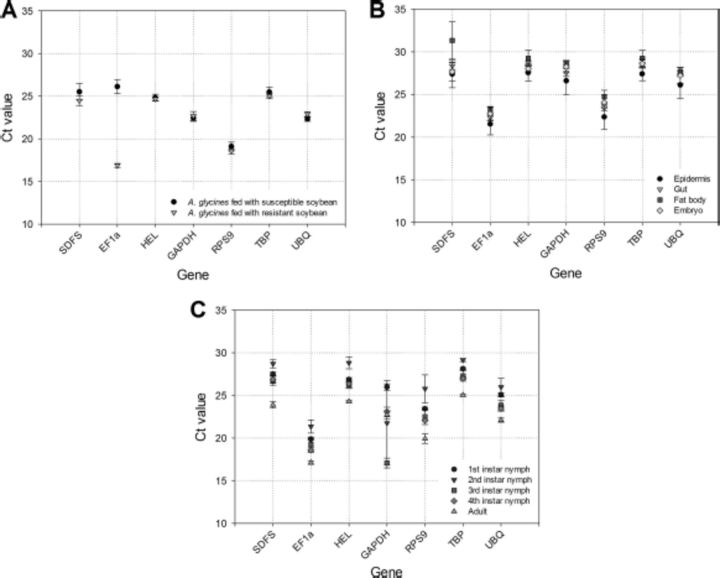
We profiled seven commonly used HKGs (SDFS, EF1a, HEL, GAPDH, RPS9, TBP and UBQ) from a transcriptomic database of A. glycines. The preliminary screening of these HKGs presented a range of Ct values across the treatments (16.87–26.11, A. glycines fed with resistant and susceptible host plants), tissues (21.52–31.33, epidermis, gut, fat body, and embryo), and developmental stages (17.08–29.15, first–fourth-instar nymphs and adults) of A. glycines (Fig. 1). Our qRT-PCR analysis was highly optimized as amplification efficiencies (E) for various primer pairs ranged from 90.44 to 108.16 and R2 were >0.94 (Table 2) (Schmittgen and Livak 2008). We included the melting curve analysis (65–95°C in increments of 0.5°C every 5 s) in qRT-PCR reaction that revealed the lack of any nonspecific product amplification (Supp. Fig. 1 [online only]). To determine a suitable reference gene for gene expression studies in A. glycines, we used GeNorm (version 3.5) and NormFinder that are freely available software (Vandesompele et al. 2002, Andersen et al. 2004).
Fig. 1.
Ct values (±SE) obtained for different HKGs under different experimental conditions. (A) Insects fed with susceptible and resistant plants (B) Various tissues. (C) Different developmental stages of A. glycines.
GeNorm Analysis.
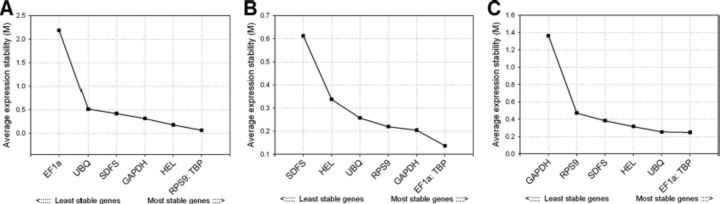
Among the seven reference genes (SDFS, EF1a, HEL, GAPDH, RPS9, TBP, and UBQ), GeNorm revealed TBP to be stably expressed across the treatments, tissues, and developmental stages (Fig. 2A–C, respectively). Although EF1a showed stable expression in both tissues and developmental stages, it had the least stability in insects fed on resistant and susceptible host plants. GeNorm determined the gene stability measure (M) among all the reference genes tested. Except for the EF1a in treatments, all the HKGs assayed in the current study displayed M < 1.5. This trend of dissimilar ranking of reference genes across treatments, tissues, and developmental stages is quite a common phenomenon observed in several gene expression studies in insects (Hiel et al. 2009, Hornakova et al. 2010, Jiang et al. 2010, Lord et al. 2010, Mamidala et al. 2011, Rajarapu et al. 2011). Hence, it is highly recommended to determine the stably expressed reference gene for a given sample as normalization is critical and a significant component of final data interpretation.
Fig. 2.
Average expression stability (M) and ranking of HKGs as calculated by GeNorm. M values and rankings are indicated for insects fed with resistant and susceptible plants (A), various tissues (B), and different developmental stages of A. glycines (C).
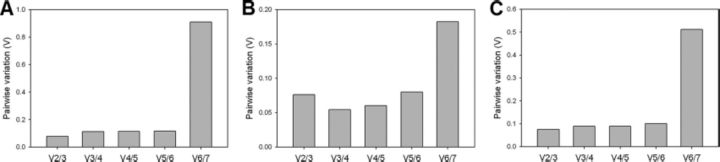
Although a single reference gene which is stable and highly expressed suffices the requirement of quantifying mRNA transcript levels for a gene of interest, it also is recommended to use at least two to three reference genes for effective normalization of gene expression data (Vandesompele et al. 2002). The requirement of optimal number of HKGs can be obtained from the pairwise variation (V), wherein Vandosomplele et al. (2002) proposed a cut-off value (0.15), below which the inclusion of other reference genes are not required. Results in the current study showed that the application of at least two stable reference genes maintains proper normalization irrespective of treatment, tissue, and development stages' samples (Fig. 3A–C, respectively). Because the pairwise variation was calculated by adding candidate genes stepwise according to rankings shown in Fig. 2, two of the most stable genes identified for a particular experimental condition (Fig. 2) should be able to be combined for normalization. The candidate genes having lower stability in a given treatment (Fig. 2) should be avoided for normalization even if it is to be used in combination with other reference gene given the suitability and availability of other reference genes. For example, GeNorm analysis suggests that for qRT-PCR normalization in A. glycines fed with resistant and susceptible soybean, both TBP and RPS9 could be used together. Similarly, for normalization among different developmental stages and among tissues of A. glycines, TBP and EF1a could be combined. It is interesting to note that various HKGs (RPS9, GAPDH, HEL, and UBQ) were not consistently ranked in terms of their stability under different experimental conditions, that is also the case observed in other insect studies (Hiel et al. 2009, Hornakova et al. 2010, Jiang et al. 2010, Lord et al. 2010, Mamidala et al. 2011). Therefore, care should be taken in determining which HKG to use depending on experimental conditions.
Fig. 3.
Pairwise variation (V) analysis of the HKGs using GeNorm. The pairwise variation (Vn/n+1) between the normalization factors NFn and NFn+1 (shown along x-axis) is calculated to determine the optimal number of HKGs for normalization in insects fed with susceptible and resistant plants (A), various tissues (B), and different developmental stages of A. glycines (C). Each bar indicates change in normalization when adding reference genes stepwise according to rankings in Fig. 2.
NormFinder Analysis.
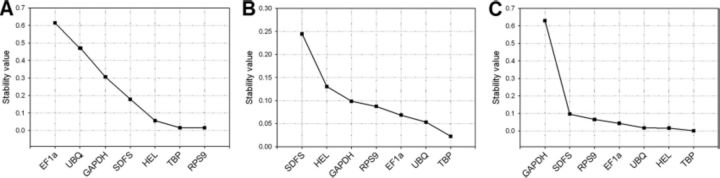
NormFinder showed similar results to GeNorm, wherein TBP was shown to have low stability value (lower variation of gene expression) across all the samples (Fig. 4), further indicating the potential for this gene as a HKG for gene expression studies in A. glycines. Although there are discrepancies between the GeNorm and Normfinder output, TBP was shown to be the best ranking reference gene with both methods among treatments, tissues, and developmental stages (Fig. 2 and 4). Also, both of these analyses displayed RPS9 as an appropriate HKG across the treatments (Figs. 2A and 4A). The different ranking of other HKGs (EF1a, GAPDH, HEL, and UBQ) by both GeNorm and NormFinder clearly demonstrates the expression stability of HKGs is influenced spatially, temporally, and also on experimental treatment.
Fig. 4.
Stability values of HKGs as calculated by Normfinder. Stability values are indicated for insects fed with susceptible and resistant plants (A), various tissues (B), and different developmental stages of A. glycines (C).
In conclusion, the HKG gene TBP showed stable expression across all the samples and did not seem to be influenced by treatments (A. glycines fed with resistant and susceptible host plants), among the various tissues and developmental stages included. Therefore, it could be used to normalize the mRNA transcript levels of candidate genes in A. glycines. The TBP is a transcription factor that binds specifically to a DNA sequence called the TATA box and is well documented for its use as reference gene in several studies (Radonic et al. 2005, Jung et al. 2007, Nygard et al. 2007). In addition, for experiments with A. glycines fed with resistant and susceptible host plants, RPS9 may be considered. Similarly, for normalization among tissues and developmental stages, EF1a also can be used along with TBP. The identified reference genes in the current study may potentially serve as ideal internal controls in other closely related aphid species. However, the order of the gene stability should be revised for using a species specific study.
Acknowledgments
This work was supported by a grant from North Central Soybean Research Program and The Ohio Soybean Council (11-2-04). The CFX-96 thermocycler system was purchased, in part, by an equipment grant from Department of Entomology and Ohio Agricultural Research and Development Center.
References Cited
- Andersen C. L., Jensen J. L., Orntoft T. F. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- Bai X., Zhang W., Orantes L., Jun T. H., Mittapalli O., Mian M.A.R., Michel A. M. 2010. Combining next-generation sequencing strategies for rapid molecular resource development from an invasive aphid species, Aphis glycines. PLoS ONE 5(6): e11370. (doi: 10.1371/journal.pone.0011370). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal R., Hulbert S., Schemerhorn B., Reese J. C., Whitworth R. J., Stuart J. J., Chen M. S. 2011. Hessian fly-associated bacteria: transmission, essentiality, and composition. PLoS ONE 6(8): e23170. (doi: 10.1371/journal.pone.0023170). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A. 2010. Developments in real-time PCR research and molecular diagnostics. Expert Rev. Mol. Diagn. 10: 713–715. [DOI] [PubMed] [Google Scholar]
- Butte A. J., Dzau V. J., Glueck S. B. 2001. Further defining housekeeping, or “maintenance,” genes focus on “a compendium of gene expression in normal human tissues”. Physiol. Genomics 7: 95–96. [DOI] [PubMed] [Google Scholar]
- Conesa A., Götz S. 2008. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- de Boer M., de Boer T., Marien J., Timmermans M., Nota B., van Straalen N., Ellers J., Roelofs D. 2009. Reference genes for QRT-PCR tested under various stress conditions in Folsomia candida and Orchesella cincta (Insecta, Collembola). BMC Mol. Biol. 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espy M. J., Uhl J. R., Sloan L. M., Buckwalter S. P., Jones M. F., Vetter E. A., Yao J. D., Wengenack N. L., Rosenblatt J. E., Cockerill F. R., et al. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19: 165–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S., Arnold R., Sebastián-León P., Martín-Rodríguez S., Tischler P., Jehl M. A., Dopazo J., Rattei T., Conesa A. 2011. B2G-FAR, a species centered GO annotation repository. Bioinformatics 27: 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S., Garcia-Gomez J. M., Terol J., Williams T. D., Nagaraj S. H., Nueda M. J., Robles M., Talón M., Dopazo J., Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36: 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L., Mauriat M., Guenin S., Pelloux J., Lefevre J. F., Lovet R., Rusterucci C., Moritz T., Guerineau F., Bellini C., et al. 2008. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6: 609–618. [DOI] [PubMed] [Google Scholar]
- Heimpel G. E., Frelich L. E., Landis D. A., Hopper K. R., Hoelmer K. A., Sezen Z., Asplen M. K., Wu K. M. 2010. European buckthorn and Asian soybean aphid as components of an extensive invasional meltdown in North America. Biol. Invasions 12: 2913–2931. [Google Scholar]
- Hiel M.B.V., Wielendaele P. V., Temmerman L., Soest S. V., Vuerinckx K., Huybrechts R., Broeck J. V., Simonet G. 2009. Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol Biol. 10: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. B., Li Y., Hartman G. L. 2004. Resistance to the soybean aphid in soybean germplasm. Crop Sci. 44: 98–106. [Google Scholar]
- Hill C. B., Crull L., Herman T., Voegtlin D. J., Hartman G. L. 2010. A new soybean aphid (Hemiptera: Aphididae) biotype identified. J. Econ. Entomol. 103: 509–515. [DOI] [PubMed] [Google Scholar]
- Hornakova D., Matouskova P., Kindl J., Valterova I., Pichova I. 2010. Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal Biochem. 397: 118–120. [DOI] [PubMed] [Google Scholar]
- Jiang H., Liu Y., Tang P., Zhou A., Wang J. 2010. Validation of endogenous reference genes for insecticide-induced and developmental expression profiling of Liposcelis bostsrychophila (Psocoptera: Liposcelidae). Mol. Biol. Rep. 37: 1019–1029. [DOI] [PubMed] [Google Scholar]
- Jung M., Ramankulov A., Roigas J., Johannsen M., Ringsdorf M., Kristiansen G., Jung K. 2007. In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC Mol. Biol. 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Hill C. B., Hartman G. L., Mian M.A.R., Diers B. W. 2008. Discovery of soybean aphid biotypes. Crop Sci. 48: 923–928. [Google Scholar]
- Klie M., Debener T. 2011. Identification of superior reference genes for data normalisation of expression studies via quantitative PCR in hybrid roses (Rosa hybrida). BMC Res. Notes 4: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord J. C., Hartzer K., Toutges M., Oppert B. 2010. Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J. Microb. Methods 80: 219–221. [DOI] [PubMed] [Google Scholar]
- Mamidala P., Rajarapu S. P., Jones S. C., Mittapalli O. 2011. Identification and validation of reference genes for quantitative real-time PCR in the bed bug. J. Med. Entomol. 48: 947–951. [DOI] [PubMed] [Google Scholar]
- Nygard A. B., Jorgensen C. B., Cirera S., Fredholm M., M. 2007. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol. Biol. 8: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonic A., Thulke S., Bae H. G., Muller M. A., Siegert W., Nitsche A. 2005. Reference gene selection for quantitative real-time PCR analysis in virus infected cells: SARS corona virus, Yellow fever virus, Human Herpesvirus-6, Camelpox virus and Cytomegalovirus infections. Virol. J. 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale D. W., Landis D. A., Brodeur J., Heimpel G. E., Desneux N. 2011. Ecology and management of the soybean aphid in North America. Annu. Rev. Entomol. 56: 375–399. [DOI] [PubMed] [Google Scholar]
- Rajarapu S. P., Mamidala P., Mittapalli O. 2011. Validation of reference genes for gene expression studies in emerald ash borer (Agrilus planipennis). Insect Sci. 19: 41–46. [Google Scholar]
- Scharlaken B., de Graaf D. C., Goossens K., Brunain M., Peelman L. J., Jacobs F. J. 2008. Reference gene selection for insect expression studies using quantitative real-time PCR: the head of the honeybee, Apis mellifera, after a bacterial challenge. J. Insect Sci. 8: 33. [Google Scholar]
- Schmittgen T. D., Livak K. J. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3: 1101–1118. [DOI] [PubMed] [Google Scholar]
- Song F., Swinton S. M. 2009. Returns to integrated pest management research and outreach for soybean aphid. J. Econ. Entomol. 102: 2116–2125. [DOI] [PubMed] [Google Scholar]
- Tilmon K. J., Hodgson E. W., O'Neal M. E., Ragsdale D. W. 2011. Biology of the soybean aphid, Aphis glycines (Hemiptera: Aphididae) in the United States. J. Integr. Pest Manag. 2(2). (doi: 10.1603/IPM10016). [DOI] [Google Scholar]
- Tinsley N. A., Steffey K. L., Estes R. E., Heeren J. R., Gray M. E., Diers B. W. 2012. Field-level effects of preventative management tactics on soybean aphids (Aphis glycines Matsumura) and their predators. J. Appl. Entomol. 136: 361–371. [Google Scholar]
- Vandesompele J., Preter K. D., Pattyn F., Poppe B., Roy N. V., Paepe A. D., Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: research0034.1–0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. L., Medrano J. F. 2005. Real-time PCR for mRNA quantitation. Biotechniques 39: 75–85. [DOI] [PubMed] [Google Scholar]