Abstract
To express the 3′‐region (1152 bp) of the cag7 gene of Helicobacter pylori 51 strain, encoding the C‐terminal 383 amino acid (ct383 aa) region of Cag7 protein that is known to cover the needle region of T4SS, in a live delivery vehicle Lactococcus lactis, the cag7‐ct383 gene was amplified by PCR. DNA sequence analysis revealed that the amino acid sequence of Cag7‐ct383 of H. pylori 51 shared 98.4% and 97.4% identity with H. pylori 26695 and J99, respectively. Intramuscular injection of the GST‐Cag7‐ct383 fusion protein into a rat could raise the anti‐Cag7 antibody, indicating the immunogenicity of the Cag7‐ct383 protein. When the cag7‐ct383 gene was cloned in Escherichia coli–L. lactis shuttle vector (pMG36e) and transformed into L. lactis, the transformant could produce the Cag7‐ct383 protein, as evidenced by Western blot analysis. The Cag7‐ct383 protein level in the L. lactis transformant reached a maximum at the early stationary phase without extracellular secretion. The oral administration of the L. lactis transformant into mice generated anti‐Cag7 antibody in serum in five of five mice. These results suggest that L. lactis transformant expressing Cag7‐ct383 protein may be applicable as an oral vaccine to induce mucosal and systemic immunity to H. pylori.
Keywords: oral vaccine, mucosal and systemic immunity, Helicobacter pylori Cag7, recombinant Lactococcus lactis
Introduction
As infection with the Gram‐negative bacterium Helicobacter pylori causes the most common gastroduodenal human diseases, antibiotic therapy is used to eradicate H. pylori infection (Crespo & Suh, 2001; Czinn, 2004). However, current therapeutic regimens have produced unsatisfactory results due to the emergence of antibiotic resistance of H. pylori and a high reinfection rate after successful eradication (Zendehdel et al., 2005). This has led to a requirement to develop a vaccination method to prevent infection by H. pylori. In this context, several cellular components of H. pylori, including the cytotoxin‐associated gene A (CagA) (Prinz et al., 2003), adhesin (Xu et al., 2005), urease (Lee et al., 2001), vacuolating cytotoxin A (Liu et al., 2004), neutrophil‐activating protein (Satin et al., 2000), catalase (Chen et al., 2003), and Cag12 (Kim et al., 2006b), have been selected as putative antigens for the vaccination against H. pylori. Among these putative antigens, CagA and Cag12 are known to be the protein components of the type IV secretion system (T4SS) that builds a needle‐like structure on the H. pylori. Significant advances in our understanding of the pathogenic role of the cag pathogenicity island (cag‐PAI) in H. pylori have revealed that the type 1 H. pylori strains, which are mainly associated with several severe forms of gastric diseases, can be distinguished from type II strains by the presence of cag‐PAI (2008, 2003). The cag‐PAI consists of 32 genes and encodes the T4SS. Helicobacter pylori uses T4SS to inject the CagA protein and possibly other factors into gastric epithelial cells, in a manner similar to T4SS of Agrobacterium tumefaciens, which transfers oncogenic DNA and effector proteins to host cells during infection (2004, 2003). Although the protein components of H. pylori T4SS, as well as the precise mechanisms by which T4SS mediates its diverse effects on host cells, remain obscure, it has been shown by microscopic techniques that the VirB10‐homologous protein HP0527 (Cag7) covers a central hollow needle region of T4SS of H. pylori as a major component of T4SS‐associated needle‐like structure surface filament (Rohde et al., 2003). Helicobacter pylori Cag7 likely has the potential to be used as a vaccine antigen, in that Cag7 is localized on the surface of T4SS, which is detected in all strains of type I H. pylori in association with their pathogenicity. However, Cag7 of the T4SS has not been tested as a vaccine antigen that induces protective immunity against H. pylori.
Lactococcus lactis, which is a Gram‐positive, noninvasive, nonpathogenic, and food‐grade bacterium, can be used as a live delivery vector system of heterologous proteins for oral vaccination, irrespective of their cellular localization (cytoplasmic, cell surface, or secreted) (Nouaille et al., 2003). Although L. lactis lacks the ability to colonize in vivo, this bacterium has been shown to deliver heterologous antigens to the mucosal and systemic immune system via mucosal routes (Steidler et al., 2000). Lactococcus lactis has approximately the same size as biodegradable microparticles that can be taken up by M cells in gut‐associated immune tissues, thus supporting the capability of L. lactis to act as an effective oral vaccine vehicle. Furthermore, the phosphopolysaccharide produced by L. lactis is nontoxic but can function as a B‐cell mitogen (Kitazawa et al., 1996). Recently, several research groups have reported that L. lactis can be genetically engineered to express bacterial or viral antigens, including the HIV Env antigen (Xin et al., 2003), Brucella abortus L7/L12 antigen (Ribeiro et al., 2002), human papillomavirus Type 16 E7 antigen (Bermudez‐Humaran et al., 2002), Plasmodium falciparum merozoite surface protein MSP3 antigen (Theisen et al., 2004), Erysipelothrix rhusiopathiae SpaA antigen (Cheun et al., 2004), severe acute respiratory syndrome coronavirus nucleocapsid antigen (Pei et al., 2005), and tetanus toxin fragment C (Robinson et al., 2004).
In an attempt to develop an oral vaccine against H. pylori, in the present study, we have performed PCR‐based amplification of the cag7 gene from H. pylori 51 that was previously isolated from a Korean patient (Lee et al., 1999), and compared its DNA sequence with those of H. pylori 26695 (UK) (Tomb et al., 1997) and H. pylori J99 (USA) (Alm et al., 1999). The 1152 bp of the 3′‐region (3874–5025 nt) of the cag7 (HP0527) gene of H. pylori, which encodes the C‐terminal 383 amino acid (ct383 aa, 41.4 kDa) of Cag7 protein, including the VirB10‐homology region (VHR, 373 aa) and an additional 10 aa of the 3′‐conserved region (TCR), appeared to be highly conserved among these three strains. It was therefore expressed in the live delivery vehicle L. lactis using the Escherichia coli–L. lactis shuttle vector (pMG36e). Sequentially, the L. lactis transformant expressing the Cag7‐ct383 protein was orally delivered into mice to examine whether it could elicit a systemic humoral immune response against Cag7 protein of H. pylori.
Materials and methods
Microorganism, vector plasmid, and growth conditions
The genomic DNAs purified from H. pylori 51 (Lee et al., 1999), H. pylori 26695 (Tomb et al., 1997), and H. pylori J99 (Alm et al., 1999) were obtained from Dr Kwang Ho Rhee (Department of Microbiology, College of Medicine, Gyeongsang National University, Jinju, Korea). Lactococcus lactis ssp. MG1363 and the E. coli–L. lactis shuttle vector pMG36e plasmid were provided by Dr Jeong Hwan Kim (Department of Food Technology, College of Agriculture, Gyeongsang National University). Escherichia coli Rosetta (DE3) pLysS and the protein expression vector pGEX‐2T were purchased from Amersham (Arlington Heights, IL). Lactococcus lactis was grown in M17 Medium (Difco Laboratories, Detroit, MI) supplemented with 1% glucose (M17G) at 30 °C, and E. coli Rosetta (DE3) pLysS was grown in Luria–Bertani (LB) broth at 37 °C. When necessary, antibiotics were added to the culture medium at the following concentrations: ampicillin, 50 μg mL−1, and erythromycin, 200 μg mL−1 for E. coli and 5 μg mL−1 for L. lactis. The recombinant plasmid pGEX‐2T harboring H. pylori cag7 gene was designated as pGEX‐2T/cag7, and the recombinant plasmid pMG36e inserted with the H. pylori cag7 gene and H. pylori cag7‐ct383 were designated as pMG36e/cag7 and pMG36e/cag7‐ct383, respectively.
PCR procedure and cloning of cag7 gene from H. pylori 51
To amplify the cag7 gene of H. pylori 51, PCR was performed in the presence of H. pylori 51 genomic DNA and both cag7‐forward primer (5′‐ATGAATGAAGAAAACGATAAACTT‐3′) and cag7‐reverse primer (5′‐GGTGATTTCTTCATGTTCTCTAG‐3′). The target DNA was amplified in 50 μL of a reaction mixture containing 5 μL of 10 × Ex Taq buffer, 4 μL of 2.5 mM dNTP, 1 μL of target DNA (40 ng), 2 pmol of each forward and reverse primer, and 1 U of Ex Taq polymerase (Takara Bio Inc., Shiga, Japan). The PCR for amplification of the cag7 gene was performed under the following conditions: 35 cycles of 20 s at 94 °C, 1 min at 59 °C, and 6 min at 68 °C. The amplified cag7 gene (5025 bp) was purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and was then used for DNA sequence analysis.
Expression of GST‐Cag7‐ct383 in E. coli
To insert H. pylori cag7‐ct383 gene encoding the C‐terminal region of Cag7 into the BamHI–EcoRI site of pGEX‐2T, the BamHI site and the EcoRI site were created on each end of the coding region of Cag7 by PCR in the presence of H. pylori genomic DNA and the synthetic primers, Cag7‐2T forward primer 5′‐GCGGATCCATGGATTTCAAAAA‐3′ and Cag7‐2T reverse primer 5′‐CGAATTCTTAATGGCCACCTTT‐3′ (BamHI and EcoRI sites are underlined). The target DNA was amplified in 50 μL of reaction mixture containing 5 μL of 10 × buffer (100 mM Tris‐HCl, 15 mM KCl, 1 mg mL−1 gelatin, pH 8.3), 4 μL of 1.25 mM dNTP, 1 μL of target DNA (100 ng), 50 pmol of each forward and reverse primer and 1 U of Taq polymerase (Promega, Madison, WI). PCR for amplification of the cag7‐ct383 gene was carried out under the following conditions: 30 cycles of 1 min at 94 °C, 1 min at 55 °C, and 1 min 30 s at 72 °C. The amplified cag7 gene was cloned in the BamHI–EcoRI site of pGEX‐2T. The recombinant plasmid was then introduced into E. coli Rosetta (DE3) pLysS and the transformants were selected on LB plates containing ampicillin (50 μg mL−1) and chloramphenicol (34 μg mL−1). The synthesis of the GST‐Cag7 fusion protein in the E. coli transformants was induced by isopropyl‐1‐thio‐d‐galacto‐pyranoside (IPTG), as described previously (Jun et al., 2003).
Construction of recombinant plasmid pMG36e/cag7‐ct383
To insert the 3′‐1152 bp of the cag7 gene, encoding the ct383 aa of Cag7, of H. pylori 51 strain into the SacI/PstI site of the pMG36e vector, PCR was performed in the presence of H. pylori genomic DNA as well as both SacI‐forward primer 5′‐CGAGCTCCAATGGATTTCAAAA‐3′ and PstI‐reverse primer 5′‐ACTGCAGTTAATGGCCACCTTT‐3′ (SacI/PstI sites are underlined). PCR was carried out under the following conditions: 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min 30 s. This PCR product was cloned in the SacI/PstI site of pMG36e, resulting in the plasmid pMG36e/cag7‐ct383, in which the cag12‐ct383 gene was placed under a P32 promoter in sense orientation. The plasmid was then transformed into E. coli DH5α or L. lactis. For the transformation of L. lactis, electroporation was performed as described previously (Holo & Nes, 1989). Briefly, L. lactis was cultured in an M17 medium until the OD600 nm reached 0.5–0.7; it was then washed in ice‐cold water and resuspended in 1/200 volume of 0.5 M sucrose containing 10% glycerol. Competent cells were added to the ligation mixture and the mixture was treated using the Gene Pulser Apparatus (BioRad, Richmond, CA) according to the manufacturer's instructions. The electroporated mixture was immediately diluted with 1 mL of M17 broth and incubated for 2 h at 30 °C and then plated onto M17 plates containing 0.5 M sucrose and erythromycin (5 μg mL−1). The transformant, designated L. lactis pMG36e/cag7‐ct383, was visible after 48 h of incubation at 30 °C.
DNA sequence analysis
Individual recombinant plasmid DNA constructs purified from the transformants of E. coli or L. lactis were subjected to DNA sequencing. The DNA sequence analysis was performed using the cycle sequencing method with the ABI BigDye terminator, with an ABI Prism 3700 DNA Analyzer (Applied Biosystems, Foster City, CA).
Immunization of rat with GST‐Cag7‐ct383 fusion protein
Escherichia coli pGEX‐2T/cag7‐ct383 was cultivated with shaking at 37 °C. When the growth OD600 nm reached 0.4, 0.1–1 mM IPTG was added and cultivation was continued for an additional 4 h. The GST‐Cag7‐ct383 fusion proteins accumulated in the soluble fraction of the E. coli extract were purified using Glutathione‐Sepharose 4B (Amersham) according to the manufacturer's instructions. GST‐Cag7‐ct383 (30 μg) 200 μL was mixed with an equal volume of Complete Freund's adjuvant, and injected into the thigh muscles of the rear legs of a 10‐week‐old Sprague–Dawley male rat. For secondary, tertiary, and quaternary immunization, protein mixed with Incomplete Freund's adjuvant was injected into the rat in the same manner every 2 weeks. The rat was cared for in compliance with an approved protocol from the Animal Care and Use Committee of Kyungpook National University. Bleeding was carried out 7 days after each immunization to test the antibody titer.
Cell lysate, protein quantitation, and Western blot analysis
Bacterial cell lysates were prepared as described elsewhere (Jun et al., 2003). Briefly, the bacterial cells of E. coli or L. lactis were suspended in phosphate‐buffered saline (PBS), disrupted by sonication, and then incubated at 4 °C for 30 min. After centrifugation at 16 000 g for 20 min, the supernatant and the pellet were obtained as the soluble fraction and the inclusion body fraction of the cell lysates, respectively. Protein quantitation was carried out using the Micro BCA kit (Pierce, Rockford, IL). Equivalent amounts of cell extracts (15 μg) were electrophoresed on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) or a 4–12% gradient SDS‐PAGE, and electrotransferred to an Immobilon‐P membrane (Millipore Corporation, Bedford, MA). The membrane was probed with a primary antibody, and then with a horseradish peroxidase‐linked secondary antibody. Anti‐GST antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Detection of each protein was performed using the ECL Western blotting detection system, according to the manufacturer's instructions (Amersham).
Oral administration of recombinant L. lactis into mice
Either recombinant L. lactis MG36e/cag7‐ct383 or L. lactis pMG36e was cultured at 30 °C for 24 h in an M17 medium containing 1% glucose and harvested by centrifugation at 800 g for 10 min. The cell pellets were washed twice with cold PBS, and then resuspended in 300 μL of PBS to a concentration of 1 × 1011 CFU mL−1. For oral immunization, recombinant L. lactis (pMG36e/cag7‐ct383) or L. lactis pMG36e was administered into 8‐week‐old C57BL/6 male mice via the orogastric route, using an oral sonde. Mice were not provided with food or water for 1 h before the challenge, but food and water were given 2 h after the challenge. Vaccination was administered at weeks 0, 1, 2, 3, 4, and 5. This study was performed according to an animal experimental procedure approved by the Animal Care and Use Committee of Kyungpook National University. Bleeding was carried out 4 days after final oral administration and the presence of anti‐Cag7‐ct383 antibody in the serum was evaluated by Western blot analysis.
Preparation of subcellular fractions
Subcellular fractionation was performed as described previously (Tai et al., 1997). Briefly, recombinant L. lactis expressing Cag7‐ct383, Cag12, or HpaA was cultured at 30 °C for 24 h in an M17 medium containing 1% glucose and harvested by centrifugation at 10 000 g for 10 min. The culture fluid obtained was concentrated to 1/10 volume using Centricon‐10 (Amicon, Danvers, MA) according to the manufacturer's instructions, and then used as the extracellular secretion fraction. The cell pellets were washed twice with PBS, resuspended in 1/10 volume of PBS, and then disrupted by sonication. After centrifugation at 120 000 g for 1 h, the supernatant was used as the soluble extract containing a mixture of the soluble portion of cell wall and cytoplasm. The pellet was washed once with PBS and resuspended in 1/10 volume of PBS and then used as the envelope fraction, composed of the insoluble portion of the cell wall and cytoplasmic membrane.
Results
Sequence analysis of the cag7 gene of H. pylori 51
To investigate whether there is genetic variation in the cag7 gene between the Korean strain H. pylori 51 and two previous isolates 26695 (UK) and J99 (USA), the cag7 gene of H. pylori 51 was amplified using the PCR method, and then the PCR product was purified for the DNA sequence analysis. As shown in Fig. 1, the amino acid sequence of Cag7 of H. pylori 51, which was deduced from the DNA sequence of the cag7 gene of H. pylori 51, showed 82.4% and 74.6% identity with the individual amino acid sequences of H. pylori 26695 and J99 counterparts, both of which were previously published in the GenBank databases. When the amino acid sequence of Cag7 (1674 aa) of H. pylori 51 was aligned with those of H. pylori 26695 and J99, the 5′‐repeat region and the middle repeat region showed a significant difference among these three strains. In contrast, the 5′‐conserved region (FCR), the TCR, and the VHR showed a high degree of identity, with the highest degree of identity in the FCR being ∼98.9% between 51 and 26695, and ∼98.9% between 51 and J99. On the other hand, when the entire deduced amino acid sequence of Cag7 was analyzed for hydrophobicity by the method of Kyte & Doolittle (1982), the major hydrophobic regions appeared to be located within the FCR and the VHR, and the rest of the molecule appeared to be hydrophilic (Supporting Information, Fig. S1a and b). It should be noted that three independent PCR products for the cag7 gene were sequenced to exclude a possible occurrence of sequence errors due to the poor fidelity of the Taq polymerase and that there were no sequence differences among three independent PCR‐amplified DNA products of the cag7 gene. The DNA sequence data of cag7 gene of H. pylori 51 were submitted to the GenBank databases under the accession number FJ998174.
Figure 1.

Comparison of amino acid sequences of Cag7 protein of Helicobacter pylori 51, 26695, and J99. Amino acids are displayed in a single‐letter abbreviation after alignment of Cag7 among three H. pylori strains for maximal identity by the clustalw program.
The current data have shown that the VHR of Cag7 was well conserved among three H. pylori strains in their amino acid sequences and that this region appeared to contain the obvious hydrophobic part between both the hydrophilic ends. In addition, previous studies have reported that the amphipathic α‐helix protein molecules possessing a hydrophobic side and a hydrophilic side separated in space were likely to serve as the immunodominant sites for helper T cells (1988, 1987). On the basis of these findings, as the antigen to raise the anti‐Cag7 antibody, we decided to use the ct383 aa region of Cag7, which includes the VHR (373 aa) and an additional 10 aa of the TCR located upstream from the VHR.
Generation of polyclonal anti‐Cag7 using GST‐Cag7‐ct383 fusion protein as the antigen
To express the N‐terminal 383 aa region (41.4 kDa) of Cag7 of H. pylori 51 as the GST‐fusion protein in E. coli, the 1152 bp of the 3′‐region (3874–5025 nt; 1152 bp) were amplified by PCR with chromosomal DNA of H. pylori 51 in the presence of BamHI‐forward primer 5′‐GCGGATCCATGGATTTCAAAAA‐3′ and EcoRI‐reverse primer 5′‐CGAATTCTTAAT‐GGCCACCTTT‐3′ (BamHI and EcoRI sites are underlined). The PCR product obtained was ligated with the pGEX‐2T vector plasmid, and then the ligation mixture was used for the transformation of E. coli Rosetta (DE3) pLysS. The recombinant plasmid pGEX‐2T/cag7‐ct383 was purified from the transformant selected on an LB plate containing ampicillin (50 μg mL−1) and chloramphenicol (34 μg mL−1), and then the nucleotide sequence of the 1152 bp encoding the ct383 aa region of Cag7 was confirmed by DNA sequence analysis. The deduced amino acid sequence of Cag7‐ct383 of H. pylori 51 showed 98.4% and 97.4% identity with 26695 and J99 counterparts, both of which were previously published in the GenBank databases.
The nucleotide sequence data of the 3′‐1152 bp of cag7 gene of H. pylori 51 were submitted to the GenBank databases under the accession number GQ118157. To exclude any possible sequence errors due to the poor fidelity of the Taq polymerase, three independent recombinant plasmids were sequenced; there were no sequence differences. The recombinant plasmid obtained was designated as pGEX‐2T/cag7‐ct383, and its transformant was designated as E. coli pGEX‐2T/cag7‐ct383.
The optimal conditions for the production of the GST‐Cag7‐ct383 fusion protein (67.4 kDa) using E. coli pGEX‐2T/cag7‐ct383 were determined to be 0.5 mM IPTG, 30 °C, and a 4‐h induction period. Under these conditions, the majority of GST‐Cag7‐ct383 fusion protein produced in the E. coli transformant appeared to remain in a soluble form without secretion (data not shown). Subsequently, the soluble form of the GST‐Cag7‐ct383 fusion protein contained in the cell lysate of the E. coli transformant was purified using Glutathione‐Sepharose 4B. As shown in Fig. 2a, the synthesis of the GST‐Cag7‐ct383 fusion protein with a molecular mass of 67.4 kDa was detected on 10% SDS‐PAGE. Approximately 120 μg of GST‐Cag7‐ct383 fusion protein was obtained from 60 mL of culture fluid under the above purification conditions. To raise a rat polyclonal antibody against the Cag7 of H. pylori 51, the purified GST‐Cag7‐ct383 fusion protein (30 μg) was mixed with Freund's complete or incomplete adjuvant and injected intramuscularly into the rat. To evaluate generation of the anti‐Cag7 antibody by Western blot analysis, the rat was bled 7 days after quaternary immunization. The antiserum, in 10 000‐fold dilution, appeared to detect the GST‐Cag7‐ct383 fusion protein (67.4 kDa) in the cell lysate as well as the purified protein that was used as the antigen (Fig. 2b). In addition, the antiserum was able to recognize specifically the individual intact Cag7 proteins (∼219 kDa) derived from the three H. pylori strains (26695, J99, and 51) (Fig. 2c). Under these conditions, the rat preimmune serum failed to recognize the GST‐Cag7‐ct383 or the intact Cag7 protein (data not shown). It is worth noting that the antiserum could detect a minor protein band with a molecular mass of 61 kDa. As this additional protein band was predicted to be a cleaved or truncated GST‐Cag7‐ct383 protein, the membrane was stripped and reprobed with anti‐GST antibody to examine this prediction. As shown in Fig. 2d, the 61‐kDa protein band detected by the rat antiserum was also recognized by anti‐GST antibody, confirming that it was a truncated or cleaved GST‐Cag7 fusion protein. These results indicated that the rat antiserum, raised against the GST‐Cag7‐ct383 fusion protein, could specifically recognize the Cag7 protein. In addition, these results demonstrated that the ct383 aa of Cag7, including the VHR (373 aa), was able to stimulate a humoral immunity to generate anti‐Cag7 antibody.
Figure 2.
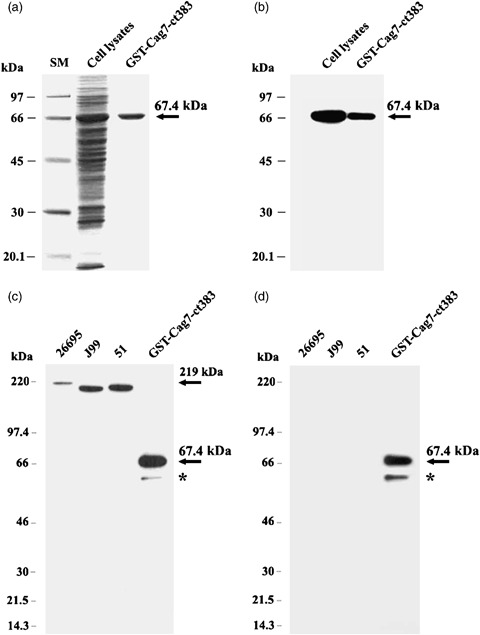
Confirmation of the GST‐Cag7‐ct383 fusion protein (67.4 kDa) on 10% SDS‐PAGE (a), and by Western blot analysis using rat polyclonal anti‐Cag7‐ct383 (b), and the specific recognition of the Cag7 protein in the cell extracts of three Helicobacter pylori strains (51, 26695, and 99) on 4–12% gradient SDS‐PAGE followed by Western blot analysis using either rat polyclonal anti‐Cag7‐ct383 (c) or anti‐GST (d). *Truncated or cleaved GST‐Cag7 fusion protein band.
Cloning and expression of cag7 gene of H. pylori 51 in E. coli and L. lactis using the shuttle vector plasmid pMG36e
To express the Cag7‐ct383 protein of H. pylori 51 in the oral vaccine delivery vehicle L. lactis, the cag7‐ct383 gene (1152 bp) was inserted into the E. coli–L. lactis shuttle vector pMG36e and the recombinant plasmid was then transformed into E. coli DH5α. The recombinant plasmid pMG36e/cag7‐ct383, isolated from the E. coli transformant, was subjected to DNA sequencing to confirm that the cag7‐ct383 gene was properly cloned in the E. coli–L. lactis shuttle vector pMG36e (data not shown). Subsequently, the plasmid pMG36e/cag7‐ct383 was transformed into L. lactis to obtain the transformant L. lactis pMG36e/cag7‐ct383. To examine whether the L. lactis transformant possessing the plasmid pMG36e/cag7‐ct383 could successfully produce the Cag7‐ct383 protein (41.4 kDa), and whether its production level in the L. lactis transformant could be differential during cultivation for 120 h, Western blot analysis for the cell lysates of the transformant obtained at various time points was performed using the rat antiserum. As shown in Fig. 3a and b, the L. lactis transformant was able to produce the 41.4‐kDa Cag7‐ct383 protein as a soluble form, and its expression level reached a maximum at the early stationary phase, although cells at either the early log phase or the late stationary phase appeared to express the Cag7‐ct383 protein to ∼80% of the maximum level. These results demonstrated that the expression of the cag7‐ct383 gene of H. pylori 51 was successfully induced to synthesize the C‐terminal region (41.4 kDa) of Cag7 protein in L. lactis. These results also indicated that the expression level of Cag7‐ct383 in the L. lactis was not significantly affected by the difference in the population growth phases, from the log phase to the stationary phase, although the maximum level occurred at the early stationary phase.
Figure 3.
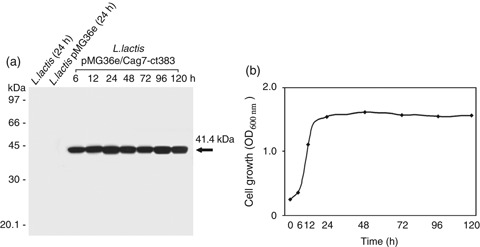
Kinetic analysis of the production of Cag7‐ct383 (a) during cultivation of Lactococcus lactis transformant for 120 h (b). For the kinetic analysis of the expression level of Cag7‐ct383 in L. lactis pMG36e/cag‐ct383 during the culture period (6–120 h), the strain was cultured in an M17 medium supplemented with 1% glucose, 40 mM dl‐threonine, and 5 μg mL−1 erythromycin. As the control, the cell lysate obtained from recipient L. lactis as well as L. lactis transformant harboring the pMG36e empty vector plasmid following cultivation for 24 h was used. Equivalent amounts of the cell extracts (15 μg) prepared from L. lactis transformants were electrophoresed on 10% SDS‐PAGE, and then electrotransferred to the Immobilon‐P membrane, and the blot was probed with the rat polyclonal anti‐Cag7‐ct383. Detection of Cag7‐ct383 was visualized using the ECL Western blotting detection system.
Antibody response against orally immunized recombinant L. lactis expressing Cag7‐ct383
To examine whether orally delivered L. lactis expressing the Cag7‐ct383 protein could induce a systemic humoral immune response against the Cag7 protein, two groups of male C57BL/6 mice (five mice in each group) were intragastrically inoculated six times at 1‐week intervals (Fig. 4a). Bleeding was carried out 4 days after the final oral administration. Thereafter, the immune response to the 41.4‐kDa Cag7‐ct383, the purified GST‐ct383 fusion protein (67.4 kDa), or the intact Cag7 protein (∼219 kDa) was evaluated by Western blot analysis using the antisera of the individual mice. As shown in Fig. 4b and c, the antisera of all five mice at a 1000‐fold dilution could detect the Cag7‐ct383 protein as well as the purified GST‐Cag7‐ct383 protein. At the same time, the antisera of all five mice at a 1000‐fold dilution were able to detect the Cag7 protein expressed in the three H. pylori strains 51, 26695, and J99. In contrast, the antisera obtained from the control group mice inoculated with L. lactis pMG36e failed to recognize the Cag7‐ct383 protein and the Cag7 protein. After the Western blots used for the data in Fig. 4b and c were stripped off the mouse antiserum, the blots were reprobed with the rat antiserum raised against the Cag7‐ct383 to investigate whether the 67.4‐kDa GST‐Cag‐ct383 protein and the entire Cag7 protein bands recognized by the mouse antiserum could also be recognized by the rat antiserum. The protein bands were recognized by the rat antiserum, confirming that the mouse antiserum could detect the ct383 aa (41.4 kDa) of Cag7 and thus the entire Cag7 protein (∼219 kDa) of H. pylori (data not shown). Consequently, these results demonstrated that the oral administration of the L. lactis expressing Cag7‐ct383 protein resulted in the induction of a systemic humoral immune response to Cag7 in five of five mice. This suggests that the recombinant L. lactis expressing Cag7‐ct383 might have the potential, as an oral vaccine candidate, to induce a protective immune response to H. pylori.
Figure 4.
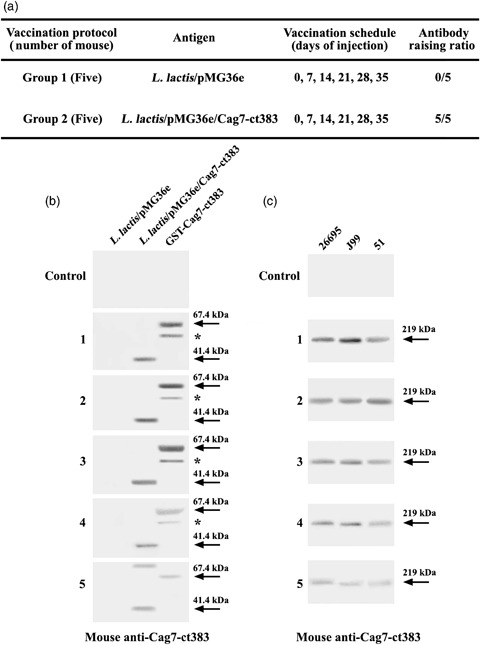
Schedule for oral administration of Lactococcus lactis transformant harboring pMG36e/cag7‐ct383 or pMG36e empty vector into mice (a), Western blot analysis of the Cag7‐ct383 protein (41.4 kDa) produced in L. lactis transformants and purified GST‐Cag7‐ct383 protein (67.4 kDa) (b), and the Cag7 protein (∼219 kDa) produced in three Helicobacter pylori strains (c) using the antiserum obtained from the mice orally immunized by L. lactis transformant expressing the Cag7‐ct383. Equivalent amounts of the cell extracts (15 μg) prepared from L. lactis transformant harboring empty vector pMG36e or recombinant plasmid pMG36e/cag7‐ct383, 0.1 μg of the purified GST‐Cag7‐ct383 fusion protein, and the cell extracts (30 μg) of individual H. pylori strains were electrophoresed on 4–12% gradient SDS‐PAGE, and then electrotransferred to the Immobilon‐P membrane. The blot was probed with the individual mice antisera at 1000 × dilution. Cag7‐ct383 and Cag7 proteins were visualized using the ECL Western blotting detection system. *Truncated or cleaved GST‐Cag7 fusion protein band.
Cellular localization of recombinant Cag7‐ct383 protein expressed in L. lactis transformant
It is generally accepted that L. lactis, which expresses heterologous proteins irrespective of their cellular localization (cytoplasmic, cell surface, or secreted), can be used as a live delivery vector system for oral vaccination (Nouaille et al., 2003). In our previous studies, we have expressed H. pylori adhesin A (hpaA) gene and H. pylori cag12 gene in L. lactis using the vector plasmid pMG36e to obtain the recombinant L. lactis that produced HpaA or Cag12 protein. Whereas the oral administration of L. lactis transformant producing Cag12 in mice resulted in the successful generation of the anti‐Cag12 antibody in serum in two of five cases (Kim et al., 2006b), the oral administration of L. lactis transformant producing HpaA in five mice failed to generate the anti‐HpaA antibody in serum (Kim et al., 2006a).
As the current results demonstrated that the oral administration of the L. lactis transformant synthesizing Cag7‐ct383 protein could generate the anti‐Cag7 antibody in the serum of five of five cases, we decided to examine whether these differences in the L. lactis transformants in their capability of inducing humoral immune response against the heterologous proteins were accompanied by a difference in their solubility or cellular localization. As shown in Fig. 5, none of these recombinant proteins expressed in the L. lactis transformants was secreted extracellularly. At the same time, the amount of Cag7‐ct383 protein located in the soluble fraction appeared to be fivefold higher than that located in the insoluble envelope fraction, whereas approximately half of the Cag12 protein produced in the L. lactis transformant was located in the soluble fraction. However, the HpaA protein produced in the L. lactis transformant was mostly located in the insoluble fraction, which was previously reported to consist of the insoluble portion of the cell wall and cytoplasmic membrane (Tai et al., 1997). These results suggested that the antigenic protein expressed in a soluble form in the recombinant L. lactis might be better than that expressed in an insoluble form at inducing the systemic humoral immune response when the recombinant bacterial cells are orally administered.
Figure 5.
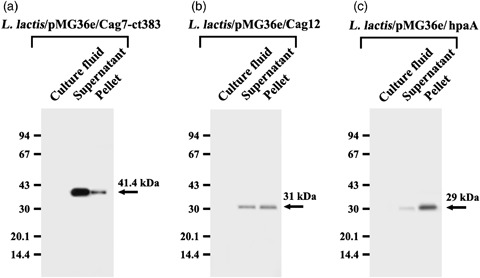
Cellular localization of Cag7‐ct383 (a), Cag12 (b), and HpaA (c) in Lactococcus lactis transformant. After cultivation of L. lactis transformant expressing Cag7‐ct383, Cag12, or HpaA for 24 h, the culture fluid was concentrated to obtain the extracellular secretion fraction, and the cells were subjected to fractionation into the soluble fraction and the envelope fraction as described in Materials and methods. After equivalent amounts of the individual fractions were electrophoresed on 10% SDS‐PAGE, and electrotransferred to the Immobilon‐P membranes, the membranes were probed with individual antibodies. Cag7‐ct383, Cag12, and HpaA protein were visualized using the ECL Western blotting detection system.
Discussion
Helicobacter pylori is known to cause gastritis, gastric and duodenal ulcers, mucosal‐associated lymphoid tissue lymphoma, and finally the development of gastric adenocarcinoma in humans (Crespo & Suh, 2001; Czinn, 2004). Recently, in relation to the pathogenicity of H. pylori, the cag pathogenicity island (cag‐PAI), which is ∼40 kb in length, has been investigated (2008, 2003). The 32 genes, encoding components of the T4SS that builds a needle‐like structure on the H. pylori surface, are known to span the cag‐PAI. The Cag7 protein encoded by the cag7 gene located in the cag‐PAI is known to cover the surface of T4SS of H. pylori. In the present study, the ct383 aa region (∼41.4 kDa) of H. pylori Cag7 has been selected as the putative vaccine antigen and is expressed in L. lactis, which can deliver heterologous proteins to mucosal as well as systemic immune systems via mucosal routes. The Cag7 protein used in this study is thought to fulfill the criteria that are important for a vaccine antigen, i.e. it is a cell surface molecule as a component of the T4SS by which H. pylori injects virulence factors, including the CagA protein, into gastric epithelial cells (2004, 2003). Furthermore, comparison of the deduced amino acid sequence of Cag7 of H. pylori 51 with those of H. pylori 26695 and J99 counterparts revealed that the FCR, the TCR, and the VHR of Cag‐7 protein possessed a high degree of identity (>95.5%) among these three strains. This highly conserved nature in the amino acid sequence of Cag7 protein among different H. pylori isolates also supports the potential of the Cag7 protein as a vaccine antigen against H. pylori. Several studies have revealed that the B‐cell epitopes on native antigen proteins are predominantly composed of hydrophilic amino acids rather than hydrophobic amino acids, because protruding regions on the surface of the antigen protein are generally recognized as the epitopes (2006, 2008). On the other hand, the immunodominant sites for helper T cells tend to be regions that can be folded as α‐helices and helices that are amphipathic, possessing a hydrophobic side and a hydrophilic side separated in space (1988, 1987). These two sides may serve to interact with the major histocompatibility molecule on the antigen‐presenting cell and with the T‐cell receptor, respectively. As hydrophobicity analysis revealed that the ct383 aa region of H. pylori Cag7 protein had a hydrophobic part between both hydrophilic ends in its primary structure, a decision was made to choose the Cag7‐ct383 aa region as the vaccination antigen.
To express the H. pylori Cag7‐ct383 protein in the oral vaccine delivery vehicle L. lactis, the PCR‐amplified cag7‐ct383 gene of H. pylori 51 was cloned in the E. coli–L. lactis shuttle vector (pMG36e). The vector plasmid pMG36e contains a promoter, a ribosomal binding site, the start of an ORF, and a transcriptional terminator, and is known to be a constitutive expression vector for the inserted gene in L. lactis (Van De Guchte et al., 1989). When the expression level of the Cag7‐ct383 protein in the L. lactis transformant was compared by Western blot analysis during the culture period for 120 h, the expression level of the Cag7‐ct383 protein (41.4 kDa) exhibited no significant variation between the early log phase and the late log phase, with the maximum level expression occurring at the early stationary phase, thus demonstrating its constitutive expression regardless of the population growth phases. In our previous studies, the expression level of the H. pylori HpaA protein by the vector plasmid pMG36e in the L. lactis transformant remained constant irrespective of the population growth phases (Kim et al., 2006a), whereas the expression level of the Cag12 protein appeared to vary during the growth phases in the L. lactis transformant, reaching a maximum level at the early log phase and decreasing to the basal level at the stationary phase (Kim et al., 2006b). As the vector plasmid pMG36e is a constitutive expression vector, these previous and current results suggest that the Cag12 protein, but not HpaA and Cag7‐ct383, produced in L. lactis may be unstable during the stationary phase. The oral administration of the L. lactis transformant expressing the Cag7‐ct383 protein into mice generated the anti‐Cag7 antibody in the serum of five of five mice, demonstrating that the systemic humoral immune response was successfully induced against the Cag7‐ct383 protein delivered by L. lactis via the mucosal route. The experimental approaches including the optimal vaccination condition and protection experiment against H. pylori infection still remain to be determined.
When we compared the solubility and cellular localization of Cag7‐ct383 (41.4 kDa) with those of HpaA (29 kDa) or Cag12 (31 kDa) generated in L. lactis transformants, none of these proteins appeared to be secreted extracellularly. The Cag7‐ct383 protein was detected mainly in the soluble fraction, whereas half of the Cag12 protein was located in the soluble fraction. In contrast, HpaA protein was mainly detected in the insoluble envelope fraction. As our previous studies revealed that oral administration of the L. lactis transformant producing Cag12 in mice resulted in successful induction of a humoral immune response to Cag12 in two of five cases (Kim et al., 2006b) and the oral administration of L. lactis transformant producing HpaA in five mice failed to induce humoral immune response to HpaA (Kim et al., 2006a), these previous and current results suggested that the antigenic protein expressed in the recombinant L. lactis in an intracellular soluble form might be more efficient in eliciting a humoral immune response compared with the antigenic protein expressed as an intracellular insoluble form upon oral administration of the recombinant bacterial cells. However, the possibility cannot be excluded that the difference in their capability of inducing a humoral immune response might simply reflect the fact that these Cag7, Cag12, and HpaA are completely different proteins.
In summary, the current results have demonstrated that the ct383 aa region of the Cag7 protein of H. pylori was highly conserved among the Korean strain (51) and the two previous isolates (26695 and J99). In addition, L. lactis transformant possessing the recombinant plasmid pMG36e/cag7‐ct383 was able to synthesize the 41.4‐kDa ct383 aa region of the Cag7 protein without extracellular secretion. Oral administration of the recombinant L. lactis expressing Cag7‐ct383 to mice appeared to generate the anti‐Cag7 antibody in serum, suggesting that it might be used as an oral vaccine to induce protective immunity to H. pylori.
Supporting information
Fig. S1. Hydrophobicity profile (a) and schematic representation of various domains (b) of full length of Cag7 protein from Helicobacter pylori 51.
Please note: Wiley‐Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Acknowledgement
This research was supported by Kyungpook National University Research Fund (2008).
Editor: Peter Timms
References
- Alm RA, Ling LS, Moir DT et al (1999) Genomic‐sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori . Nature 397: 176–180. [DOI] [PubMed] [Google Scholar]
- Backert S & Selbach M (2008) Role of type IV secretion in Helicobacter pylori pathogenesis. Cell Microbiol 10: 1573–1581. [DOI] [PubMed] [Google Scholar]
- Bermudez‐Humaran LG, Langella P, Miyoshi A, Gruss A, Guerra RT, Montes de Oca‐Luna R & Le Loir Y (2002) Production of human papillomavirus type 16 E7 protein in Lactococcus lactis . Appl Environ Microb 68: 917–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzofsky JA (1988) Features of T‐cell recognition and antigen structure useful in the design of vaccines to elicit T‐cell immunity. Vaccine 6: 89–93. [DOI] [PubMed] [Google Scholar]
- Berzofsky JA, Cease KB, Cornette JL, Spouge JL, Margalit H, Berkower IJ, Good MF, Miller LH & DeLisi C (1987) Protein antigenic structures recognized by T cells: potential applications to vaccine design. Immunol Rev 98: 9–52. [DOI] [PubMed] [Google Scholar]
- Chen M, Chen J, Liao W, Zhu S, Yu J, Leung WK, Hu P & Sung JJ (2003) Immunization with attenuated Salmonella typhimurium producing catalase in protection against gastric Helicobacter pylori infection in mice. Helicobacter 8: 613–625. [DOI] [PubMed] [Google Scholar]
- Cheun HI, Kawamoto K, Hiramatsu M, Tamaoki H, Shirahata T, Igimi S & Makino SI (2004) Protective immunity of SpaA‐antigen producing Lactococcus lactis against Erysipelothrix rhusiopathiae infection. J Appl Microbiol 96: 1347–1353. [DOI] [PubMed] [Google Scholar]
- Crespo A & Suh B (2001) Helicobacter pylori infection: epidemiology, pathophysiology, and therapy. Arch Pharm Res 24: 485–498. [DOI] [PubMed] [Google Scholar]
- Czinn SJ (2004) Helicobacter infection: pathogenesis. Curr Opin Gastroenterol 20: 10–15. [DOI] [PubMed] [Google Scholar]
- Dong XN, Qi Y, Ying J, Chen X & Kim YH (2006) Candidate peptide vaccine induced potent protection against CSFV and identified a principal sequential neutralizing determinant on E2. Vaccine 24: 426–434. [DOI] [PubMed] [Google Scholar]
- Holo H & Nes IF (1989) High‐frequency transformation by electroporation of Lactococcus lactis subsp. cemoris grown with glycine in osmotically stabilized media. Appl Environ Microb 55: 3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun DY, Rue SW, Kim BW & Kim YH (2003) Detection of mitotic centromere‐associated kinesin (MCAK) during cell cycle progression of human Jurkat T cells using polyclonal antibody raised against its N‐terminal region overexpressed in E. coli . J Microbiol Biotechn 13: 912–918. [Google Scholar]
- Khan AA, Babu JP, Gupta G & Rao DN (2008) Identifying B and T cell epitopes and studying humoral, mucosal and cellular immune response of V antigen of Yersinia pestis . Vaccine 26: 316–332. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Jun DY, Yang CA & Kim YH (2006a) Cloning and expression of hpaA gene of Korean strain Helicobacter pylori 51 in oral vaccine delivery vehicle Lactococcus lactis subsp. lactis MG1363. J Microbiol Biotechn 16: 318–324. [Google Scholar]
- Kim SJ, Jun DY, Yang CH & Kim YH (2006b) Expression of Helicobacter pylori cag12 gene in Lactococcus lactis MG1363 and its oral administration to induce systemic anti‐Cag12 immune response in mice. Appl Microbiol Biot 72: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa H, Itoh T, Tomioka Y, Mizugaki M & Yamaguchi T (1996) Induction of IFN‐gamma and IL‐1 alpha production in macrophages stimulated with phosphopolysaccharide produced by Lactococcus lactis ssp. cremoris . Int J Food Microbiol 31: 99–106. [DOI] [PubMed] [Google Scholar]
- Kyte J & Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132. [DOI] [PubMed] [Google Scholar]
- Lee MH, Roussel Y, Wiks M & Tabaqchali S (2001) Expression Helicobacter pylori urease subunit B gene in Lactococcus lactis MG1363 and its use as a vaccine delivery system against H. pylori infection in mice. Vaccine 19: 3927–3935. [DOI] [PubMed] [Google Scholar]
- Lee WK, Choi SH, Park SG et al (1999) Genomic diversity of Helicobacter pylori . J Korean Soc Microbiol 34: 519–532. [Google Scholar]
- Liu XL, Li SQ, Liu CJ, Tao HX & Zhang ZS (2004) Antigen epitope of Helicobacter pylori vacuolating cytotoxin A. World J Gastroentero 10: 2340–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M & O'Callaghan D (2004) Euroconference on the biology of type IV secretion processes: bacterial gates into the outer world. Mol Microbiol 53: 1–8. [DOI] [PubMed] [Google Scholar]
- Nouaille S, Ribeiro LA, Miyoshi A, Pontes D, Le Loir Y, Oliverira SC, Langella P & Azevedo V (2003) Heterologous protein production and delivery systems for Lactococcus lactis . Genet Mol Res 2: 102–111. [PubMed] [Google Scholar]
- Pei H, Liu J, Cheng Y, Sun C, Wang C, Lu Y, Ding J, Zhou J & Xiang H (2005) Expression of SARS‐coronavirus nucleocapsid protein in Escherichia coli and Lactococcus lactis for serodiagnosis and mucosal vaccination. Appl Microbiol Biot 68: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz C, Hafsi N & Voland P (2003) Helicobacter pylori virulence factors and the host immune response: implications for therapeutic vaccination. Trends Microbiol 11: 134–138. [DOI] [PubMed] [Google Scholar]
- Ribeiro LA, Azevedo V, Le Loir Y, Oliveira SC, Dieye Y, Piard JC, Gruss A & Langella P (2002) Production and targeting of the Brucella abortus antigen L7/L12 in Lactococcus lactis : a first step towards food-grade live vaccines against brucellosis. Appl Environ Microb 68: 910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K, Chamberlain LM, Lopez MC, Rush CM, Marcotte H, Le Page RW & Wells JM (2004) Mucosal and cellular immune responses elicited by recombinant Lactococcus lactis strains expressing tetanus toxin fragment C. Infect Immun 72: 2753–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde M, Puls J, Buhrdorf R, Fischer W & Haas R (2003) A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol 49: 219–234. [DOI] [PubMed] [Google Scholar]
- Satin B, Del Giudice G, Della Bianca V, Dusi S, Laudanna C, Tonello F, Kelleher D, Rappuoli R, Montecucco C & Rossi F (2000) The neutrophil‐activating protein (HP‐NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J Exp Med 191: 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, Fiers W & Remaut E (2000) Treatment of murine colitis by Lactococcus lactis secreting interleukin‐10. Science 289: 1352–1355. [DOI] [PubMed] [Google Scholar]
- Tai SS, Wang TR & Lee CJ (1997) Characterization of hemin binding activity of Streptococcus pneumoniae . Infect Immun 65: 1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J, Suzuki T, Mimuro H & Sasakawa C (2003) Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol 5: 395–404. [DOI] [PubMed] [Google Scholar]
- Theisen M, Soe S, Brunstedt K, Follmann F, Bredmose L, Israelsen H, Madsen SM & Druilhe P (2004) A Plasmodium falciparum GLURP‐MSP3 chimeric protein; expression in Lactococcus lactis, immunogenicity and induction of biologically active antibodies. Vaccine 22: 1188–1198. [DOI] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR et al (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori . Nature 388: 539–547. [DOI] [PubMed] [Google Scholar]
- Van De Guchte M, Jos M, Der Vossen BM, Kok J & Venema G (1989) Construction of a Lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microb 55: 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin KQ, Hoshino Y, Toda Y et al (2003) Immunogenicity and protective efficacy of orally administered recombinant Lactococcus lactis expressing surface‐bound HIV Env. Blood 102: 223–228. [DOI] [PubMed] [Google Scholar]
- Xu C, Li ZS, Du YQ, Tu ZX, Gong YF, Jin J, Wu HY & Xu GM (2005) Construction of a recombinant attenuated Salmonella typhimurium DNA vaccine carrying Helicobacter pylori hpaA. World J Gastroentero 11: 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zendehdel N, Nasseri‐Moghaddam S, Malekzadeh R, Massarrat S, Sotoudeh M & Siavoshi F (2005) Helicobacter pylori reinfection rate 3 years after successful eradication. J Gastroen Hepatol 20: 401–404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Hydrophobicity profile (a) and schematic representation of various domains (b) of full length of Cag7 protein from Helicobacter pylori 51.
Please note: Wiley‐Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


