Abstract
Penicillium marneffei is the etiologic agent of a severe systemic disease in immunocompromised hosts in Southeast Asia. In the present study, a novel method, known as loop-mediated isothermal amplification (LAMP), is described for the rapid and specific detection of the species, using a primer set derived from the internal transcribed spacer (ITS) region of the rRNA gene. Amplification products can be detected macroscopically by visual inspection in vials using SYBR Green I as well as by electrophoresis on agarose gel. The LAMP assay resulted in specific amplification of P. marneffei ITS using pure cultures after a 1-h reaction at 65 °C in a water bath; no cross-reactivity with other fungi including other biverticillate penicillia was observed. The detectable DNA limit was two copies. In addition, specific amplification was achieved using paraffin wax-embedded tissue samples from patients with penicilliosis marneffei and tissue samples from bamboo rats. The method provides a powerful tool for rapid diagnostics in the clinical lab, and has potential for use in ecological studies.
Keywords: Penicillium marneffei, penicilliosis, rapid diagnosis, LAMP
Introduction
Penicillium marneffei is the agent of a life-threatening systemic mycosis known as penicilliosis marneffei, occurring in patients infected with HIV in Southeast Asia (Supparatpinyo et al., 1994; Wong et al., 1998; Liyan et al., 2004) and now recognized as an AIDS-defining disease (Lee, 2008). Cases were particularly frequent in endemic zones of northern Thailand (Watanabe et al., 2008), but the disease has also been observed in China (Fisher et al., 2005). Since the first reported Chinese case in 1985 (Wei, 1985), there has been a drastic increase in the incidence of the infection, concomitant with the emergence of the AIDS pandemic. More than 100 cases of AIDS with penicilliosis marneffei were reported between 2003 and 2006 in a single hospital in Guangzhou (Linghua Li & Weiping, 2008). Clinical diagnosis may be hampered by the fact that major manifestations of the mycosis in HIV-infected patients are not specific for P. marneffei. As a result, many patients do not receive timely and appropriate antifungal treatment, and their prognosis is poor. Traditionally, penicilliosis marneffei is diagnosed by a microscopic observation of fungal fission yeast cells in alveolar macrophages and by culturing the etiologic agent. These procedures may be time-consuming (Ukarapol et al., 1998; Mo et al., 2002), and there is a need for experimental diagnostic methods. Serological diagnosis (Panichakul et al., 2002) is tedious because it requires paired, acute- and convalescent-phase sera, and the results may be influenced by contamination or cross-reaction. Several molecular methods have been proposed, such as nested or semi-nested PCR (LoBuglio & Taylor, 1995; Vanittanakom et al., 2002; Prariyachatigul et al., 2003), PCR-enzyme immunoassays (Lindsley et al., 2001) and PCR hybridization (Vanittanakom et al., 1998). All have been developed on the basis of cultured material, and require a fully equipped molecular laboratory. Thus, there is still a need for a rapid and simple technique that is able to deliver an unambiguous identification within a single day.
Loop-mediated isothermal amplification (LAMP) was introduced for the detection of hepatitis B virus DNA by Notomi (2000). This novel technique is able to amplify DNA with high specificity, efficiency and rapidity under isothermal conditions. The assay is based on the use of Bst DNA polymerase, performing autocycling strand displacement DNA synthesis using a set of four or six specially designed primers that recognize six or eight distinct sequences on the target DNA. The cycling reactions result in the accumulation of 109- to 1010-fold amplification of the target in less than an hour. Amplification products can be detected easily by visual assessment of turbidity in Eppendorf vials or by electrophoresis. The sensitivity of LAMP does not appear to be affected by the presence of nontarget DNA in samples, and there is no interference by known PCR inhibitors such as blood, serum, plasma or heparin (Notomi et al., 2000; Enosawa et al., 2003; Poon et al., 2005). These properties of high specificity, selectivity, simplicity and speed made LAMP attractive for the diagnosis of bacteria (Iwamoto et al., 2003; Yoshida et al., 2005; Aoi et al., 2006), viruses (Poon et al., 2004; Hagiwara et al., 2007; Cai et al., 2008) and parasites (Ikadai et al., 2004; Iseki et al., 2007). However, very few papers have appeared on the use of LAMP with fungi (Endo et al., 2004; Ohori et al., 2006; Inacio et al., 2008). We recently developed a protocol for LAMP detection for Fonsecaea agents of chromoblastomycosis (Sun, 2009). In the present study, we introduce LAMP diagnostics for P. marneffei in paraffin wax-embedded human tissue and in bamboo rat tissue samples.
Materials and methods
Strains and biopsy specimens
Forty strains of P. marneffei isolated from human patients and 46 reference strains used in this study are listed in Table 1. All isolates were cultured on Sabouraud's glucose agar plates at 25 °C for 1 week; Escherichia coli was cultured in flasks shaken at 250 r.p.m. with Luria–Bertani at 37 °C overnight. About 0.5 g of mycelium or conidia, or precipitate of E. coli, respectively, were harvested for DNA extraction.
Table 1.
Sampling data of the isolates used in this study
| Species | Accession no. | GenBank | Source |
| Penicillium marneffei | CBS101038 | Assam, India | |
| CBS555.90T | Australia | ||
| CBS388.87 | Vietnam | ||
| CBS122.89 | Indonesia | ||
| CBS440.88 | USA | ||
| SUMS0266 | FJ009553 | Bamboo rat spleen, Jiangxi, China | |
| SUMS0267 | FJ009551 | Bamboo rat spleen, Jiangxi, China | |
| SUMS0264 | FJ009566 | Bamboo rat kidney, Jiangxi, China | |
| SUMS0268 | FJ009565 | Bamboo rat spleen, Jiangxi, China | |
| SUMS0272 | FJ009555 | Bamboo rat lung, Jiangxi, China | |
| SUMS0344 | FJ009554 | Bamboo rat spleen, Shaoguan, China | |
| SUMS0345 | FJ009559 | Bamboo rat kidney, Shaoguan, China | |
| SUMS0346 | FJ009560 | Bamboo rat lung, Shaoguan, China | |
| SUMS0347 | FJ009564 | Bamboo rat liver, Fujian, China | |
| SUMS0348 | FJ009563 | Bamboo rat lung, Fujian, China | |
| SUMS0349 | FJ009552 | Bamboo rat liver, Shaoguan, China | |
| SUMS0350 | FJ009557 | Bamboo rat lung, Shaoguan, China | |
| SUMS0351 | FJ009562 | Bamboo rat lung, Shaoguan, China | |
| SUMS0352 | FJ009558 | Bamboo rat spleen, Shaoguan, China | |
| SUMS0353 | FJ009561 | Bamboo rat lung, Shaoguan, China | |
| SUMS0354 | FJ009556 | Bamboo rat lung, Shaoguan, China | |
| IFM47289 | AB298957 | Chiba University, Japan | |
| IFM47288 | AB298956 | Chiba University, Japan | |
| IFM47287 | AB298955 | Chiba University, Japan | |
| IFM47286 | AB298954 | Chiba University, Japan | |
| IFM47285 | AB298953 | Chiba University, Japan | |
| SUMS0152 | AB353913 | Human blood and bone marrow, Guangdong, China | |
| SUMS0112 | AB353909 | Human, Jiangxi, China | |
| SUMS0165 | AB353908 | Human blood, Guangdong, China | |
| SUMS0186 | AB353917 | Human face and blood, Guangdong, China | |
| SUMS0178 | AB353916 | Human blood, Guangdong, China | |
| SUMS0174 | AB353915 | Human neck, Guangdong, China | |
| SUMS0164 | AB353914 | Human face, Guangdong, China | |
| SUMS0096 | AB353912 | Human dialysate, Guangdong, China | |
| SUMS0050 | AB353911 | Animal, Guangxi, China | |
| SUMS0051 | AB353910 | Human, Guangxi, China | |
| SUMS0111 | AB353919 | Human, Guangdong, China | |
| SUMS0187 | AB353918 | Human blood, Guangdong, China | |
| SUMS0107 | AB353907 | Human legs, Guangdong, China | |
| SUMS0047 | AB353906 | Human legs, Guangdong, China | |
| Aspergillus fumigatus | SUMS0106 | FJ011537 | Human, Shanghai, China |
| SUMS0317 | FJ011543 | ||
| A. flavus | SUMS0060 | FJ011539 | Human sputum, Guangdong, China |
| SUMS0062 | FJ011545 | Human lung, Guangdong, China | |
| A. niger | SUMS0061 | FJ011541 | Human acoustic meatus, Guangdong, China |
| SUMS0037 | FJ011542 | Human, Guangdong, China | |
| A. terreus | SUMS0191 | FJ011538 | Human acoustic meatus, Guangdong, China |
| SUMS0113 | FJ011536 | Human sputum, Guangdong, China | |
| Penicillium griseofulvum | SUMS0392 | FJ011548 | Human face, Guangzhou, China |
| Paecilomyces variotii | SUMS0303 | FJ011547 | Human, Guangzhou, China |
| Penicillium janthinellum | IFM40620 | IAM 7018 | |
| P. chrysogenum | IFM5338 | MTU7003 | |
| P. purpurogenum | IFM40627 | IAM7095 | |
| P. citrinum | IFM40616 | IAM7003 | |
| P. duclauxii | CBS187.89 | NRRL 2020 | |
| P. verruculosum | CBS101366 | Soil, Hong Kong | |
| P. minioluteum | CBS442.89 | Soil, Lyngby, Denmark | |
| P. crustosum | IFM47479 | IFO31913 silk-worm foods | |
| P. funiculosum | IFM57310-11 | CBS235.94, ATCC11797 MD | |
| P. pinohilum | IFM57309-L1 | CBS631.66, IMI114933, ATCC36839 | |
| Talaromyces flavus | IFM42233 | F-S-1 | |
| T. trachyspermus | IFM42251 | M-2143 | |
| T. stipitatus | IFM42240 | NHL 6092 | |
| T. thermophis | IFM52998 | ATCC10518, CBS236.58, IMI48593 | |
| T. derxii | CBS413.89 | Cultivated soil, Okayama Prefecture, Kurashiki City, Higashitomii, Japan | |
| T. intermedius | CBS152.65 | Alluvial pasture and swamp soil, Nottingham, Attenborough, UK | |
| Blastomyces dermatitidis | IFM40753 | From Jyunntenndo University | |
| Coccidioides immitis | IFM45811 | Patient, San Jose, AZ | |
| Paracoccidioides brasiliensis | IFM41620 | Patient, E. Burger | |
| Sporathrix schenckii | SUMS0382 | FJ011549 | Human, Guangzhou, China |
| SUMS0383 | FJ011550 | Human, Guangzhou, China | |
| Fonsecaea pedrosoi | CBS272.37T | Brazil | |
| F. monophora | CBS269.37T | AY857511 | South America |
| Histoplasmo capsulatum | SUMS0035 | AB353921 | |
| H. duboisii | IFM5417 | MTU 16024, TIMM0738, IP638 | |
| H. farciminosum | IFM41335 | CDC B-22, L. Ajiello | |
| Cryptococcus neoformans | SUMS0167 | AB436650 | Human cerebrospinal fluid, Guangzhou, China |
| SUMS0042 | AB436636 | Human cerebrospinal fluid, Guangzhou, China | |
| Candida albicans | ATCC90028 | AB049119 | Unknown |
| C. tropicalis | SUMS0125 | FJ011533 | Human blood, Guangzhou, China |
| C. parapsilosis | ATCC22019 | AB105209 | Unknown |
| C. krusei | ATCC6258 | AB105208 | Unknown |
| C. guilliermondii | ATCC6260 | AF022717 | Unknown |
| C. glabrata | ATCC2001 | AF134719 | Unknown |
| C. dubliniensis | SUMS0393 | FJ011546 | Human sputum, Guangzhou, China |
| Escherichia coli | ATCC25922 | DQ360844 | Shangdong, China |
CBS, Centraalbureau voor Schimmelcultures, Baarn, the Netherlands; SUMS, Sun Yat-Sen University Medical Science, Guangzhou, China; IFM, Research Center for Pathogenic Fungi and Microbial Toxicoses, Chiba University, Chiba, Japan; ATCC, American type culture collection, Rockville, MD; TType strain; IAM, Institute of Applied Microbiology, University of Tokyo, Tokyo, Japan; MTU, Department of Bacteriology, Faculty of Medicine, University of Tokyo, Tokyo, Japan; IFO, Institute for Fermentation, Osaka, Japan; IMI, CAB International Mycological Institute, Kew, UK; NHL, National Collection of Pathogenic Fungi, Mycological Reference Laboratory, Central Public Health Laboratory, London, UK; TIMM, Research Center for Medical Mycology, Teikyo University, Hachioji, Tokyo, Japan; IP, Unite de Mycologie, Institut Pasteur, Paris, France; CDC, Communicable Disease Centers, United States Public Health Services, Atlanta, GA; NRRL, Northern Regional Research Laboratory, Peoria, IL.
Twenty-three tissue samples from 23 patients (Zeng et al., 2009) were selected. These included 12 samples from patients with proven penicilliosis marneffei, three from chromoblastomycosis, three from sporotrichosis, one from histoplasmosis, one from cryptococcosis, one from candidiasis, one from pulmonary aspergillosis and one from visually healthy human skin. Cases from human patients were confirmed by routine and molecular identification methods. Penicillium marneffei was also isolated from 10 of 11 bamboo rat tissue samples; one (bamboo rat liver) was used as a negative control. The time that elapsed after paraffin embedding of the tissue samples ranged between one day and 13 years. About 10-µg sectioned paraffin material was used for DNA extraction.
DNA extraction and quality test
Fungal DNA from pure culture was extracted using 6% InStaGeneTMMatrix (Bio-Rad, CA) as described previously (Xi et al., 2009). Crude DNA of paraffin wax-embedded tissue was extracted from approximately 10-µg sections of paraffin wax-embedded tissue using the QIAamp® FFPE Tissue Kit (Qiagen, Hilden, Germany) according to Zeng (2009). DNA concentrations were measured spectrophotometrically at 260 nm (Shimadzu Corp., Japan). DNA quality was confirmed by successful PCR amplification using universal fungal primers internal transcribed spacer (ITS)4 and ITS5 (Zeng et al., 2009). PCR was performed as follows: 95 °C for 5 min; 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min, with a final extension at 72 °C for 10 min. Amplicons were detected by electrophoresis (Bio-Rad) on a 2% agarose gel (NuSieve, Rockland, ME).
Design of LAMP primers
Four sets of 24 species-specific primers were designed based on the rRNA gene ITS region of P. marneffeiSUMS0152 (AB353913) (Liu et al., 2007; Xi et al., 2007) using primerexplorer v4 software (http://primerexplorer.jp). A set of six species-specific LAMP primers was selected as follows: forward outer primer (F3): CCG AGC GTC ATT TCT GCC, reverse outer (B3): AGT TCA GCG GGT AAC TCC T, forward inner primer (FIP): TCG AGG ACC AGA CGG ACG TCT TTT TCA AGC ACG GCT TGT GTG, reverse inner (BIP): TAT GGG GCT CTG TCA CTC GCT CTT TTA CCT GAT CCG AGG TCA ACC, loop forward (LF): GTT GGT CAC CAC CAT ATT TAC CA and loop reverse (LB): TGC CTT TCG GGC AGG TC.
LAMP reaction
LAMP was performed in 25-µL reaction volumes containing 0.25 µM of F3 and B3 each, 1.0 µM of FIP and BIP each, 0.5 µM of LF and LB each, 1.0 mM dNTPs, 1 M betaine (Sigma), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 4 mM MgSO4, 0.1% Triton X-100 and 8 U of Bst DNA large fragment polymerase (New England Biolabs), with 2 µL of crude DNA extract as the template. The reaction mixture, except Bst DNA polymerase, was denatured at 95 °C for 5 min and cooled on ice, followed by the addition of 1 µL Bst polymerase and incubation at 65 °C in a water bath for 60 min and final heating at 85 °C for 2 min to terminate the reaction.
DNAs of 40 P. marneffei and 46 reference strains were used as templates to evaluate the specificity of the LAMP assay. DNA of strain SUMS0152 was used as a positive control; reaction mixtures without P. marneffei DNA, i.e. healthy human skin DNA, healthy bamboo rat DNA and DNAs from Penicillium purpurogenum, Penicillium funiculosum and other biverticillate penicillia taxonomically close to P. marneffei were used as negative controls.
A recombinant plasmid (pT-IT12) was constructed as a template for establishing the detection limit of the LAMP assay. The ITS region of P. marneffei (603 bp) was amplified from SUMS0152 genomic DNA using primers ITS4 and ITS5 and subcloned into the pGEM-T Easy vector (Promega) according to the manufacturer's instructions. Detection limits were evaluated using 10-fold serial dilutions of plasmid pT-IT12. The plasmid DNA (0.32 µg µL−1, equivalent to 8.067 × 1010 copies µL−1) was 10-fold serially diluted and 2 µL of each dilution was used as a template for the LAMP reaction. DNA of P. marneffeiSUMS0152 was used as a positive control; the reaction mixture without DNA was used as a negative control. To evaluate the inhibition of nontarget DNA in the LAMP assay, 2 µL crude DNA extract each of P. marneffei was added to the LAMP-negative samples, and then tested by LAMP again.
Visualization
Amplified products were analyzed by electrophoresis on 1% agarose gels, stained with ethidium bromide and photographed. A 100-bp DNA ladder was used as the molecular weight standard. LAMP reaction products were made visible by the addition of 2.0 µL of 10-fold diluted SYBR Green I (Cambrex Bio Science, Wokingham, UK) to each reaction tube separately; the change in the color of the solution was observed directly by the naked eye or using a UV transilluminator.
Application of LAMP to paraffin wax-embedded tissues
Crude-extracted DNA of 2 µL each from 34 paraffin wax-embedded tissues' samples was used as a template for LAMP assays. The amplified products were analyzed by the naked eye or by electrophoresis.
Results
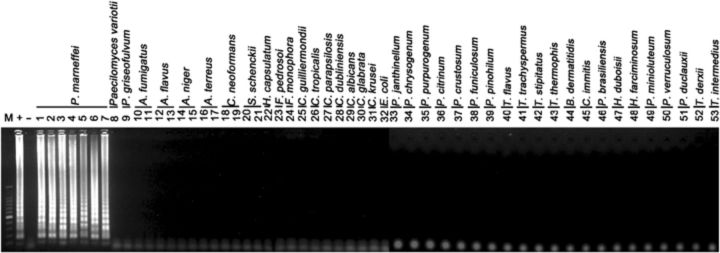
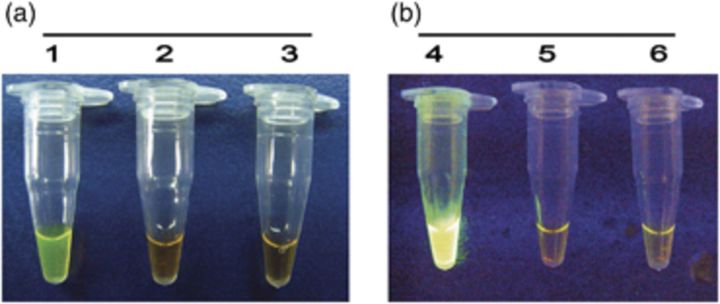
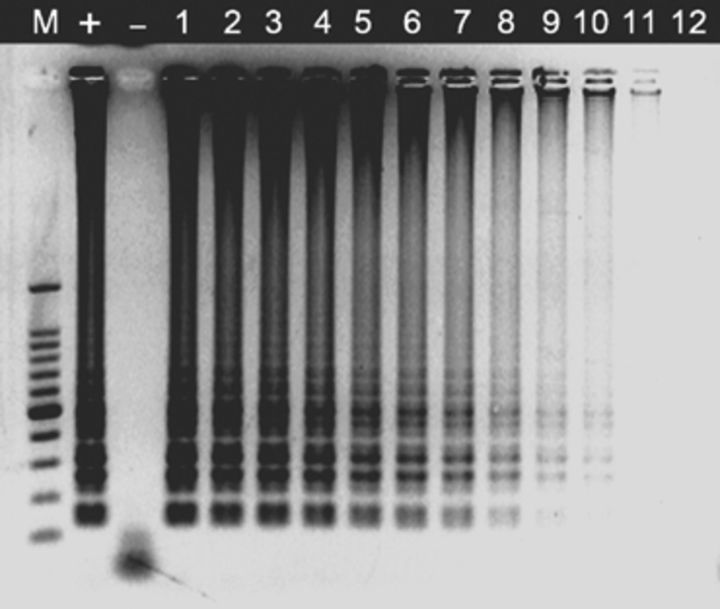
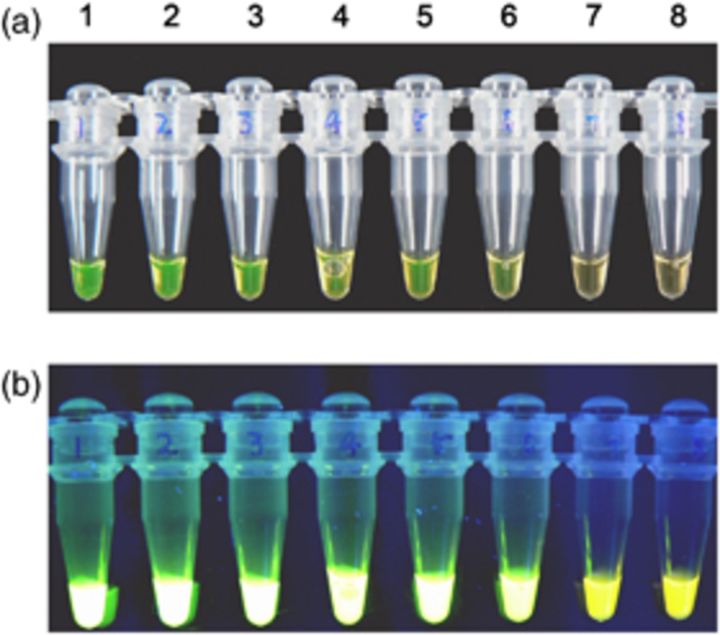
LAMP assays using a set of six species-specific LAMP primers yielded positive results in all P. marneffei strains, but remained negative in all isolates used for reference, including related biverticillate penicillia (Table 1). Amplification was completed within 1 h isothermally at 65 °C in a water bath. The products of the LAMP reaction could be detected by electrophoresis on 1% agarose gels and showed ladder-like patterns (Fig. 1). The products could also be made visible to the naked eye directly in Eppendorf vials or under UV transillumination after adding SYBR Green I dye. Positive reactions showed bright green fluorescence, whereas negative reactions remained light orange (Fig. 2). The detection limit of P. marneffei DNA by the LAMP assay was found to be two copies by electrophoresis (Fig. 3). The visual sensitivity obtained after adding SYBR Green I correlated with the sensitivity established on agarose gel (Fig. 4).
Figure 1.
Agarose gel electrophoresis of LAMP products from tested strains obtained using the primer set designed in this work. Left to right: lane ‘M’, 100-bp DNA marker; lane ‘+’, SUMS0152 positive control; lane ‘−’, negative control without DNA; lanes1–7, CBS101038, CBS555.90, IFM47287, SUMS0266, SUMS0347, SUMS0112, SUMS0051, respectively; lanes 8–53, SUMS0303, SUMS0392, SUMS0106, SUMS0317, SUMS0060, SUMS0062, SUMS0061, SUMS0037, SUMS0191, SUMS0113, SUMS0167, SUMS0042, SUMS0382, SUMS0383, SUMS0035, CBS272.37, CBS269.37, ATCC6260, SUMS0125, ATCC22019, SUMS0393, ATCC90028, ATCC2001, ATCC6258, ATCC25922, IFM40620, IFM5338, IFM40627, IFM40616, IFM47479, IFM57310-11, IFM57309-L1, IFM42233, IFM42251, IFM42240, IFM52998, IFM40753, IFM45811, IFM4162, IFM5417, IFM41335, CBS442.89, CBS101366, CBS187.89, CBS413.89, CBS152.65, respectively.
Figure 2.
Visual appearance of LAMP reactions from isolates after addition of SYBR Green I. (a) Positive reaction (tube 1), negative reaction (tube 2) and tube without DNA templates (tube 3). (b) Under UV transillumination, positive reaction (tube 4), negative reaction (tube 5) and tube without DNA templates (tube 6).
Figure 3.
Analytical sensitivity of LAMP for detection of the ITS1–5.8S–ITS2 rRNA gene. Left to right: Lane ‘M’, 100-bp DNA ladder; lane ‘+’, SUMS0152 positive control; lane ‘−’, negative control without DNA; lanes 1–12, 2 × 109, 2 × 108, 2 × 107, 2 × 106, 2 × 105, 2 × 104, 2 × 103, 2 × 102, 2 × 101, 2 × 100, 2 × 10−1 and 2 × 10−2 copies per tube, respectively.
Figure 4.
Visual sensitivity of LAMP reactions using SYBR Green I. (a) Direct detection by the naked eye, (b) under UV transillumination. Tubes 1–7, 2 × 105, 2 × 104, 2 × 103, 2 × 102, 2 × 101, 2 × 100, 2 × 10−1 copies per tube, respectively; tube 8, negative control without DNA.
All 12 proven P. marneffei-positive tissue samples and 10 samples of bamboo rat tissue tested positive, whereas samples of unaffected human skin and the remaining tissue samples affected by other fungi and tested for comparison yielded a negative response (Table 2). The correspondence between the LAMP assays and the cultural and molecular results of the same tissue samples proved to be 100%. In the inhibition test, it was found that all LAMP-negative samples became positive after the addition of 2 µL crude DNA extract of P. marneffei.
Table 2.
Biopsy specimens used in this study and the results of different detection methods
| Detection method | ||||
| Sample no. | Source | Culture and ITS sequencing | PAS | LAMP |
| Paraffin wax-embedded tissues | ||||
| 1 | Human, skin | Penicillium marneffei | + | + |
| 2 | Human, skin | P. marneffei | + | + |
| 3 | Human, skin | P. marneffei | − | + |
| 4 | Human, skin | P. marneffei | − | + |
| 5 | Human, lung | P. marneffei | + | + |
| 6 | Human, skin | P. marneffei | + | + |
| 7 | Human, lymph node | P. marneffei | + | + |
| 8 | Human, skin | P. marneffei | + | + |
| 9 | Human, skin | P. marneffei | + | + |
| 10 | Human, skin | P. marneffei | + | + |
| 11 | Human, skin | P. marneffei | + | + |
| 12 | Human, lung | P. marneffei | + | + |
| 13 | Human, skin | Sporathrix schenckii | − | − |
| 14 | Human, skin | S. schenckii | − | − |
| 15 | Human, face | S. schenckii | − | − |
| 16 | Human, lung | Aspergillus sp. | − | − |
| 17 | Human, leg | Fonsecaea pedrosoi | − | − |
| 18 | Human, leg | F. pedrosoi | − | − |
| 19 | Human, abdominal skin | F. pedrosoi | − | − |
| 20 | Human, face | Cryptococcus neoformans | − | − |
| 21 | Human, leg | Histoplasma capsulatum | + | − |
| 22 | Human, skin | Candida albicans Candida parapsilosis | + | − |
| 23 | Human normal skin | – | − | − |
| Bamboo rat tissues | ||||
| 1 | Bamboo rat, kidney | P. marneffei | − | + |
| 2 | Bamboo rat, spleen | P. marneffei | − | + |
| 3 | Bamboo rat, lung | P. marneffei | − | + |
| 4 | Bamboo rat, liver | P. marneffei | − | + |
| 5 | Bamboo rat, lung | P. marneffei | − | + |
| 6 | Bamboo rat, liver | P. marneffei | − | + |
| 7 | Bamboo rat, lung | P. marneffei | − | + |
| 8 | Bamboo rat, spleen | P. marneffei | − | + |
| 9 | Bamboo rat, lung | P. marneffei | − | + |
| 10 | Bamboo rat, liver | P. marneffei | − | + |
| 11 | Bamboo rat, lung | P. marneffei | − | + |
| 12 | Bamboo rat, liver | – | − | − |
Discussion
LAMP is a powerful innovative gene amplification technique providing a simple and rapid tool for early detection and identification of microbial diseases. Most developments in molecular diagnostics published recently concerned improvements in PCR methodology on DNA extracted from pure cultures or from clinical specimens. This had led to changes in the primer design and reaction temperature (Boehme et al., 2007; Inacio et al., 2008) and to integration with hybridization and enzyme-linked immunosorbent assay techniques (Nagamine et al., 2002; Lee et al., 2009). In the present study, we further developed and evaluated the LAMP assay, exemplified by the detection and identification of P. marneffei in DNA from pure cultures as well as in paraffin wax-embedded tissues. Compared with any detection method applied thus far, the method is very fast, as it can be carried out within 1 h. It also does not require expensive laboratory equipment, because the method can be carried out isothermally at 65 °C in a water bath. Further, it is simple to use in a routine laboratory, as the results can be observed directly by the naked eye (Fig. 3). In addition, the detection limit is very low. With only two DNA copies, it has a higher sensitivity than the currently applied molecular methods, such as semi-nested PCR (10 pg) (Prariyachatigul et al., 2003), PCR enzyme immunoassay (3.2 pg) (Lindsley et al., 2001), PCR hybridization (0.1 pg) (Vanittanakom et al., 1998) and nested PCR (0.07 pg) (Zeng et al., 2009).
The results of P. marneffei detection by LAMP in 23 paraffin wax-embedded clinical samples and 11 bamboo rat tissues were also highly specific. The etiologic agents of the 23 clinical samples were verified previously using culture and sequencing data. Twelve samples were histopathologically positive; all molecular identifications matched with the clinical diagnoses. Samples from penicilliosis and from the natural bamboo rat host were positive with LAMP, whereas all others, including healthy human skin, proved to be negative. Test results were not inhibited by nontarget DNA. This makes the LAMP technique highly promising for evaluation and application in problematic clinical samples such as blood, urine and sputum.
In this study, we have proved with the example of P. marneffei that LAMP is a very efficient method for the quick and sensitive identification of fungal pathogens and opportunists. The method can be applied not only to cultures but also to a variety of clinical samples. This can be of great significance to organisms that cause invasive or disseminated infections that are difficult to cultivate from such samples, such as the zygomycete species. A further application may be for detection without isolation of the fungi in the environment.
In summary, in the current study, we proved that the LAMP technique enables specific detection of P. marneffei and excludes related biverticillate penicillia and Talaromyces teleomorphs. Similar results were obtained in Paracoccidioides (Endo et al., 2004), Candida (Inacio et al., 2008) and Ochroconis (Ohori et al., 2006). However, in Fonsecaea, identification was possible only at the generic level (Najafzadeh, 2009). An explanation for this phenomenon may be found in the fact that Penicillium species are relatively distant from each other, with ITS barcoding gaps well over 1%, whereas in Fonsecaea ITS, interspecific differences are a few bases only, species delimitations being based on multilocus analyses.
Acknowledgements
We thank Prof. Yokoyama (Center for Pathogenic Fungi and Microbial Toxicoses Chiba University, Chiba, Japan) for providing the reference strains taxonomically close to P. marneffei included in this paper. This study was supported partly by a grant (30770121/2007) from the National Natural Science Foundation of China.
References
- Aoi Y., Hosogai M., Tsuneda S. (2006) Real-time quantitative LAMP (loop-mediated isothermal amplification of DNA) as a simple method for monitoring ammonia-oxidizing bacteria. J Biotechnol 125: 484–491. [DOI] [PubMed] [Google Scholar]
- Boehme C.C., Nabeta P., Henostroza G., et al. (2007) Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J Clin Microbiol 45: 1936–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T., Lou G., Yang J., Xu D., Meng Z. (2008) Development and evaluation of real-time loop-mediated isothermal amplification for hepatitis B virus DNA quantification: a new tool for HBV management. J Clin Virol 41: 270–276. [DOI] [PubMed] [Google Scholar]
- Endo S., Komori T., Ricci G., Sano A., Yokoyama K., Ohori A., Kamei K., Franco M., Miyaii M., Nishimura K. (2004) Detection of gp43 of Paracoccidioides brasiliensis by the loop-mediated isothermal amplification (LAMP) method. FEMS Microbiol Lett 234: 93–97. [DOI] [PubMed] [Google Scholar]
- Enosawa M., Kageyama S., Sawai K., Watanabe K., Notomi T., Onoe S., Mori Y., Yokomizo Y. (2003) Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J Clin Microbiol 41: 4359–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M.C., Hanage W.P., De Hoog S., Johnson E., Smith M.D., White N.J., Vanittanakom N. (2005) Low effective dispersal of asexual genotypes in heterogeneous landscapes by the endemic pathogen Penicillium marneffei. PLoS Pathog 1: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara M., Sasaki H., Matsuo K., Honda M., Kawase M., Nakagawa H. (2007) Loop-mediated isothermal amplification method for detection of human papillomavirus type 6, 11, 16, and 18. J Med Virol 79: 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikadai H., Tanaka H., Shibahara N., Matsuu A., Uechi M., Itoh N., Oshiro S., Kudo N., Igarashi I., Oyamada T. (2004) Molecular evidence of infections with Babesia gibsoni parasites in Japan and evaluation of the diagnostic potential of a loop-mediated isothermal amplification method. J Clin Microbiol 42: 2465–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inacio J., Flores O., Spencer-Martins I. (2008) Efficient identification of clinically relevant Candida yeast species by use of an assay combining panfungal loop-mediated isothermal DNA amplification with hybridization to species-specific oligonucleotide probes. J Clin Microbiol 46: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseki H., Alhassan A., Ohta N., Thekisoe OMM, Yokoyama N., Inoue N., Nambota A., Yasuda J., Igarashi I. (2007) Development of a multiplex loop-mediated isothermal amplification (mLAMP) method for the simultaneous detection of bovine babesia parasites. J Microbiol Meth 71: 281–287. [DOI] [PubMed] [Google Scholar]
- Iwamoto T., Sonobe T., Hayashi K. (2003) Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol 41: 2616–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.F., Chen Y.H., Peng C.F. (2009) Evaluation of reverse transcription loop-mediated isothermal amplification in conjunction with ELISA-hybridization assay for molecular detection of Mycobacterium tuberculosis. J Microbiol Meth 76: 174–180. [DOI] [PubMed] [Google Scholar]
- Lee N. (2008) Penicilliosis: an AIDS-defining disease in Asia. Hong Kong Med J 14: 88–89. [PubMed] [Google Scholar]
- Lindsley M.D., Hurst S.F., Iqbal N.J., Morrison C.J. (2001) Rapid identification of dimorphic and yeast-like fungal pathogens using specific DNA probes. J Clin Microbiol 39: 3505–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linghua Li X.T., Weiping C. (2008) A clinical study on 101 AIDS cases complicated with penicilliosis marneffei. Chin J AIDS STD 114: 12–20. [Google Scholar]
- Liu H., Xi L., Zhang J., Li X., Liu X., Lu C., Sun J. (2007) Identifying differentially expressed genes in the dimorphic fungus Penicillium marneffei by suppression subtractive hybridization. FEMS Microbiol Lett 270: 97–103. [DOI] [PubMed] [Google Scholar]
- Liyan X., Changming L., Xianyi Z., Luxia W., Suisheng X. (2004) Fifteen cases of penicilliosis in Guangdong, China. Mycopathologia 158: 151–155. [DOI] [PubMed] [Google Scholar]
- LoBuglio K.F., Taylor J.W. (1995) Phylogeny and PCR identification of the human pathogenic fungus Penicillium marneffei. J Clin Microbiol 33: 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo W., Deng Z., Li S. (2002) Clinical blood routine and bone marrow smear manifestations of disseminated penicilliosis marneffei. Chin Med J (England) 115: 1892–1894. [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. (2002) Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probe 16: 223–229. [DOI] [PubMed] [Google Scholar]
- Najafzadeh M.J., Sun J., Vicente V., Xi L., Gerrits van den Ende AHG, De Hoog G.S. (2009) Fonsecaea nubica, a new species of agent of human chromoblastomycosis revealed using molecular data. Med Mycol, in press. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28: E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohori A., Endo S., Sano A., Yokoyama K., Yarita K., Yamaguchi M., Kamei K., Miyaii M., Nishimura K. (2006) Rapid identification of Ochroconis gallopava by a loop-mediated isothermal amplification (LAMP) method. Vet Microbiol 114: 359–365. [DOI] [PubMed] [Google Scholar]
- Panichakul T., Chawengkirttikul R., Chaiyaroj S.C., Sirisinha S. (2002) Development of a monoclonal antibody-based enzyme-linked immunosorbent assay for the diagnosis of Penicillium marneffei infection. Am J Trop Med Hyg 67: 443–447. [DOI] [PubMed] [Google Scholar]
- Poon L.L., Leung C.S., Tashiro M., Chan K.H., Wong BWY, Yuen K.Y., Guan Y., Peiris JSM. (2004) Rapid detection of the severe acute respiratory syndrome (SARS) coronavirus by a loop-mediated isothermal amplification assay. Clin Chem 50: 1050–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Wong B.W., Chan K.H., Ng S.S., Yuen K.Y., Guan Y., Peiris J.S. (2005) Evaluation of real-time reverse transcriptase PCR and real-time loop-mediated amplification assays for severe acute respiratory syndrome coronavirus detection. J Clin Microbiol 43: 3457–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prariyachatigul C., Chaiprasert A., Geenkajorn K., Kappe R., Chuchottaworn C., Termsetjaroen S., Srimuang S. (2003) Development and evaluation of a one-tube seminested PCR assay for the detection and identification of Penicillium marneffei. Mycoses 46: 447–454. [DOI] [PubMed] [Google Scholar]
- Sun J., Najafzadeh M.J., Vicente V., Xi L., De Hoog G.S. (2009) Rapid detection of pathogenic fungi using loop-mediated isothermal amplification, exemplified by Fonsecaea agents of chromoblastomycosis. J Microbiol Meth DOI: 10.1016/j.mimet.2009.1010.1002. [DOI] [PubMed] [Google Scholar]
- Supparatpinyo K., Khamwan C., Baosoung V., Nelson K.E., Sirisanthana T. (1994) Disseminated Penicillium marneffei infection in southeast Asia. Lancet 344: 110–113. [DOI] [PubMed] [Google Scholar]
- Ukarapol N., Sirisanthana V., Wongsawasdi L. (1998) Penicillium marneffei mesenteric lymphadenitis in human immunodeficiency virus-infected children. J Med Assoc Thailand 81: 637–640. [PubMed] [Google Scholar]
- Vanittanakom N., Merz W.G., Sittisombut N., Khamwan C., Nelson K.E., Sirisanthana T. (1998) Specific identification of Penicillium marneffei by a polymerase chain reaction/hybridization technique. Med Mycol 36: 169–175. [PubMed] [Google Scholar]
- Vanittanakom N., Vanittanakom P., Hay R.J. (2002) Rapid identification of Penicillium marneffei by PCR-based detection of specific sequences on the rRNA gene. J Clin Microbiol 40: 1739–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Asoh N., Kobayashi S., et al. (2008) Clinical and microbiological characteristics of community-acquired pneumonia among human immunodeficiency virus-infected patients in northern Thailand. J Infect Chemother 14: 105–109. [DOI] [PubMed] [Google Scholar]
- Wei X.G. (1985) Report of the first case of penicilliosis marneffei in China. Zhonghua Yi Xue Za Zhi 65: 533–534. [PubMed] [Google Scholar]
- Wong K.H., Lee S.S., Chan K.C., Choi T. (1998) Redefining AIDS: case exemplified by Penicillium marneffei infection in HIV-infected people in Hong Kong. Int J STD AIDS 9: 555–556. [PubMed] [Google Scholar]
- Xi L., Xu X., Liu W., Li X., Liu Y., Li M., Zhang J., Li M. (2007) Differentially expressed proteins of pathogenic Penicillium marneffei in yeast and mycelial phases. J Med Microbiol 56: 298–304. [DOI] [PubMed] [Google Scholar]
- Xi L., Sun J., Lu C., Liu H., Xie Z., Fukushima K., Takizawa K., Najafzadeh M.J., E Hoog G.S. (2009) Molecular diversity of Fonsecaea (Chaetothyriales) causing chromoblastomycosis in southern China. Med Mycol 47: 27–33. [DOI] [PubMed] [Google Scholar]
- Yoshida A., Nagashima S., Ansai T., Tachibana M., Kato H., Watari H., Notomi T., Takehara T. (2005) Loop-mediated isothermal amplification method for rapid detection of the periodontopathic bacteria Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola. J Clin Microbiol 43: 2418–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Li X., Chen X., Zhang J., Sun J., Xie Z., Xi L. (2009) Identification of Penicillium marneffei in paraffin-embedded tissue using nested PCR. Mycopathologia 168: 31–35. [DOI] [PubMed] [Google Scholar]