Abstract
A genome scan was conducted to seek evidence for polymorphic genes that influence cause of death in mice produced by a cross between CB6F1 females and C3D2F1 males. Loci on chromosomes 1 and 4 were found to modulate risk of lymphoma. A locus on chromosome 4 influenced risk of mammary adenocarcinoma among multiparous female mice, but had no significant effect in virgin females. A chromosome 4 locus was found to modulate risk of death from either hemangiosarcoma or fibrosarcoma. A suggestive linkage was noted (at p =.09) between a marker on chromosome 11 and hepatocellular carcinoma. Lastly, a locus on chromosome 6 was noted to influence the likelihood that pulmonary adenocarcinoma would be present at death. The collection of normal and neoplastic tissues from 1004 terminal necropsies, together with genetic information, provides a valuable resource for further studies of the genetic influences on late-life diseases in mice.
MOST analyses of the genetic basis of cancer risk in mice begin with an inbred strain with an unusually high rate of occurrence of a specific tumor type, and involve examination of back-crosses and congenic stocks to try to map the gene or genes responsible for the index lesion. Such an approach makes excellent use of the inbred nature of most laboratory mouse lines, in which uniform homozygosity often reveals the effects of recessive alleles at loci whose wild-type alleles prevent neoplastic transformation. The use of inbred stocks to screen for cancer-prone alleles has some limitations, however, including the possibility that the phenotype may depend upon a specific combination of recessive alleles that may be unlikely to occur together except in the context of deliberate inbreeding. The historical development of mouse genetics, linked for many years to the retention and selection of mouse stocks with unusually high tumor incidence, is also likely to have favored the detection of rare allele combinations of this kind.
For several years, we have been seeking evidence for polymorphic genes that modulate life span and age-sensitive traits in a four-way cross between four of the common laboratory inbred stocks. The mice of this UM-HET3 (UM-HET stands for “University of Michigan genetically Heterogeneous”) stock are the progeny of (BALB/c × C57BL/6J)F1 females and (C3H/HeJ × DBA/2J)F1 males. The construction of this stock ensures that each of the test mice receives 25% of its genes from each of the four inbred grandparents, but none of the test mice receives two copies of any allele from any of the four progenitors. Thus, the test mice will not be homozygous at any loci, except those loci where by chance two of the four progenitors carry identical alleles. A genome scan of this population thus has the potential to reveal evidence for loci where a polymorphism modulates risk of a specific illness to an extent that is relatively independent of any one specific set of background genes.
In three successive studies of the UM-HET3 population, we have accumulated comprehensive necropsy data for over 1000 individual genotyped mice, of which 886 cases were judged to have died from a specific illness. We report here the results of genome scan calculations that reveal loci on mouse chromosomes 1, 4, and 6, and possibly chromosome 11, which modulate the risk of specific forms of neoplasia diagnosed at terminal necropsy.
Methods
Mice
All of the mice used derived from a four-way cross among four inbred strains: BALB/cJ (C), C57BL/6J (B6), C3H/HeJ (C3), and DBA/2J (D2). These experimental animals are the progeny of (C × B6)F1 females and (C3 × D2)F1 males and are referred to as UM-HET3 mice. The F1 breeding animals were purchased from The Jackson Laboratories (Bar Harbor, ME). Animals were housed segregated by sex (except for the BCA [Breast Cancer and Aging] females from 3 to 6 months of age; see below) in a single suite of specific pathogen-free (SPF) rooms and were exposed to identical environmental conditions (12:12 hour light:dark cycle, 23°C). Mice were given ad libitum access to water and Purina (St. Louis, MO) laboratory mouse chow. The cages were covered with microisolator tops to minimize the spread of infectious agents. Sentinel mice were tested every 3 months to verify the pathogen-free status. The test battery always includes titers for Sendai, minute virus of mice (MVM), and coronavirus (mouse hepatitis virus), and one of the four quarterly tests includes, in addition, assessment of titers for PVM (pneumonia virus of mice), GD-7, Reo-3 (reovirus), Mycoplasma pulmonis, LCMV (lymphocytic choriomeningitis virus), ectromelia, K virus, polyoma virus, mouse adenovirus, and parvovirus. All such tests were negative throughout the course of the study. This work was approved by the Animal Care and Use Committee at the University of Michigan.
Necropsy data were pooled for analysis from three independent cohorts. The LAG1 (Longevity Assurance Gene) cohort consisted of mice born between 9/93 and 3/95, and included 131 virgin males and 147 virgin females. (The numbers given refer to those animals that were submitted for necropsy.) The BCA cohort consisted of mice born between 9/94 and 7/95, and included 293 female mice. All mice in the BCA cohort were caged with males (not part of the life span study) between 2 and 6 months of age, and nearly all of them gave birth to multiple litters at these ages, which were removed from the cages at age 21 days. The LAG2 cohort consisted of mice born between 3/98 and 10/99, and included 179 virgin males and 254 females.
Mice were culled from the colony for several reasons: (a) when male mice were found to have been seriously injured by bite wounds, all mice in the cage (regardless of health status) were culled, typically at age 6–14 months; (b) a small number of mice were removed from the database because technical errors resulted either in their escape from captivity or loss of information about exact date of death.
Necropsy
Mice were inspected at least daily. Mice suspected to be ill (because of weight loss, poor grooming, or visible tumor) were observed twice daily except on weekends. Mice judged by an experienced technician to be so severely ill that survival for more than a few additional days was unlikely were taken to the necropsy suite and humanely euthanized using CO2 asphyxiation; this group made up 59% of the total. The set of criteria used for this decision include: (a) large and/or bleeding tumor mass; (b) inability to eat or to drink; (c) loss of more than 10% of body weight in a week; (d) lethargy and lack of responsiveness to gentle prodding; and (e) other signs of inanition, such as poor grooming and hunched posture. Mice found dead were also submitted for necropsy. The necropsy protocol has been described in detail elsewhere (1), and involved both gross inspection and histological examination of sections from 37 organs. Histopathological evaluation of cases in the LAG1 and BCA cohorts was conducted by the late Clarence Chrisp. Evaluation of cases in the LAG2 cohort was performed by Ruth Lipman. A selection of cases with a provisional diagnosis of fibrosarcoma (from the LAG1 and BCA cohorts) or hemangiosarcoma (from the LAG2 cohort) were directly compared to ascertain that the difference in diagnosis reflected physical differences in the lesion and not merely a difference in nomenclature. Although it is difficult to determine the cause of death in mice found dead, or to assign a cause of death in cases where severe morbidity prompted euthanasia of the animal, for this study, a lesion was deemed to be the cause of death if it was the sole serious lesion present, or if the lesion was so severe that it was likely to have led to the death of the mouse or to the symptoms that led the husbandry staff to submit the animal for euthanasia. Mice with no severe lesions or with multiple severe lesions were classified as “unknown” in the cause of death tabulation.
Genotyping Assays
Genomic DNA was prepared from 1 cm sections of tail from 4-week-old animals using a standard phenol extraction method (2). Final DNA preparations were tested for concentration, ability to sustain polymerase chain reaction (PCR) amplification under standard conditions, and electrophoretic size distribution. DNA was genotyped using an ALFexpress automated sequence analyzer (Pharmacia, Piscataway, NJ); the details of this genotyping method have been described previously (3). Primer pairs were purchased from MWG Biotech, Inc. (High Point, NC). In total, 185 markers were examined from 99 genetic loci. Of the 99 loci, 86 markers were informative for both the maternal- and paternal-derived alleles and 13 loci were only informative for maternal or paternal. The selection of genetic loci was described previously in detail (4). Chromosomal localization and order of markers were calculated using the MapMaker QTX program package (Whitehead Institute, MIT, Cambridge, MA).
Qualitative Trait Locus Mapping
Qualitative Trait Loci (QualTL) is a maximum likelihood-based method for interval mapping analysis of discrete phenotypes from experimental genetic crosses. From the conceptual point of view, QualTL is an example of a latent class model in which the categorical outcomes (phenotypes) are modeled as a function of unknown (unobservable) putative genotype at a putative marker. The computational algorithm is outlined in Galecki and colleagues (5), and the method has been used successfully in a previous study of fitness and life span in Caenorhabditis elegans (6). In the context of the current article, single marker analysis was used instead of interval mapping, so a simplified version of the QualTL algorithm corresponding to a standard logistic regression was used. The analysis performed is designed to evaluate the effects of genetic differences on the proportions of mice dying of specific causes, or with specific lesions. This approach is biased to an unknown extent by a potential confound between the cause of death and the life span of the mice; the data set cannot provide information about risks for specific diagnosis in mice at times later than the age at death.
Results
Three life span experiments were conducted using genetically heterogeneous mice of the UM-HET3 stock, bred from a cross between CB6F1 dams and C3D2F1 sires. The LAG1 and LAG2 experiments each involved a mixture of virgin male and virgin female animals, and the BCA experiment involved female mice that had been repeatedly pregnant from age 2 months to 6 months. Previous publications have focused on the genetic control of T-cell subsets (3,7), life span (8,9), hormone levels (10), cataracts (11), and body weight (12) in one or more of these populations. Table 1 summarizes key life table statistics for this set of longevity experiments.
Table 1.
Summary of Life Span Statistics for Three Cohorts of UM-HET3 Mice.
| Cohort | Number of Males/Females | Mean Life Span | Mean Life Span: Males | Mean Life Span: Females | Mean Age of Oldest 5 | Age of Oldest |
|---|---|---|---|---|---|---|
| LAG1 | 131/147 | 746 | 678 | 807 | 1108 | 1169 |
| BCA | 0/293 | 775 | NA | 775 | 1106 | 1182 |
| LAG2 | 179/252 | 891 | 909 | 878 | 1269 | 1306 |
Note: NA = not available.
A surveillance system, described in (1), was employed to try to increase the proportion of mice that produced useful diagnostic necropsies. Key features included at least daily inspection for recent deaths (typically twice a day for 5 days each week), and a willingness to euthanize mice that appeared so severely ill that survival for more than a few additional days seemed unlikely. This system allowed us to obtain informative necropsies on more than 90% of the mice in this study (excluding those culled for fighting or because of errors in record-keeping or mouse husbandry). An attempt was made to infer a most likely cause of death in each case, and this was possible in 886 of the 1004 cases evaluated. In the other 118 cases, no single cause of death could be assigned, either (a) because advanced tissue autolysis prevented evaluation, (b) because two or more lesions were judged to have contributed to death or moribund state, or (c) in a small number of cases, because no lesion serious enough to have caused death could be identified.
Table 2 summarizes the cause of death diagnoses in each of the three independent cohorts, further stratified by sex for the LAG1 and LAG2 series. Table 2 also lists (right column) the total number of cases in the series with the diagnoses indicated. The “miscellaneous nonneoplastic” disease category included a wide range of illnesses, such as peritonitis, brain infarct, glomerulonephritis, inanition secondary to enamel organ dysplasia, myocardial degeneration, ovarian hemorrhage, polyarteritis nodosa, and various localized infections. The “miscellaneous neoplastic” category included diagnoses of mast cell tumor, rhabdomyosarcoma, islet cell carcinoma, pheochromocytoma, sperm granuloma, myxosarcoma, c-cell adenocarcinoma, mesothelioma, nerve sheath tumor, thyroid adenocarcinoma, leiomyosarcoma, osteosarcoma, and others.
Table 2.
Cause of Death Diagnoses in Three Cohorts of UM-HET3 Mice.
| Cause of Death | Percentage of Diagnosed Cases |
Number of Cases | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LAG1 Virgin Females | LAG1 Virgin Males | BCA Multiparous Females | LAG2 Virgin Females | LAG2 Virgin Males | All Combined | |||||
| Endometrial sarcoma | 3 | 0 | 6 | 2 | 0 | 25 | ||||
| Fibrosarcoma | 18 | 5 | 10 | 3 | 1 | 64 | ||||
| Hemangiosarcoma | 3 | 1 | 3 | 13 | 10 | 56 | ||||
| Hepatocellular carcinoma | 1 | 12 | 2 | 1 | 8 | 34 | ||||
| Histiocytic sarcoma | 3 | 3 | 5 | 6 | 5 | 40 | ||||
| Lymphoma | 34 | 8 | 18 | 35 | 25 | 217 | ||||
| Mammary adenocarcinoma | 7 | 0 | 24 | 9 | 0 | 91 | ||||
| Pituitary adenoma | 6 | 0 | 2 | 1 | 1 | 16 | ||||
| Pulmonary adenocarcinoma | 2 | 14 | 6 | 10 | 25 | 96 | ||||
| Miscellaneous, neoplastic | 6 | 4 | 8 | 14 | 12 | 81 | ||||
| Congestive heart failure | 2 | 6 | 5 | 0 | 0 | 23 | ||||
| Mouse urinary syndrome | 0 | 32 | 0 | 0 | 0 | 37 | ||||
| Miscellaneous, nonneoplastic | 15 | 15 | 13 | 6 | 13 | 106 | ||||
| Diagnosable Cases (n) | 136 | 117 | 268 | 208 | 157 | 886 | ||||
| Total Cases (n) | 147 | 131 | 293 | 254 | 179 | 1004 | ||||
Notes: The bottom two lines of the Table show, for each combination of sex and cohort, the total number of cases submitted for necropsy and the number of cases for which the pathologist was able to infer a most likely cause of death (i.e., “diagnosable” cases). The first five columns in the body of the Table present the percentage of the diagnosable cases with the indicated cause of death. For example, in the LAG1 population of virgin females, 2% of the 136 diagnosable cases were found to have died because of congestive heart failure. The rightmost column lists the total number of cases with the indicated diagnosis, pooled across all five of the sex/cohort combinations.
UM-HET = University of Michigan genetically HETerogeneous; LAG = Longevity Assurance Gene; BCA = Breast Cancer and Aging.
Several features of Table 2 should be noted because they influenced the gene mapping strategy. There is evidence of unanticipated differences in pathology between the LAG1 and LAG2 cohorts. Mouse urinary syndrome, for example, was common among males in the LAG1 group, but not noted in the LAG2 males. In addition, we found unexpectedly that fibrosarcoma was frequently a cause of death in the LAG1 and BCA mice (18% of virgin females and 5% of virgin males), but was infrequent in the LAG2 series (3% of virgin females and 1% of virgin males). Conversely, hemangiosarcoma was rare in the LAG1 group (3% and 1% in females and males, respectively), but much more common in LAG2 (13% and 10%, respectively). These variations in lesion prevalence may reflect subtle environmental differences under which the cohorts of mice were raised. It is also apparent that the proportion of mice dying of mammary adenocarcinoma was far higher among multiparous females (24% of diagnosable cases) than for virgin females (9%), consistent with previous assessments of the effects of hormonal environment on mammary cancer in mice.
A qualitative trait locus (QualTL) mapping procedure was then used to seek evidence for loci at which allelic differences modulated the likelihood of specific causes of death. Results for which experiment-wise p <.10 are summarized in the first five lines of Table 3. Four of these met the criterion of p <.05, the preselected significance threshold for the protocol, and one (the maternal allele of D11Mit156 as a predictor of hepatocellular carcinoma) was suggestive but not definitive at p =.09. For hepatocellular carcinoma and lymphoma, the calculations involved data from both male and female mice pooled together. Because the risk of mammary adenocarcinoma differs between multiparous and virgin females, the calculation for this cause of death was done three times, once for multiparous females (BCA cohort only), once for virgin females (LAG1 and LAG2 cohorts), and once for all females regardless of mating status. Only the multiparous females showed a significant genetic effect, which is therefore listed in Table 3. The genome scan found no significant QualTL for hemangiosarcoma or for fibrosarcoma, but did find a strong association between D4Mit170p on mouse chromosome 4 and the likelihood that the mouse would die either of fibrosarcoma or hemangiosarcoma, considering these two histologically distinct lesions as a single combined category.
Table 3.
Summary of Quantitative Trait Locus Locations.
| Traita | Locusb | Group/Nc | Allelesd | p Valuee | Sf | Pos (cM)g | Pos (mb) | MGI |
|---|---|---|---|---|---|---|---|---|
| Hepatocellular carcinoma | D11Mit156m | All/873 | B6: 6% C: 1% | .09 | 34 | 64 | 89041 | |
| Lymphoma | D1Mit206p | All/876 | D2: 30% C3: 18% | .003 | * | 95.8 | 174 | 91585 |
| Lymphoma | D4Mit84p | All/848 | D2: 29% C3: 18% | .02 | * | 37.7 | 75 | 92920 |
| Hemangiosarcoma or fibrosarcoma | D4Mit170p | All/842 | C: 17% B6: 8% | .009 | * | 66.6 | 136 | 92798 |
| Mammary adenocarcinoma | D4Mit55p | Multiparous females/212 | C3: 34% D2: 12% | .018 | * | 19.8 | 45 | 92889 |
| Pulmonary adenocarcinoma (incidental) | D6Mit198m | All/869 | C: 23% B6: 13% | .007 | * | 68 | 140 | NA |
Notes: a Trait is the diagnosed cause of death, except for pulmonary adenocarcinoma, in which the trait was the presence of this lesion at necropsy, regardless of cause of death.
b Locus indicates the SSLP marker most closely linked to the QTL. An “m” in the locus designation indicates maternal inheritance, and a “p” indicates paternal inheritance.
c Indicates the group of mice evaluated (all mice, or just multiparous females), and the number of mice included in the calculation. Values of “N” differ slightly because the amount of missing genotype data varied among the SSLP marker loci.
d Gives the proportion of cases with indicated trait for each of the two relevant genetic alleles.
e The permutation-based experiment-wise probability level for the association.
f An asterisk indicates those associations that reach the p <.05 significance criterion.
g Pos (cM), Pos (mb), and MGI indicate, respectively, the position of the SSLP marker in centimorgans from the centromere, the position of the SSLP marker in millions of base pairs, and the identification code in the Mouse Genome Informatics database.
SSLP = simple sequence length polymorphism; NA = not available; QTL = quantitative trait locus.
Figure 1 illustrates the strength of association between alleles of D1Mit206 and D4Mit84 and the risk of dying because of lymphoma. For each of these loci, inheriting the D2 allele gives a substantially increased risk of death by lymphoma. The two effects are additive; as shown in the bottom panel of the figure, mice that inherit the D2 allele at both loci have a 36% chance of dying from lymphoma, compared to a 13% chance for mice that inherit the C3 allele at both loci.
Figure 1.
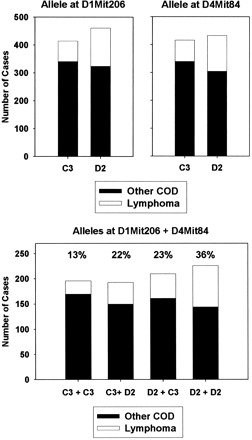
Each bar corresponds to the number of mice with the indicated allele (top panels) or combination of alleles (bottom panel), pooled across all three experimental populations. The top portion of each bar corresponds to the number of cases for which lymphoma was the cause of death; the bottom portion represents all other diagnoses. The percentages in the bottom figure show the proportion of lymphoma cases compared to the total number of mice with each of the four possible allele combinations. In the bottom figure, the allele at D1Mit206 is listed followed by the allele at D4Mit84. COD = cause of death
Figure 2 illustrates the strength of association between alleles at D4Mit55 and the risk of mammary adenocarcinoma death in multiparous female mice. The C3 allele at this locus increases the risk of death from breast cancer from 12% to 34%. When the outcome was “incidental” mammary adenocarcinoma, that is, the presence of this lesion at necropsy regardless of inferred cause of death, the incidence rates were 15% and 45%, respectively, a difference of three-fold. No locus reached significance when the calculations included only virgin female mice, or when the group of virgin and multiparous female animals was evaluated together. For virgin females, the risk of death from mammary adenocarcinoma was 6.8% (12/176) in mice with the C3 allele at D4Mit55, and 6.7% (13/193) in mice with the D2 allele. Thus, the D4Mit55-linked locus seemed to modulate breast cancer risk in multiparous mice only.
Figure 2.
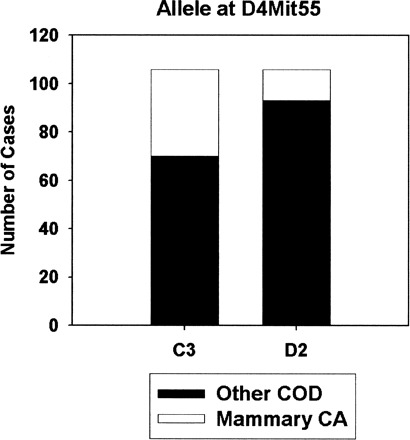
Each bar corresponds to the number of multiparous female mice with the indicated allele. The top portion of each bar corresponds to the number of cases for which mammary adenocarcinoma was the cause of death; the bottom portion represents all other diagnoses. COD = cause of death; CA = carcinoma
A genome scan was also used to seek loci that influenced the likelihood of “incidental” lesions, that is, lesions noted at necropsy whether or not they were thought to have led to death or moribund status. Only one new association was detected, using an experiment-wise significance criterion of p <.05, with this set of outcome measures: As shown at the bottom of Table 3, the C allele at locus D6Mit198 was found to be associated with the appearance of pulmonary adenocarcinoma at necropsy (p =.007). This lesion was found in 23% of the mice with the C allele at D6Mit198, and in 13% of those with the B6 allele. Pulmonary adenocarcinoma was more common in males than in females (see Table 2), but the effect of the locus linked to D6Mit198 was apparent both in male mice (36% vs 20%; p <.003 by chi-square test) and in female mice (17% vs 9%; p =.012). Although the association between D6Mit198m and lethal pulmonary adenocarcinoma did not reach experiment-wise significance in the genome scan, a post hoc analysis by chi-square testing found higher rates of lethal pulmonary adenocarcinoma associated with this locus in males (22% vs 12%, p <.03), though not in females (7% vs 4%, p <.16).
Discussion
Our previous work has found evidence, in the LAG1 population, for genetic loci on chromosomes 2, 9, 10, 12, and 16 with significant effects on life span in male mice, female mice, or both (8,9), and has shown that the effects of at least three of these loci modulate life expectancy both in mice whose death is caused by neoplasia and those that die of a nonneoplastic disease (9). The current study does not focus on length of life, but instead on genes that modulate the cause of death rather than its time of occurrence.
Necropsy cases from three separate studies of the same genetic cross were combined to increase statistical power, even though this strategy runs the risk that differences among the studies may have diminished the strength of gene/trait association in the pooled data set. Some of these differences are matters of design, such as the inclusion of virgin mice in LAG1 and LAG2 and the use of multiparous females for the BCA population. It is clear, though, that the studies also differ in ways that were unanticipated. For example, males in the LAG1 group had a high incidence of the mouse urinary syndrome (13,14), which for unknown reasons was not observed among the LAG2 males. Fibrosarcoma, though commonly judged to be the cause of death among the LAG1 mice (particularly among the virgin females), was much less common in the LAG2 series; conversely, hemangiosarcoma was a frequent cause of death in the LAG2 mice only. Because each population was produced by breeding (BALB/c × C57BL/6J)F1 females to (C3H/HeJ × DBA/2J)F1 males, we can only speculate that these differences reflect some uncontrolled factor(s), such as unknown contaminants in water or food or subtle alterations in environmental noise, air quality, or the like.
The genome scan results for lymphoma are the most straightforward. In the UM-HET3 population, lymphoma is the most frequent single cause of death, responsible for the deaths of about 35% of virgin females, 18% of multiparous females, and 8%−25% of virgin males. Mice inheriting the D2 allele at D1Mit206 or the D2 allele at D4Mit84 were at greatest risk of dying from lymphoma, with each of these two loci contributing about an equal risk in an additive fashion.
The C3 allele at D4Mit55 led to a nearly three-fold increase in risk that a multiparous female would die of mammary adenocarcinoma, but this effect was not seen in virgin females of the LAG1 or LAG2 studies, for which the two alleles were associated with mammary cancer incidence rates of 6.8% and 6.7%. The basis for the increased risk of mammary adenocarcinoma in multiparous as compared to virgin female mice is not understood, and contrasts with the relation between mammary cancer risk and child-bearing in humans. It seems likely that the locus on chromosome 4 modulates the factors, presumably hormonal, that increase mammary cancer risk in multiparous female mice, although it is noteworthy that mating increases the risk of breast cancer (from 6.7% to 12%) even in mice that inherit the low-risk allele at D4Mit55. The proneoplastic effect of mammary tumor virus transmitted in milk from C3H/HeJ females to their offspring does not influence tumor risk in UM-HET3 mice, in which the C3H/HeJ alleles are contributed by the paternal grandfather.
The B6 allele at D11Mit156 seems to lead to a six-fold increase in the risk of hepatocellular carcinoma in these mice. This tumor is fairly rare as a cause of death, however, with only 34 cases seen among the 886 diagnosable cases, and the association with the chromosome 11 allele did not reach our criterion for experiment-wise significance.
Pulmonary adenocarcinoma was seen at necropsy primarily in those mice that inherited the C allele at D6Mit198. This is a lesion much more common in male than in female UM-HET3 mice, and post hoc analyses showed that this C allele was associated with an 80% increase in the risk of lethal pulmonary tumors in male mice, with an insignificant effect in females. We can postulate that the sex-specific factors that increase risk of lung cancer in these males may themselves be modulated by this chromosome 6 polymorphism. These data also provide a new perspective on a prior study (15) of strain differences in tumor susceptibility in mice given a single oral dose of the potent carcinogen 7,12-dimethylbenz[a]anthracene (DMBA). This earlier work included analyses of induced tumor incidence in BALB/cJ, C3H/HeJ, C57BL/6J, and DBA/2J mice, the grandparental stocks used to produce UM-HET3 mice. Among the four inbred stocks, the highest incidence of pulmonary adenocarcinoma was seen in the BALB/c mice, and it is thus tempting to note the provisional association of the C allele at D6Mit198 with this lesion in the UM-HET3 stock.
The results for hemangiosarcoma and fibrosarcoma are the more difficult to explain. There are two potential confounding factors: (a) slides from the LAG2 mice were not evaluated by the same individual responsible for evaluating the LAG1 and BCA cohorts; and (b) the two groups of animals were produced and raised several years apart, and might therefore have been exposed, inadvertently, to different dietary or environmental influences. Comparison of a sample of cases with these lesions from each of the two necropsy series confirmed that they were indeed histologically distinct. Angiosarcoma in humans has been noted to exhibit microscopic heterogeneity; in one series, for example (16), all cases were reported to have focal vasoformative components similar to those in hemangiosarcomas in the LAG2 cohort, but 7% of the cases also contained solid components resembling the fibrosarcoma seen in the LAG1/BCA mice. Although it is possible that sampling at necropsy resulted in systematic differences in lesion presentation between the two series of cases, we do not consider this a likely explanation for the disparity. The second possibility is that the difference in lesion presentation was due to unknown environmental factor(s) that were different for the two groups of mice. We suspect that some unknown variation in environmental conditions between 1995 and 1998, such as an inadvertent change in the composition of the natural products diet or some alteration in water quality, may have led to the reciprocal increase in vascular neoplasia and reduction in fibrosarcoma risk, but we cannot put this idea to any immediate test. Our genetic results show that an allele linked to D4Mit170 in the BALB/c genome leads to a two-fold increase in risk of hemangiosarcoma and fibrosarcoma compared to the corresponding B6 allele. The experiment-wise p value <.009 makes it unlikely that this is a chance association. We thus hypothesize that the vascular tumors and sarcomas may, in UM-HET3 mice, share a common underlying basis, modulated by this chromosome 4 locus.
It is noteworthy that a previous study of the LAG1 mice, using a separate computational approach, found evidence for two loci on chromosome 4 with an influence on fibrosarcoma risk (17). The strongest association was with a locus mapping to the proximal portion of chromosome 4, between 0–20 cM from the centromere. A second, weaker, association was seen for paternal alleles mapping between 50–92 cM. The current study found no evidence for the proximal gene, but did detect a paternal allele linked to marker D4Mit170 at 67 cM. These two results, though consistent for the distal marker, cannot be considered as independent confirmation, because the LAG1 mice evaluated by Xu and colleagues were included in the current, much larger data set. It is of interest that the proximal quantitative trait locus for LAG1 mice did not produce a detectable signal in the current population pooled across all three study groups.
Our earlier papers, in which UM-HET3 mice were used to map polymorphic loci for age-sensitive traits including T-cell subsets (3,7), life span (8,9), hormone levels (10), and body weight (12) used traditional quantitative trait locus methods, in which the outcome, age at death, was a continuous quantitative trait. In contrast, the current data set was evaluated using an algorithm adapted for mapping of qualitative traits, in which the phenotype takes on one of a small number of discrete values, in the current instance, either the presence or the absence of a specific lethal lesion.
This necropsy series provides a valuable foundation for future studies of the genetic and molecular factors that influence disease in a genetically heterogeneous mouse population. Each mouse in these three cohorts has been genotyped at > 150 biallelic markers, and DNA has been archived that would support fine-scale mapping of chromosomal regions of interest in later studies. Tissue blocks and stained sections are available from each of the necropsy cases as well, which could promote follow-up studies of questions concerning the histopathology of spontaneous late-life tumors, and, potentially, of their molecular constituents as evaluated by tissue microarrays or related immunohistochemical techniques. Each mouse in the series has also been tested for levels of several serum hormones (10,18) and for age-sensitive T-cell patterns (3,7). The combination of genetic, physiological, and terminal histopathological resources for these mice provides a unique resource for future studies of tumor biology in aging animals.
Acknowledgments
Supported by National Institute on Aging grants AG08808 and AG11687. We thank Amanda Pilling, Gretchen Buehner, Maggie Vergara, Steve Pinkosky, and Steve Durkee for their technical assistance.
Footnotes
Decision Editor: James R. Smith, PhD
References
- 1.Chrisp CE, Turke P, Luciano A, Swalwell S, Peterson J, Miller RA. Lifespan and pathology in genetically heterogeneous (four-way cross) mice: a new model for aging research. Vet Pathol. 1996;33:735-743. [DOI] [PubMed] [Google Scholar]
- 2.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989.
- 3.Jackson AU, Fornes A, Galecki A, Miller RA, Burke DT. Multiple-trait quantitative trait loci analysis using a large mouse sibship. Genetics. 1999;151:785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volkman SK, Galecki AT, Burke DT, et al. Quantitative trait loci for femoral size and shape in a genetically heterogeneous mouse population. J Bone Miner Res. 2003;18:1497-1505. [DOI] [PubMed] [Google Scholar]
- 5.Galecki AT, Ten Have TR, Molenberghs G. A simple and fast alternative to the EM algorithm for incomplete categorical data and latent class models. Comput Stat Data Anal. 2001;35:265-281. [Google Scholar]
- 6.Ayyadevara S, Ayyadevara R, Vertino A, Galecki A, Thaden JJ, Reis RJS. Quantitative trait loci affecting fitness and life span in caenorhabditis elegans: categorical trait interval mapping in CL2a4Bergerac-BO recombinant-inbred worms. Genetics. 2002;163:557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller RA, Jackson AU, Galecki AT, Burke DT. Genetic polymorphisms in mouse genes regulating age-sensitive and age-stable T cell subsets in mice. Genes Immun. 2003;4:30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson AU, Galecki AT, Chrisp C, Burke DT, Miller RA. Mouse loci associated with life span exhibit sex-specific and epistatic effects. J Gerontol Biol Sci. 2002;57A:B9-B15. [DOI] [PubMed] [Google Scholar]
- 9.Miller RA, Chrisp C, Jackson AU, Galecki AT, Burke DT. Coordinated genetic control of neoplastic and nonneoplastic diseases in mice. J Gerontol Biol Sci. 2002;57A:B3-B8. [DOI] [PubMed] [Google Scholar]
- 10.Harper JM, Galecki AT, Burke DT, Pinkosky SL, Miller RA. Quantitative trait loci for insulin-like growth factor-I, leptin, thyroxine, and corticosterone in genetically heterogeneous mice. Physiol Genom. 2003;15:44-51. [DOI] [PubMed] [Google Scholar]
- 11.Wolf N, Galecki A, Lipman RD, et al. Quantitative trait locus mapping for age-related cataract severity and synechia prevalence using four-way cross mice. Invest Ophthalmol Vis Sci. 2004;45:1922-1929. [DOI] [PubMed] [Google Scholar]
- 12.Miller RA, Harper JM, Galecki A, Burke DT. Big mice die young: early-life body weight predicts longevity in genetically heterogeneous mice. Aging Cell. 2002;1:22-29. [DOI] [PubMed] [Google Scholar]
- 13.Everitt JI, Ross PW, Davis TW. Urologic syndrome associated with wire caging in AKR mice. Lab Anim Sci. 1988;38:609-611. [PubMed] [Google Scholar]
- 14.Tuffery AA. Urogenital lesions in laboratory mice. J Pathol Bacteriol. 1966;91:301-309. [DOI] [PubMed] [Google Scholar]
- 15.Lipman RD. Effect of calorie restriction on mortality kinetics in inbred strains of mice following 7,12-dimethylbenz[a]anthracene treatment. J Gerontol Biol Sci. 2002;57A:B153-B157. [DOI] [PubMed] [Google Scholar]
- 16.Neuhauser TS, Derringer GA, Thompson LD, et al. Splenic angiosarcoma: a clinicopathologic and immunophenotypic study of 28 cases. Mod Pathol. 2000;13:978-987. [DOI] [PubMed] [Google Scholar]
- 17.Xu S, Yi N, Burke D, Galecki A, Miller RA. An EM algorithm for mapping binary disease loci: application to fibrosarcoma in a four-way cross mouse family. Genet Res. 2003;82:127-138. [DOI] [PubMed] [Google Scholar]
- 18.Harper JM, Wolf N, Galecki AT, Pinkosky SL, Miller RA. Hormone levels and cataract scores as sex-specific, mid-life predictors of longevity in genetically heterogeneous mice. Mech Ageing Devel. 2003;124:801-810. [DOI] [PubMed] [Google Scholar]


