Abstract
BackgroundThe clinical features and incidence of human coronavirus (HCoV) infections in chronically ill older adults need better definition
MethodsHCoV infection was determined on the basis of a 4-fold increase in serum antibody and the detection of HCoV by reverse-transcription polymerase chain reaction. Laboratory-documented influenza (LDI) was detected by serologic assay and culture. HCoV illnesses were compared with other acute respiratory illnesses identified by active surveillance, during the 1998–99 winter respiratory-virus season, of 2215 patients with chronic obstructive pulmonary disease who were ⩾50 years old and who received influenza vaccines
ResultsHCoV-229E and HCoV-OC43 were associated with 90 (14%) of 665 illnesses (HCoV-229E in 22, HCoV-OC43 in 67, and both in 1), LDI with 107 (16%) of 678 illnesses. In multivariate logistic regression analysis, myalgia was less likely with HCoV infection than with LDI (OR, 0.27 [95% confidence limit, 0.13–0.58]). A majority of these HCoV and LDI illnesses exhibited each of 11 symptoms and signs of acute respiratory illness. Spirometric results worsened most often with LDI, and many acute respiratory illnesses, regardless of etiology, were associated with hospitalization. A total of 8 illnesses were associated with HCoV-NL63, 1 with HCoV-HKU1
ConclusionsThe frequencies of HCoV and LDI illnesses were similar. HCoV illness was less severe than LDI illness, was accompanied by multiple respiratory and systemic symptoms, and was associated with hospitalization
Human coronavirus (HCoV) is an enveloped, single-stranded, positive-sense RNA virus that since the mid-1960s has been known to be a cause of the common cold [1]. The earliest strains identified—HCoV-229E and HCoV-OC43—belong to groups I and II, respectively. The emergence of severe acute respiratory syndrome (SARS) [2, 3] necessitated a rethinking of the role that coronaviruses play as a potential cause of more-severe respiratory illness. Even before the emergence of SARS, as well thereafter, HCoV infection has been viewed as a contributor to exacerbations of underlying chronic obstructive pulmonary disease (COPD), asthma, congestive heart failure, and severe illnesses requiring emergency care and hospitalization of patients with chronic medical conditions [4–16]
The renewed interest in coronaviruses has led to the identification of 2 new coronavirus strains, HCoV-NL63 and HCoV-HKU1 [17–33]. HCoV-NL63 is a group I strain that is more readily culturable from clinical specimens in tissue culture, whereas HCoV-HKU1 is a group II coronavirus; both have been reported from geographically diverse regions but were first described in the Netherlands and Hong Kong, respectively [17–33], and they have been associated with a range of illnesses, from mild, febrile upper-respiratory-tract illnesses to severe illness, in hospitalized patients with bronchiolitis and pneumonia. Reports describe these newer strains as occurring predominantly in children but also in adults [23–25, 34]. Among hospitalized patients in Hong Kong who have acute respiratory-tract infections, coronaviruses—including (in order of decreasing frequency) HCoV-OC43, HCoV-NL63, HCoV-HKU1, and HCoV-229E—have been detected, by reverse-transcription polymerase chain reaction (RT-PCR), in 2% of patients, who had an age range of 1 month–88 years [30]. Kistler et al. [34] have reported the presence of HCoV-NL63, HCoV-HKU1, and HCoV-OC43 in adults with and without asthma who were evaluated early after the onset of common-cold symptoms
Prototype strains of HCoV groups I and II have been adapted to tissue culture, but wild-type strains are not readily culturable and therefore are not amenable to isolation from clinical specimens. Most earlier studies relied on serologic methods to detect HCoV infection. More recently, RT-PCR has been used to detect HCoV-229E and HCoV–OC43 RNA in respiratory secretions during acute infection. RT-PCR has confirmed that HCoV-229E and HCoV–OC43 can be associated with acute respiratory illnesses [15, 16]; however, comparisons of concordance between serologic results, detection of the virus by RT-PCR, and correlation with clinical symptoms are needed
In Department of Veterans Affairs Cooperative Study 448, older adults with COPD were randomized to receive trivalent inactivated influenza-virus vaccine (TIV) intramuscularly either with or without intranasal live-attenuated, cold-adapted influenza-virus vaccine (LAIV), for the 1998–1999 winter respiratory-virus season and then were monitored prospectively for the occurrence of acute respiratory illness [35]. The present study (1) compares the clinical characteristics and significance of HCoV-229E and HCoV-OC43, the predominant HCoV strains in the present study, with those of laboratory-documented influenza (LDI) and all other acute respiratory illnesses; (2) compares the frequencies of HCoV-229E and HCoV-OC43 infection detected by RT-PCR and serologic assay; and (3) characterizes illnesses that were found to be associated with either HCoV-NL63 or HCoV-HKU1
Methods
Patients ⩾50 years of age who met spirometric criteria for COPD were recruited at 20 VA Medical Centers, as described elsewhere [35]. All patients gave written informed consent, and the study received approvals by responsible committees on human experimentation and followed procedures in accordance with the recommendations found in the Helsinki Declaration of 1975 as revised in 1983
Pulmonary-function tests included forced expiratory volume in 1 s (FEV1), percentage of predicted FEV1, and the ratio of FEV1 to forced vital capacity (FEV1:FVC) [35–37]. Spirometric criteria for COPD were an FEV1 that was ⩽80% of the predicted value and an FEV1:FVC that was <0.70
Immunizations occurred from October 1998 through January 1999. All patients received TIV for the 1998–1999 influenza season. Patients were randomly assigned, in a 1:1 ratio, to receive intranasally either LAIV (MedImmune Vaccines [formerly Aviron]) (TC group) or placebo (TP group) [35]. From November 1998 through April 1999, the patients were evaluated when they had either 3 symptoms of acute respiratory illness or fever accompanied by 2 symptoms of acute respiratory illness. A nasal and oropharyngeal (NOP) swab specimen and a serum specimen were obtained for detection of virus and antibody, respectively, and a second serum specimen was obtained 3–4 weeks after the onset of acute respiratory illness; all specimens were stored at −70°C
LDI was defined as the sudden onset of respiratory illness plus (1) an NOP-swab culture positive for wild-type influenza virus A or B and/or (2) a 4-fold increase in the end-point titer of serum hemagglutination inhibition (HAI) antibodies to influenza A or B, measured as described elsewhere [35, 38, 39]. A change in titer of serum HAI antibody was not used to diagnose an LDI-caused illness if the acute respiratory illness occurred <28 days after vaccination for influenza
HCoV-associated illness was defined as the sudden onset of respiratory illness plus (1) an NOP-swab culture positive for either HCoV-229E or HCoV-OC43, by RT-PCR using the SARS/Coronaplex kit (Prodesse), which detects SARS coronavirus and HCoV-OC43 and HCoV-229E, and/or (2) a 4-fold increase in the end-point titer of serum antibody to HCoV, by ELISA. RNA for RT-PCR assay was extracted from NOP specimens according to the manufacturer’s recommended procedure (QIAamp Kit; Qiagen). Available NOP specimens from illnesses that had a 4-fold rise in antibody to HCoV-NL63 were cultured in LLC-MK2 cells (a gift from Lia van der Hoek, University of Amsterdam, the Netherlands). RNA extracted from supernatant fluids by use of the QIAmp Ultrasens Virus Kit (Qiagen) was subjected to RT-PCR by use of the GeneAmp RNA PCR Kit (Applied Biosystems) and Spike (S) gene nested primers for amplification of cDNA. The HCoV-NL63 S gene–specific outer primers comprised positions 22557–22582 (positive sense) and positions 23043–23063 (antisense), as described elsewhere [20]. The inner primers, designed by use of the Netherlands HCoV-NL63 isolate (GenBank accession number AY567487) comprised positions 22519–22538 (positive sense) and positions 22909–22928 (antisense). Specimens with viral bands confirmed by gel electrophoresis of the RT-PCR products were considered to be positive. RT-PCR products were purified by use of the QIA quick PCR Purification Kit Protocol (Qiagen) and were sequenced, with the primers, by Alpha BioLaboratory
The HCoV antigens used for the antibody ELISA were produced as follows. HCoV-229E and HCoV-OC43 were obtained from American Type Culture Collection (ATCC) and were grown in MRC-5 and HCT-8–cell monolayers (ATCC), respectively. HCoV-NL63 was grown in LLC-MK2–cell monolayers. Virus-infected cells were frozen and thawed 3 times, the supernatant fluid was clarified of cell debris by centrifugation, the virus was concentrated by overnight centrifugation, and the virus pellet was resuspended in PBS. The concentrated virus was inactivated by psoralen compound (Sigma), followed by irradiation by long-wavelength UV light, as described elsewhere [40, 41]. Mock antigen was prepared, in the same way, from uninfected cells
His6-tagged recombinant N protein of HCoV-HKU1 was used as antigen in the ELISA to detect antibody to HCoV-HKU1. Expression vector pET-28b (+) (Novagen), encoding the N protein of HCoV-HKU1 cloned into the EcoRI and NotI sites in-frame and downstream from a series of 6 histidine residues, as described elsewhere [27, 28], was a gift from K. Y. Yuen (University of Hong Kong, Hong Kong). The recombinant N protein was expressed by transformation of BL21 (DE3) Singles Competent Cells (Novagen) by the plasmid and was purified by the Ni2+ -loaded HiTrap chelating system (Amersham Biosciences), according to the manufacturer’s instructions. In brief, transformed cells were disrupted, and the protein sample was prepared by isolation of inclusion bodies via sonication and washing. The protein sample was loaded onto the HiTrap chelating HP column prepacked with precharged Ni+2 Sepharose High Performance. The protein then was purified and refolded by serial buffer washes of the column and by liquid chromatography in an FPLC system (Pharmacia LBK Biotechnology), with elution of protein, which was collected in fractions. The fractions were analyzed for the presence of 53-kDa protein by SDS-PAGE. Protein concentration was determined by the Bio-Rad Protein Assay (Bio-Rad Laboratories) and was adjusted to 3 μg/mL in the ELISA. Mock antigen was produced from the same plasmid DNA vector, but without the N protein gene sequence, by the same procedure
Each viral antigen and its respective control were used to coat flat-bottom 96-well Maxisorp Immuno-plates (Nalge Nunc International). The sequence of reagents consisted of serum in a series of 8 2-fold dilutions to generate a broad dose-response curve, mouse anti–human IgG (Fc-specific) conjugated with horseradish peroxidase (Accurate Chemical and Scientific), and peroxidase substrate (KPL). Optical density (OD) was measured at 405 nm by use of a Tecan SLT400 spectrophotometer (Research Triangle Park, North Carolina). The anti-HCoV antibody titer in the ELISA was calculated by the reference-line least-squares-fit method [42]. The cutoff OD was set at 0.3 for all viral antigens, a level that was at least twice the background OD for the respective mock antigens, and corresponded to the linear range of OD versus reciprocal-dilution curves
The first episodes of acute respiratory illness that occurred >7 days after vaccination were included in the clinical analyses and to categorize patients in terms of “illness group.” Subsequent illnesses are not described, both because the earlier illness could affect the characteristics of the later illness and because the number of later illnesses was small. The illness groups comprised those whose first episode of acute respiratory illness was associated with (1) either HCoV-229E or HCoV-OC43, (2) LDI, (3) LDI and either HCoV-229E or HCoV-OC43, or (4) neither LDI nor HCoV-229E nor HCoV-OC43. The respiratory illnesses associated with HCoV-NL63 and HCoV-HKU1 are described individually
The severity of illness was assessed by use of the self-reported, 6 symptom–based, chronic lung disease severity index (CLDSI), which was developed, as part of the Veteran’s Health Study [43–45], to evaluate functional status and well-being and the effects that chronic lung disease has on quality of life. The CLDSI score ranges from 6 (best) to 27 (most severe) [43]. Hospitalizations during the study were identified on the basis of serious-adverse-event forms. Hospitalization was considered to be temporally associated with an acute respiratory illness if it occurred either during the illness or <1 month thereafter
Descriptive statistics were used for the initial analysis of variables; 2×2 χ2 tests or Fisher exact tests were used to compare categorical characteristics of the illness groups, and the Wilcoxon&rank-sum test was used to compare continuous characteristics. P values are reported; as an arbitrary correction for multiple comparisons, P<.01 was considered to be statistically significant. An FEV1 change between study visits was considered to be clinically significant if it was ⩾15% [37, 46]. Univariate logistic regression and χ2 analysis were used to identify the univariate association of illness group with each of 11 clinical symptoms and signs of acute respiratory illness and the influenza-vaccine group. Multivariate analysis using stepwise logistic regression identified factors independently associated with HCoV infections. P<.10 was necessary for the factor to be included in the logistic model, and P<.05 was necessary for it to be retained in the model
Results
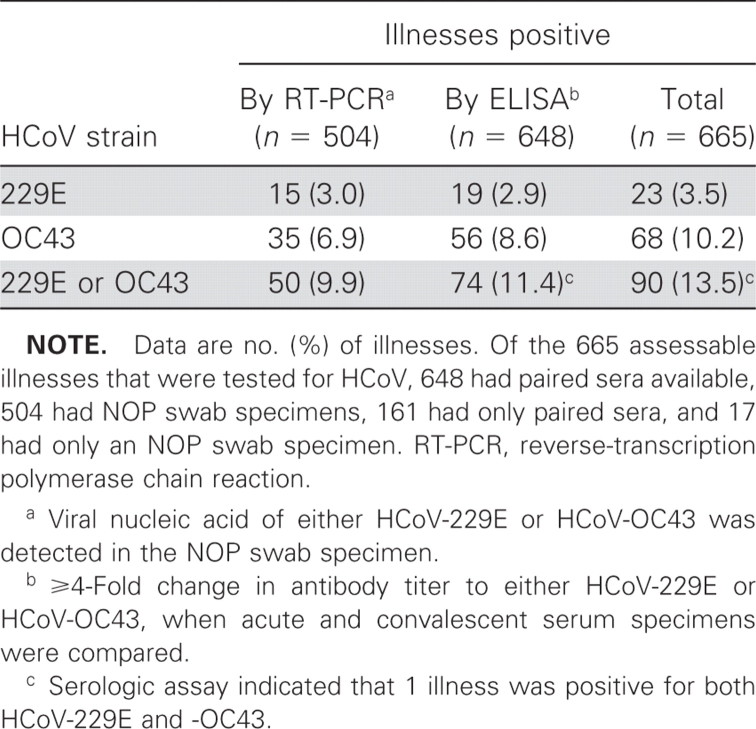
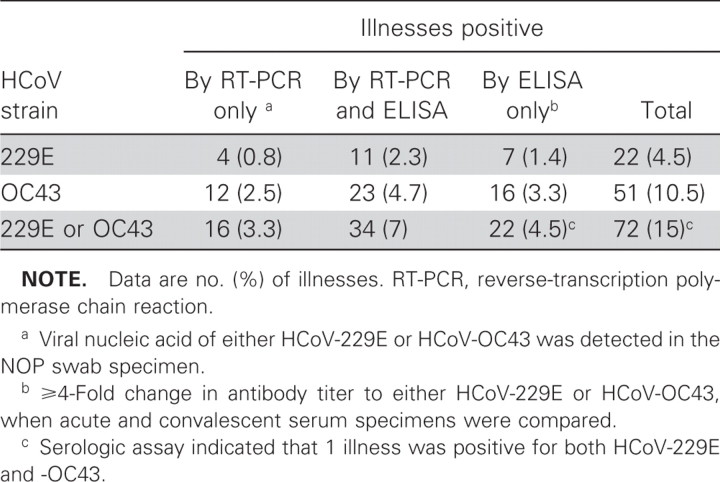
A total of 715 acute respiratory illnesses occurred among 2215 vaccinated patients; 678 of them were acute respiratory illnesses that were clinically assessable in 585 patients (503 patients had 1 acute respiratory illness and 82 had more than 1 respiratory illness). We tested for HCoV-229E and HCoV-OC43 infections in 679 illnesses among 586 patients, but only 665 among 575 patients that were tested for HCoV-229E and HCoV-OC43 infections were clinically assessable. Overall, 90 (14%) of the 665 fully assessable illnesses were positive for either HCoV-229E or HCoV-OC43 infection (table 1), and the number of illnesses associated with HCoV-OC43 was ∼3 times more than that associated with HCoV-229E. Of the 648 illnesses for which paired serum specimens were available, 74 (11%) had ⩾4-fold changes in the titer of antibody to either HCoV-229E or HCoV-OC43 (table 1). Of the 504 illnesses assessed by RT-PCR, 50 (10%) were positive for either HCoV-229E or HCoV-OC43 (table 1), and none were positive for SARS-coronavirus. None of the 17 illnesses for which only an NOP specimen was available were positive by RT-PCR. Of the 161 illnesses for which only paired serum specimens were available, 18 (11%) were positive for HCoV infection (1 for HCoV-229E and 17 for HCoV-OC43). Of the 487 illnesses for which both NOP-swab and paired serum specimens were tested, 72 (15%) were positive for either HCoV-229E or HCoV-OC43 (table 2). The proportion of these 487 illnesses that was found to be positive by antibody testing (56 [11%]) was not significantly different from that found to be positive by RT-PCR (50 [10%]), but only 34 (47%) of the 72 illnesses positive for either HCoV-229E or HCoV-OC43 were positive by both antibody testing and RT-PCR
Table 1.
Illnesses positive for human coronavirus (HCoV)–229E and HCoV-OC43 of 665 fully assessable illnesses for which both nasal-and-oropharyngeal (NOP)–swab and/or paired serum specimens were tested
Table 2.
Fully assessable illnesses positive for human coronavirus (HCoV)–229E and HCoV-OC43, of 487 illnesses for which both nasal-and-oropharyngeal (NOP)–swab and paired serum specimens were tested
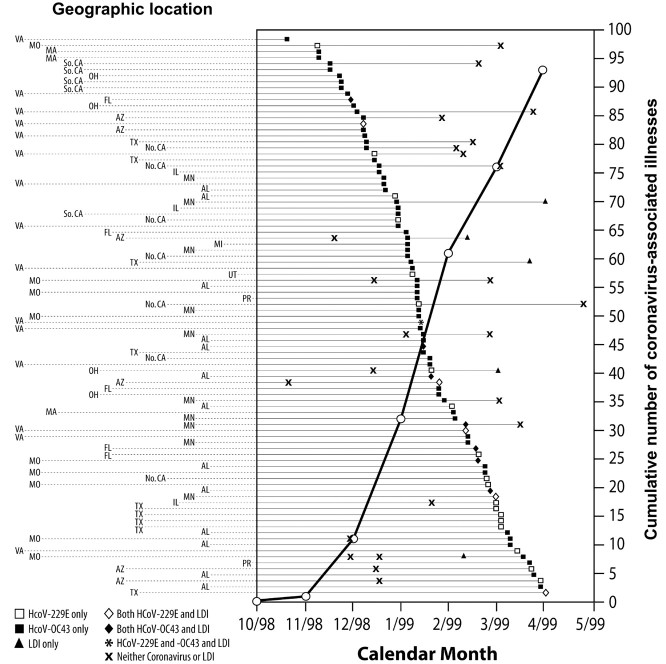
No patient experienced more than 1 HCoV-associated illness (figure 1). The HCoV-229E– and HCoV-OC43–associated illnesses occurred throughout the follow-up period at 19 of the 20 study sites. The majority of illnesses (50) associated with HCoV-229E and -OC43 occurred during December 1998 and January 1999 (figure 1). Until March 1999, the number of HCoV-OC43–associated illnesses was greater than the number of HCoV-229E–associated illnesses (figure 1)
Figure 1.
List of 93 acute respiratory illnesses associated with human coronavirus (HCoV)-229E and -OC43 virus infections. The illnesses are listed in chronological order (regardless of whether the illness is a first episode of acute respiratory illness), by calendar month, from the beginning of October 1998 to April 1999 (the end of follow-up). Included among these 93 illnesses are 3 (2 HCoV-229E and 1 HCoV-OC43) that were not among the 665 illnesses that were assessable. For the patients infected with HCoV-229E and HCoV-OC43, the other respiratory illnesses, occurring before or after the HCoV-associated illness, are also shown. Of the patients with an illness associated with either HCoV-229E or HCoV-OC43, 1 had laboratory-documented influenza (LDI) before the HCoV illness, 4 had an HCoV illness before LDI, 13 had LDI concurrent with the HCoV infection during the illness, 9 had a non-HCoV/non-LDI illness before the HCoV illness, and 12 had either an HCoV-229E or HCoV-OC43 illness before a non-HCoV/non-LDI illness. Each row represents 1 patient, and the reporting site is identified by the 2-letter code for US states and Puerto Rico (PR), to the left of the graph. Of 20 sites, 19 in 13 states and PR reported illnesses associated with HCoV (HCoV-229E in 11 states and HCoV-OC43 in 12 states and PR). HCoV-associated illnesses were reported by 2 sites in Florida, 2 in Southern and 1 in Northern California, 2 in Texas, and 2 in Virginia; study sites reporting the most HCoV-229E and -OC43-associated illnesses were in Virginia (15 illnesses), Alabama (13 illnesses), Minnesota (10 illnesses), Missouri (9 illnesses), and Texas (9 illnesses). The cumulative numbers of illnesses of HCoV-associated illnesses, regardless of whether it was a first episode of acute respiratory illness and including those associated with HCoV and those with both HCoV and LDI, are graphed by calendar month (the cumulative number is that which occurred up to the beginning of the corresponding month); 32 HCoV-associated illnesses (5 HCoV-229E and 27 HCoV-OC43) occurred by the end of 1998, and 61 occurred during 1999 (19 HCoV-229E, 41 HCoV-OC43, and 1 with both strains). The only period when HCoV-229E–associated illnesses predominated was after February 1999 (10 HCoV-229E and 7 HCoV-OC43)
Our analysis of 585 first episodes of acute respiratory illnesses included 81 that were associated with HCoV-229E and/or HCoV-OC43 and 94 that were associated with LDI; 19 (20%) of the 94 LDI-associated illnesses were confirmed by culture, and the remainder were confirmed by serologic assay; 88 LDI-associated illnesses had 1 type of influenza virus (A/H3N2 in 59, A/H1N1 in 5, and B in 24), 3 had both type A and type B of influenza virus, and 3 had both A subtypes. A total of 12 of the first episodes of acute respiratory illness were associated with both LDI and either HCoV-229E or HCoV-OC43
Demographic characteristics and health-care outcomes (hospitalization, prednisone use, and death) of the illness groups are listed in table 3 The baseline demographic characteristics of the group of 1630 patients who had no acute respiratory illness diagnosed during the study were similar to those of patients who did have such an illness diagnosed during the study (table 3). In the HCoV, LDI, and non-HCoV/non-LDI groups, prednisone use during the study, but not necessarily during the acute respiratory illness, increased from the levels at enrollment (table 3); the proportion of patients who used prednisone at some time during the study was smaller in the no-illness group and the LDI group than in the non-HCoV/non-LDI group. The proportion of patients who were hospitalized during the study was significantly smaller in the no-illness group than in the non-HCoV/non-LDI group (P<.001) (table 3). Of the patients who were hospitalized, a majority were hospitalized either during or ⩽1 month after acute respiratory illness that was associated with HCoV infection and/or LDI (table 3). All deaths in the HCoV and LDI groups occurred >1 month after the acute respiratory illness; of the 4 deaths in the non-HCoV/non-LDI group, 2 occurred ⩽1 month after the acute respiratory illness
Table 3.
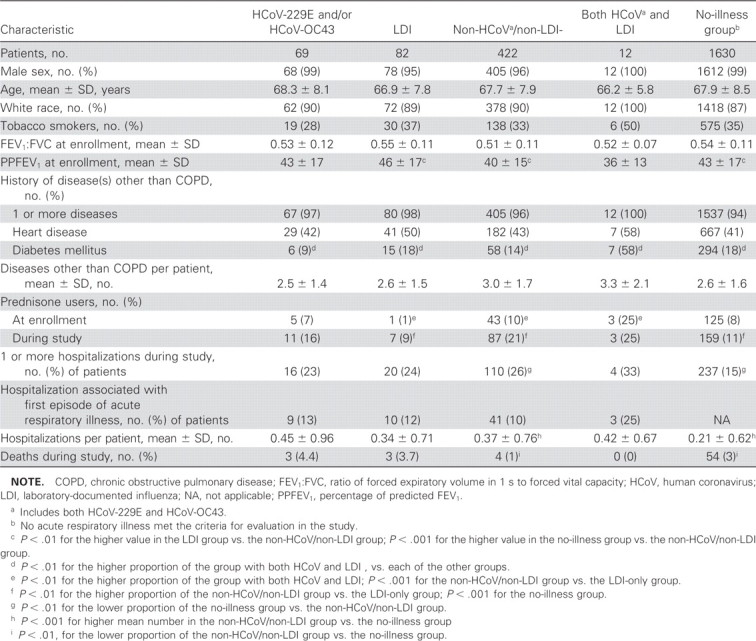
Demographic and clinical characteristics and clinical outcomes for 585 veterans with acute respiratory illnesses, categorized by first episode
Of patients with illness only related to HCoV, each of the 11 symptoms and signs of acute respiratory illness was found in >50% of patients with HCoV-related illness, and >80% of these patients had new or increased cough, sputum production, nasal congestion, and fatigue and/or malaise (table 4). In stepwise logistic regression analysis, myalgia was found to be significantly less frequently associated with HCoV illness (table 4). Patients in the LDI group were less likely to receive LAIV+TIV vaccines. For each of the 11 signs and symptoms, the proportion of patients in the HCoV group was not significantly different from that in the non-HCoV/non-LDI group (table 4)
Table 4.
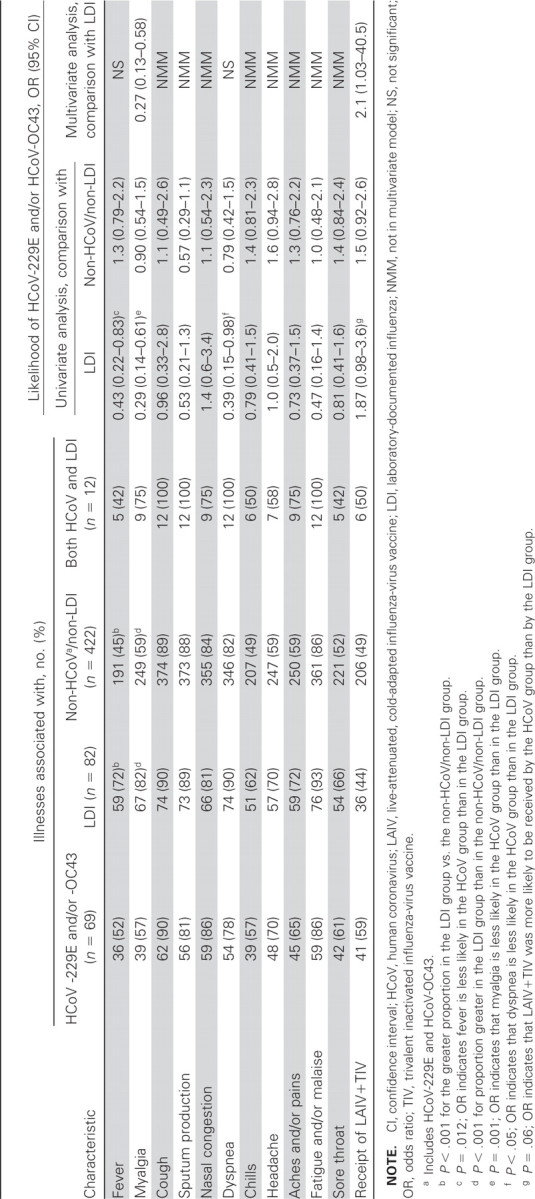
Univariate and multivariate associations between illness group and 11 symptoms/signs of acute respiratory illness and vaccine group
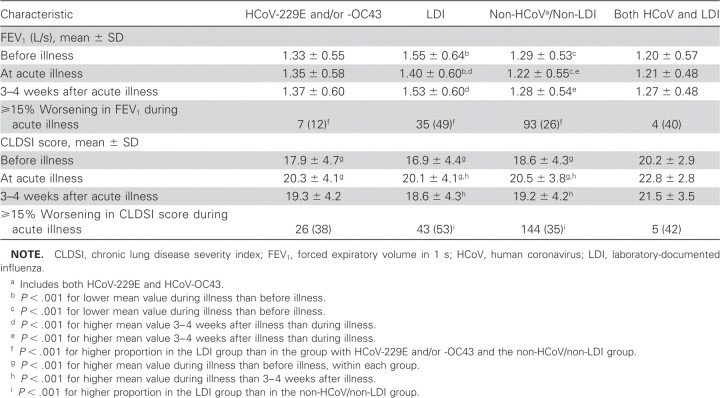
Compared with the mean FEV1 before the acute respiratory illness, both that at the visit for the first episode of acute respiratory illness and that 3–4 weeks later did not change in the HCoV group; however, in both the LDI group and the non-HCoV/non-LDI group, it was significantly lower (worse) at the visit for the first episode, improving 3–4 weeks later (table 5). At the visit for the first episode, the proportion of the LDI group that had ⩾15% worsening in FEV1 was significantly greater than both that of the HCoV group and that of the non-HCoV/non-LDI group. In the HCoV, LDI, and non-HCoV/non-LDI groups, the mean CLDSI score increased (worsened) significantly at the visit for the first episode, improving 3–4 weeks later in the LDI and non-HCoV/non-LDI groups (table 5); the proportion of patients with a ⩾15% worsening in CLDSI score at the visit for the first episode was largest in the LDI group
Table 5.
Spirometric and clinical measures of severity of acute respiratory illness, by illness group
Eight patients had a 4-fold rise in antibody titer to HCoV-NL63. In these patients, 7 of the illnesses were first episodes of acute respiratory illness, and 4 were positive, by RT-PCR, for HCoV-NL63. The sequences of the RT-PCR products were located between positions 22569 and 22882 and were very similar to that of HCoV-NL63 (GenBank accession number AY567487); there was 98%–100% similarity at the nucleotide level. One patient had a 4-fold rise in antibody titer to HCoV-HKU1 in association with the first episode, without concomitant evidence of infection by either HCoV-229E or HCoV-OC43 (table 6). All illnesses were associated with 5–10 symptoms of acute respiratory illness. Of these 9 patients, 1 was hospitalized not in association with HCoV, 3 received prednisone (including 1 who received it in association with HCoV-HKU1), and none died
Table 6.
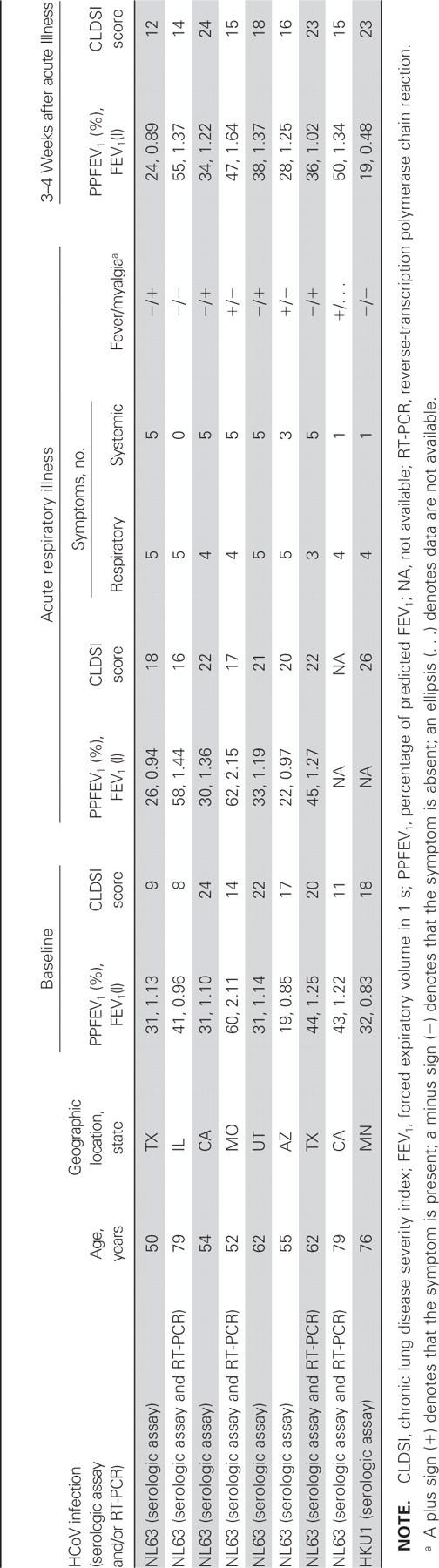
Characteristics of patients and first episodes of acute respiratory illnesses associated with either human coronavirus (HCoV)–NL63 or HCoV–HKU1
Discussion
HCoV infections were clinically significant in this population of older patients with COPD and other chronic illnesses. The frequency of HCoV-229E and HCoV-OC43 infections that were associated with acute respiratory illnesses was similar to that for LDI infection. We may have underestimated the number of HCoV-associated illnesses; illnesses not meeting our criteria for evaluation were not assessed, and an end-of-study serum sample was not obtained, so we were unable to estimate the frequency of HCoV infections in the 1630 patients who did not have an acute respiratory illness. HCoV-associated illnesses were identified at diverse geographic sites within the continental United States and Puerto Rico. More than 80% of the HCoV-associated infections were characterized by new or increased cough, sputum production, nasal congestion, and fatigue and/or malaise. In the multivariate logistic regression analysis, HCoV-associated illness was statistically less likely than LDI to be associated with myalgia. Hence, HCoV infections were difficult to distinguish from LDI in this influenza-vaccinated population, whose influenza-associated illnesses may have been less severe than those in an unvaccinated population. Illnesses not associated with either HCoV or LDI were not easily distinguishable, statistically, from HCoV. In previous analyses, influenza was more likely to be associated with fever, myalgia, and a worsening in FEV1 than were noninfluenza acute respiratory illnesses [47, 48]. Acute respiratory illness, no matter the cause, was associated with significant worsening in the CLDSI score, but, on the basis of changes in FEV1, HCoV appeared to be less severe
Serologic and RT-PCR assays were used as complementary tests for detection of infection by HCoV-229E and HCoV-OC43. More illnesses were positive for HCoV by serologic than by RT-PCR criteria. Delays in collection of NOP specimens after the onset of symptoms of acute respiratory illness, as well as the long interval (∼7 years) between collection and assay of the samples, may have resulted in numbers of viral copies that were below the threshold for detection by the RT-PCR assay
HCoV-OC43 and HCoV-229E infections were associated with more illnesses in our older, chronically ill population than might have been expected on the basis of some previous reports [4–6, 10, 11, 23, 26, 30], although rates similar to those found by the present study have been reported. For instance, 22.9% of 70 virus-associated respiratory-tract infections in patients with COPD that were reported by Greenberg et al. were due to either HCoV-229E or HCoV-OC43 [8]. In a community-based study of elderly patients conducted during 1998–1999, Graat et al. [12] reported that 17% of 108 respiratory infections were due to HCoV-229E and HCoV-OC43 and that this frequency was second to that for rhinoviruses. As in the present study, HCoV-OC43 also predominated over HCoV-229E in a number of other studies [5, 6, 8, 15, 23, 30, 49], whereas HCoV-NL63 and HCoV–HKU1 predominated in 2 recent studies [26, 50]. We identified a small number of illnesses associated with the recently described HCoV-NL63 and HCoV–HKU1. HCoV-NL63 and HCoV-HKU1 cocirculated during the 1998–1999 winter respiratory-virus season at geographically diverse sites in the United States, and the spectrum of illness was similar to that for the HCoV-229E and HCoV-OC43 strains
Respiratory infection due to HCoV has been associated with hospitalization. Glezen et al. [9] have reported that, in 26 (2.5%) of 1029 patients hospitalized because of acute respiratory conditions, the infections were related to either HCoV-229E or HCoV-OC43 and that 14 of these 26 patients were adults >45 years of age. Falsey et al. [11] have reported that 8 of 100 hospitalized elderly patients had concomitant HCoV-229E or HCoV-OC43 infection. In the present study, hospitalization was associated with respiratory infection in all illness groups, both during the month immediately after the visit for the first episode of acute respiratory illness and throughout the study follow-up. There was a lower rate of hospitalization in the group without an acute respiratory illness that qualified for evaluation by the present study; during the study follow-up, the overall mean number of hospitalizations per patient in the HCoV-infected group was more than double that in the no-illness group. In all illness groups, prednisone use increased during the study
Although commonly thought to be associated with cold months [13, 18, 20, 21, 25], HCoV infection has been reported as also occurring during other times of the year [22, 23, 50], and the seasonality of HCoV in tropical and subtropical areas is not so restricted [26, 30]. The present study’s follow-up period extended from October to April, and HCoV infections were detected throughout that period, although they were most frequent during December and January
The results of the present study underscore the clinical importance of HCoV infection in older patients with COPD. Clinical characteristics of HCoV were difficult to distinguish from those characterizing influenza, at least in our influenza-vaccinated population. Although HCoV-229E and HCoV-OC43 were the most frequent HCoV strains identified, HCoV-NL63 and HCoV-HKU1 cocirculated during the 1998-1999 season. Further study of this older, chronically ill population is necessary to determine the genetic evolution of circulating viral strains, the impact that contemporary strains have on the clinical characteristics and severity of illness, and the seasonality of infections
Acknowledgments
We thank Gira B. Patel and Nancy Krudwig, for laboratory technical assistance; Carolyn Novotny, for expert secretarial assistance, and Yolanda Blocker-Hearn, for preparation of the figure
Footnotes
Potential conflicts of interest: K.L.N. reports research funding from GlaxoSmithKline and sanofi pasteur and is a consultant to GlaxoSmithKline, MedImmune Inc., sanofi pasteur, CSL Ltd., and Novartis
Presented in part: 45th Annual Meeting of the Infectious Diseases Society of America, San Diego, California, 4–7 October 2007 (abstract 776)
Financial support: Cooperative Studies Program of the Department of Veterans Affairs Office of Research and Development (CSP448 and CSP448A to G.J.G., T.Z.O., and J.N.V.); National Institute of Allergy and Infectious Disease, National Institutes of Health (contracts N01-AI-25464, HHSN272200800003C, and N01-AI-80003 to G.J.G.); MedImmune Vaccines, Inc. (formerly Aviron) (funds to the Comparative Studies Program of the Department of Veterans Affairs Office of Research and Development)
References
- 1.McIntosh K, Anderson LJ. Coronaviruses, including severe acute respiratory syndrome (SARS)-associated coronavirus. In: Principles and practice of infectious diseases, 6th ed. Vol 2., editors; Mandell GL, Bennett JE, Dolin R, editors. Principles and practice of infectious diseases. 6th ed. Philadelphia: Elsevier Churchill Livingstone; 2005. pp. 1990–7. [Google Scholar]
- 2.Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–94. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 3.Peiris JSM, Yuen KY, Osterhaus ADME, Stöhr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–41. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 4.Gump DW, Phillips CA, Forsyth BR, McIntosh K, Lamborn KR. Role of infection in chronic bronchitis. Am Rev Respir Dis. 1976;113:465–74. doi: 10.1164/arrd.1976.113.4.465. [DOI] [PubMed] [Google Scholar]
- 5.Buscho RO, Saxtan D, Shultz PS, Finch E, Mufson MA. Infections with viruses and Mycoplasma pneumoniae during exacerbations of chronic bronchitis. J Infect Dis. 1978;137:377–83. doi: 10.1093/infdis/137.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CB, Golden CA, Kanner RE, Renzetti AD., Jr Association of viral and Mycoplasma pneumoniae infections with acute respiratory illness in patients with chronic obstructive pulmonary disease. Am Rev Respir Dis. 1980;121:225–32. doi: 10.1164/arrd.1980.121.2.225. [DOI] [PubMed] [Google Scholar]
- 7.Fagon J-Y. Chastre J. Severe exacerbations of COPD patients: the role of pulmonary infections. Semin Respir Infect. 1996;11:109–18. [PubMed] [Google Scholar]
- 8.Greenberg SB, Allen M, Wilson J, Atmar RL. Respiratory viral infections in adults with and without chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2000;162:167–73. doi: 10.1164/ajrccm.162.1.9911019. [DOI] [PubMed] [Google Scholar]
- 9.Glezen WP, Greenberg SB, Atmar RL, Piedra PA, Couch RB. Impact of respiratory virus infections on persons with chronic underlying conditions. JAMA. 2000;283:499–505. doi: 10.1001/jama.283.4.499. [DOI] [PubMed] [Google Scholar]
- 10.El-Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin Infect Dis. 2000;31:96–100. doi: 10.1086/313937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falsey AR, Walsh EE, Hayden FG. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J Infect Dis. 2002;185:1338–41. doi: 10.1086/339881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graat JM, Schouten EG, Heijnen M-LA et al. A prospective, community-based study on virologic assessment among elderly people with and without symptoms of acute respiratory infection. J Clin Epidemiol. 2003;56:1218–23. doi: 10.1016/S0895-4356(03)00171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falsey AR, Walsh EE. Viral pneumonia in older adults. Clin Infect Dis. 2006;42:518–24. doi: 10.1086/499955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallia P, Johnston SL. How viral infections cause exacerbation of airway diseases. Chest. 2006;30:1203–10. doi: 10.1378/chest.130.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Infect Dis. 2003;36:985–9. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Elden LJR, van Loon AM, van Alphen F, et al. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptase polymerase chain reaction. J Infect Dis. 2004;189:652–7. doi: 10.1086/381207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pyrc K, Berkhout B, van der Hoek L. The novel coronaviruses NL63 and HKU1. J Virol. 2007;81:3051–7. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouchier RAM, Hartwig NG, Bestebroer TM, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA. 2004;101:6212–6. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vabret A, Mourez T, Dina J, et al. Human coronavirus NL63, France. Emerg Infect Dis. 2005;11:1225–9. doi: 10.3201/eid1108.050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moës E, Vijgen L, Keyaerts E, et al. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect Dis. 2005;5:6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esper F, Weibel C, Ferguson D, Landry ML, Kahn JS. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J Infect Dis. 2005;191:492–8. doi: 10.1086/428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garbino J, Crespo S, Aubert J-D et al. A prospective hospital-based study of the clinical impact of non-severe acute respiratory syndrome (non-SARS)-related human coronavirus infection. Clin Infect Dis. 2006;43:1009–15. doi: 10.1086/507898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arden KE, Nissen MD, Sloots TP, Mackay IM. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J Med Virol. 2005;75:455–62. doi: 10.1002/jmv.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bastien N, Anderson K, Hart L, et al. Human coronavirus NL63 infection in Canada. J Infect Dis. 2005;191:503–6. doi: 10.1086/426869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu SS, Chan KH, Chu KW, et al. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005;40:1721–9. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo PCY, Lau SKP, Tsoi H-W et al. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J Infect Dis. 2005;192:1898–907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo PCY, Lau SKP, Chu C-M et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–95. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vabret A, Dina J, Gouarin S, Petitjean J, Corbet S. Detection of the new human coronavirus HKU1: A report of 6 cases. Clin Infect Dis. 2006;42:634–9. doi: 10.1086/500136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau SKP, Woo PCY, Yip CCY, et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006;44:2063–71. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esper F, Weibel C, Ferguson D, Landry ML, Kahn JS. Coronavirus HKU1 infection in the United States. Emerg Infect Dis. 2006;12:775–9. doi: 10.3201/eid1205.051316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao X, Guo X, Esper F, Weibel C, Kahn JS. Seroepidemiology of group I human coronaviruses in children. J Clin Virol. 2007;40:207–13. doi: 10.1016/j.jcv.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kistler A, Avila PC, Rouskin S, et al. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–25. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorse GJ, O’Connor TZ, Young SL, et al. Efficacy trial of live, cold-adapted and inactivated influenza virus vaccines in older adults with chronic obstructive pulmonary disease: a VA cooperative study. Vaccine. 2003;21:2133–44. doi: 10.1016/s0264-410x(02)00748-x. [DOI] [PubMed] [Google Scholar]
- 36.Medical Section of the American Lung Association. American Thoracic Society Standardization of spirometry, 1994 update. Am J Respir Crit Care Med. 1995;152:1107–36. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 37.Medical Section of the American Lung Association. American Thoracic Society Lung function testing: selection of reference values and interpretive strategies. Am Rev Respir Dis. 1991;144:1202–18. doi: 10.1164/ajrccm/144.5.1202. [DOI] [PubMed] [Google Scholar]
- 38.Atlanta, GA: Centers for Disease Control and Prevention, World Health Organization; 1975. The hemagglutination inhibition test for influenza viruses. [Google Scholar]
- 39.Dowdle WR, Kendal AP, Noble GR. Influenza viruses. In: Lennette EH, Schmidt NJ, editors. Diagnostic procedures for viral, rickettsial, and chlamydial infections. 5th ed. Washington, DC: American Public Health Association; 1979. pp. 585–609. [Google Scholar]
- 40.Hanson CV, Riggs JL, Lennette EH. Photochemical inactivation of DNA and RNA viruses by psoralen derivatives. J Gen Virol. 1978;40:345–58. doi: 10.1099/0022-1317-40-2-345. [DOI] [PubMed] [Google Scholar]
- 41.Redfield DC, Richman DD, Oxman MN, Kronenberg LH. Psoralen inactivation of influenza and herpes simplex viruses and of virus-infected cells. Infect Immun. 1981;32:1216–26. doi: 10.1128/iai.32.3.1216-1226.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reizenstein E, Hallander H-O. Blackwelder WC, Kühn I, Ljungman M. Comparison of five calculation modes for antibody ELISA procedures using pertussis serology as a model. J Immunol Methods. 1995;183:279–90. doi: 10.1016/0022-1759(95)00067-k. [DOI] [PubMed] [Google Scholar]
- 43.Selim AJ, Ren XS, Fincke G, Rogers W, Lee A. A symptom-based measure of the severity of chronic lung disease: results from the Veterans Health Study. Chest. 1997;111:1607–14. doi: 10.1378/chest.111.6.1607. [DOI] [PubMed] [Google Scholar]
- 44.Kazis LE, Miller DR, Skinner KM, et al. Patient-reported measures of health, the veterans health study. J Ambul Care Manage. 2004;27:70–83. doi: 10.1097/00004479-200401000-00012. [DOI] [PubMed] [Google Scholar]
- 45.Skinner KM, Miller DR, Spiro A III, et al. Measurement strategies designed and tested in the veterans health study. J Ambul Care Manage. 2004;27:180–9. doi: 10.1097/00004479-200404000-00014. [DOI] [PubMed] [Google Scholar]
- 46.Pellegrino R, Viegi G, Brusasco V, et al. Interpretative strategies for lung function tests. Eur Respir J. 2005;26:948–68. doi: 10.1183/09031936.05.00035205. [DOI] [PubMed] [Google Scholar]
- 47.Gorse GJ, O’Connor TZ, Young SL, et al. Impact of a winter respiratory virus season on patients with COPD and association with influenza vaccination. Chest. 2006;130:1109–16. doi: 10.1378/chest.130.4.1109. [DOI] [PubMed] [Google Scholar]
- 48.Neuzil KM, O’Connor TZ, Gorse GJ, Nichol KL. Recognizing influenza illness in influenza-vaccinated, older patients with chronic obstructive pulmonary disease. Clin Infect Dis. 2003;36:169–74. doi: 10.1086/345668. [DOI] [PubMed] [Google Scholar]
- 49.Dare RK, Fry AM, Chittaganpitch M, Sawanpanyalert P, Olsen SJ. Human coronavirus infections in rural Thailand: a comprehensive study using real-time reverse-transcription polymerase chain reaction assays. J Infect Dis. 2007;196:1321–8. doi: 10.1086/521308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuypers J, Martin ET, Heugel J, Wright N, Morrow R. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics. 2007;119:e70–6. doi: 10.1542/peds.2006-1406. [DOI] [PubMed] [Google Scholar]