Dear Editor,
Since early December 2019, a large outbreak of pneumonia caused by a novel coronavirus (COVID-19) had emerged in Wuhan, China (Wu et al.2020a, b; Zhou et al.2020; Zhu et al.2020; Jiang and Shi 2020). Similar to severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), the new coronavirus also belongs to Betacoronavirus, and shares highest sequence identity to three SARS-like CoVs of bat origin (bat_CoV_RaTG13: 96.0%, bat-SL-CoVZC45: 88.0% and bat-SL-CoVZXC21: 87.2%) (Zhou et al.2020). Although only sharing about 79.5% genomic sequence identity to SARS-CoV, the new virus was demonstrate to use the same receptor angiotensin converting enzyme II (ACE2) for human infection as SARS-CoV (Lu et al.2020; Wu 2020; Zhou et al.2020) and is officially named as SARS-CoV-2 (also known as 2019-nCoV) (Gorbalenya et al.2020). Epidemically data showed that the virus has strong human-to-human transmission ability, and it is spread by droplets produced by coughing and sneezing, infecting susceptible subjects through direct contacts and other possible transmission routes (e.g. fecal-mouth transmission) (Guan et al.2020; Li et al.2020).
As of Mar. 27, 2020, the outbreak had resulted in 533,464 laboratory-confirmed cases, including 24,097 deaths around the world. Currently, there lacks suitable antiviral drugs or vaccines for SARS-CoV-2. Early, rapid, and reliable diagnostic assays for SARS-CoV-2 is of top priority to facilitate public health interventions that can reduce or avoid further spread of SARS-CoV-2 (Dennis Lo and Chiu 2020). Quantitative Real-time PCR (qPCR) is a robust technology for reliable laboratory diagnosis, and routinely used to detect causative pathogens. Since the release of genomic sequence of SARS-CoV-2 in Jan. 11, 2020, many TaqMan probe-based RT-qPCR assays had been developed and used in confirmation of large number of SARS-CoV-2 infections during the outbreak (Chu et al.2020; Dennis Lo and Chiu 2020). Facing the on-going increase of suspected cases and potential contacts, however, the detection capacity of RT-qPCR assays have shown limitations in speed and volume because these assays require sophisticated equipment and highly trained personnel, and are relatively time-consuming (about 1.5–2 h). Therefore, there is an urgent demand for faster and sensitive Point-of-Care testing (POCT) assays to facilitate rapid detection of SARS-CoV-2 infection. However, there is no POCT assay available for SARS-CoV-2 yet. Here, we present a novel visual reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for rapid and sensitive detection of SARS-CoV-2 using mismatch-tolerant technique.
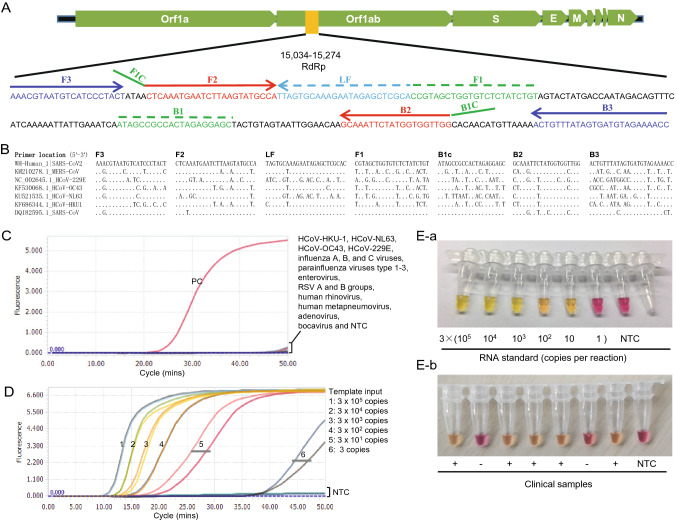
Based on SARS-CoV-2 genomic sequences, we designed six sets of LAMP primers (4, 1, and 1 in N, S and RdRp genes, respectively) using the open access Primer Explorer V.5 software tool (http://primerexplorer.jp/). We firstly excluded primer sets with non-specific amplification, and then selected the primers having high amplification efficiency using the conventional RT-LAMP method. The RdRp primers showed higher amplification efficiency, and were selected to establish the SARS-CoV-2 detection assay using the mismatch-tolerant RT-LAMP method (Li et al.2019; Zhou et al.2019). The RdRp primer information is shown in Fig. 1A, 1B. A 25 µL reaction system includes 1 × isothermal amplification buffer, 6 mmol/L MgSO4, 1.4 mmol/L dNTPs, 8 units of WarmStart Bst 3.0 DNA polymerase, 7.5 units of WarmStartR RT, 0.15 unit of Q5 High-Fidelity DNA Polymerase, 1.6 μmol/L each of primers of FIP and BIP, 0.2 μmol/L each of primers of F3 and B3, 0.8 μmol/L of loop primer FLP and BLP, and 0.4 mmol/L SYTO 9 (Life technologies, Carlsbad, CA, United States). Three types of polymerases were purchased from New England Biolabs (Beverly, MA, United States). Three microliters of RNA were used for the RT-LAMP assays. The RT-LAMP reaction was performed at 63 °C for 50 min for real-time monitoring using the Light Cycler 96 real-time PCR System (Roche Diagnostics, Mannheim, Germany).
Fig. 1.
A Location of the primers in SARS-CoV-2 genome. B Sequence comparison among seven human coronaviruses (SARS-CoV2, SARS-CoV, MERS-CoV, OC43, HKU1, NL63 and 229E). C Cross-reactivity test of the novel SARS-CoV-2 RT-LAMP assay to other common respiratory viruses. Tested common respiratory viruses include HCoV-HKU-1, HCoV-NL63, HCoV-OC43, HCoV-229E, influenza A, B, and C viruses, parainfluenza viruses type 1–3, enterovirus, respiratory syncytial virus A and B groups, human rhinovirus, human metapneumovirus, adenovirus and bocavirus. RNA from a COVID-19 patient was used as positive control (PC). NTC, non-template control. D Sensitivity test of the novel SARS-CoV-2 RT-LAMP assay. Positive amplification was defined only when all three replicates are successfully amplified. NTC: non-template control. E Visual detection of SARS-CoV-2 by the colorimetric RT-LAMP assay. E-a RNA standards; E-b Clinical samples. NTC: non-template control.
To determine the specificity of the SARS-CoV-2 RT-LAMP assay, 15 clinical samples positive for common respiratory viruses (including influenza A, B, and C viruses, parainfluenza viruses type 1–3, enterovirus, respiratory syncytial virus (RSV) A and B groups, HCoV-HKU-1, HCoV-NL63, human rhinovirus, human metapneumovirus, adenovirus and bocavirus), and two human coronavirus standard strains HCoV-OC43 (VR-1558) and HCoV-229E (VR-740) were tested. None or very weak amplification signals were observed for all 17 respiratory viruses after 50 min, indicating good specificity of the assay (Fig. 1C). Although we did not test the specificity for SARS-CoV and MERS-CoV because of the lack of clinical samples or standard strains, the sequence comparison suggests the RT-LAMP can also amplify SARS-CoV due to high sequence similarity (Fig. 1B).
To prepare RNA standard, a SARS-CoV-2 RdRp fragment (15,034–15,274 nt in Wuhan-Hu-1, GenBank: MN908947.3) was amplified from positive clinical sample with T7-promoter-containing primers, and then the products were subjected to in vitro transcription. RNA products were quantitated by Nanodrop 2000C (Thermo, USA). A tenfold serial dilution of RNA standard from 106 to 1 copies per μL was used to test the sensitivity of the SARS-CoV-2 RT-LAMP assay. The result showed that the RT-LAMP can detect as few as 3 copies of SARS-CoV-2 RNA per 25 μL reaction (Fig. 1D). In particular, amplification curves of template inputs > 300 copies appeared within 20 min, showing a fast amplification (Fig. 1D).
For the POCT diagnosis of SARS-CoV-2 infection in the resource-poor settings, we developed the assay into a visual detection using WarmStart Colorimetric LAMP 2 × Master Mix (New England Biolabs, Beverly, MA, United States). A 25 µL reaction system includes 1 × WarmStart Colorimetric LAMP buffer, 0.15 unit of Q5 High-Fidelity DNA Polymerase, 1.6 μmol/L each of primers of FIP and BIP, 0.2 μmol/L each of primers of F3 and B3, and 0.8 μmol/L of loop primer FLP and BLP. The reactions were performed at 63 °C, and the color change was observed at the 30-, 40-, and 50-min time points. The visual detection system contains a pH-sensitive indicator dye cresol red. Because LAMP amplification results in a significant pH change of buffer from an initial alkaline condition to a final acidic condition (Tanner et al.2015), the color change of cresol red from burgundy to orange or yellow indicates a positive reaction. The assay gave a clear color indication for all tested samples during 30–50 min (data not shown). We selected 40 min as the cut-off for the visual assay (Fig. 1E). The sensitivity of the colorimetric RT-LAMP assay for SARS-CoV-2 was 30 copies per reaction, slightly lower than the real-time monitoring (Fig. 1E).
To evaluate the performance of the colorimetric RT-LAMP assay for SARS-CoV-2, we compared it with the SARS-CoV-2 RT-qPCR kit (Liferiver Bio, Shanghai, China) using 24 clinical samples. All 17 positive samples as shown by the RT-qPCR assay (CT values: 28.8–40) were also positive by the novel RT-LAMP assay, demonstrating a 100% consistency (Supplementary Table S1).
The severity of SARS-CoV-2 pandemic had exceeded SARS-CoV. In Jan. 30, 2020, The COVID-19 pandemic was declared as a public health emergency of international concern by WHO. Early diagnosis and timely implementation of intervention and quarantine measures are very crucial for prevention and control of emerging infectious diseases such as COVID-19 (Dennis Lo and Chiu 2020). The rapid identification of SARS-CoV-2 and subsequent release of its genome sequence are very important in a public health perspective, and these made the development of SARS-CoV-2 diagnostic assays possible. Many RT-qPCR assays have been developed and used in the confirmation of SARS-CoV-2 infection during the pandemic (Chu et al.2020). However, high level facility requirement of RT-qPCR assays is a major factor limiting the confirmation speed and causing a large number of suspected cases waiting for tests as these assays are not available in primary care and community hospitals or health care centers where there were no real-time thermal cycler and trained professional. On the other hand, the median incubation period of COVID-19 is 4 days (range 0–24.0 days), and the longest incubation period observed is 24 days (Guan et al.2020). In particular, some infected individuals did not develop obvious clinical symptoms (Rothe et al.2020). The long incubation period and asymptomatic infections imply a huge risk of virus transmission to other people (Wu et al.2020a, b). In this respect, POCT detection tools that can be performed in fields and even patients’ own houses have a huge advantage, and should be developed.
LAMP is the most commonly used isothermal amplification technique for POCT diagnosis due to its high sensitivity, rapid reaction, simple operation, and easy observation of results. Recently, we developed a mismatch-tolerant LAMP method by adding 0.15 U of high-fidelity DNA polymerase (Zhou et al.2019). The main difference between the mismatch-tolerant LAMP and the conventional ones is the inclusion of an additional amount of high-fidelity DNA polymerase in the former, which largely improves the sensitivity and reaction speed (Li et al.2019; Zhou et al.2019). Using this mismatch-tolerant technique, we developed a one-step single-tube RT-LAMP assay for detection of SARS-CoV-2. The assay can be performed at 63 °C for 40 min in a real-time or a regular thermal cycler, or any heating block such as a dry incubator or a water bath. The result is easily judged by the color change from burgundy to orange or yellow. Compared to the conventional LAMP protocol, the novel SARS-CoV-2 RT-LAMP assay is more rapid and more sensitive. The sensitivity of the assay is 30 copies per reaction. Although we did not test the specificity for SARS-CoV and MERS-CoV due to a lack of clinical samples, we sound the assay exhibited specificity by testing 17 common human respiratory viruses, including four other human coronaviruses OC43, 229E, HKU-1 and NL63. The evaluation with 24 clinical samples showed that all 17 COVID-19 patients in Nantong city were positive for SARS-CoV-2 by both the RT-LAMP and the RT-qPCR assays, showing a full consistence.
In conclusion, the novel visual RT-LAMP assay is a simple, rapid, and sensitive approach for detection of SARS-CoV-2, and it is ready for application in primary care and community hospitals or health care centers, and even patients’ own houses in response to the current SARS-CoV-2 epidemic because the assay does not require sophisticated equipment and skilled personnel. Furthermore, it is also ready to be used in fields for screening samples from wild animals and environments to facilitate the identification of potential intermediate hosts that mediate the cross-species transmission of SARS-CoV-2 from bats to humans.
í supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the Biobank of Pathogen Discovery and Big Data Center, Institut Pasteur of Shanghai, Chinese Academy of Sciences, for storing the standard strains of hCoV-OC43 and hCoV-229E that were previously purchased from the American Type Culture Collection (ATCC). This work was supported by the grants from the National Science and Technology Major Project of China (2019YFC1200603, 2017ZX10103009-002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
The study was approved by the Ethics Committees of Nantong Third Hospital Affiliated to Nantong University (E2020003). Written informed consents were obtained from all involved patients.
Footnotes
Renfei Lu and Xiuming Wu have contributed equally to this work.
Change history
4/23/2020
The corrected legend is given below.
References
- Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020 doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis Lo YM, Chiu RWK. Racing towards the development of diagnostics for a novel coronavirus (2019-nCoV) Clin Chem. 2020 doi: 10.1093/clinchem/hvaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J, Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Shi ZL. The first disease X is caused by a highly transmissible acute respiratory syndrome coronavirus. Virol Sin. 2020 doi: 10.1007/s12250-020-00206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou Y, Ma Y, Xu R, Jin X, Zhang C. A mismatch-tolerant RT-LAMP method for molecular diagnosis of highly variable viruses. Bio-protocol. 2019;9:e3415. doi: 10.21769/BioProtoc.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Li M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JTK, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, Seilmaier M, Drosten C, Vollmar P, Zwirglmaier K, Zange S, Wolfel R, Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020 doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner NA, Zhang Y, Evans TC., Jr Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. 2015;58:59–68. doi: 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- Wu Y. Compensation of ACE2 function for possible clinical management of 2019-nCoV-induced acute lung injury. Virol Sin. 2020 doi: 10.1007/s12250-020-00205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wan Z, Yang S, Li Y, Li M, Wang B, Hu Y, Xia X, Jin X, Yu N, Zhang C. A mismatch-tolerant reverse transcription loop-mediated isothermal amplification method and its application on simultaneous detection of all four serotype of dengue viruses. Front Microbiol. 2019;10:1056. doi: 10.3389/fmicb.2019.01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus I, Research T A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.