Abstract
Traditional, culture based methods for the diagnosis of fungal infections are still considered as gold standard, but they are time consuming and low sensitive. Therefore, in order to overcome the limitations, many researchers have focused on the development of new immunological and molecular based rapid assays that could enable early diagnosis of infection and accurate identification of fungal pathogens causing superficial and invasive infection. In this brief review, we highlighted the advantages and disadvantages of conventional diagnostic methods and possibility of non-culture based assays in diagnosis of superficial fungal infections and presented the overview on currently available immunochromatographic assays as well as availability of biomarkers detection by immunodiagnostic procedures in prompt and accurate diagnosis of invasive fungal infections. In addition, we presented diagnostic efficiency of currently available molecular panels and researches in this area.
Keywords: Fungal infection, Rapid detection, Conventional assays, Immunoassays, Molecular assays
1. Introduction
The primary task of microbiological/mycological diagnostics is to provide adequate information of the infection cause and its antimicrobial susceptibility in optimal period of time. Any delay in accurate pathogen identification and appropriate treatment initiation undoubtedly affect disease outcome.
Despite significant advances in medical technology in terms of rapid accurate diagnostic laboratory tests following by new therapeutics that have had a major effect in decreasing of morbidity and mortality of many fatal diseases, we are the witnesses that invasive fungal infections (IFIs) are still one of the biggest problems in medicine [1], [2], [3]. Increasing the number of immunocompromised patients with T-cell mediated immunity deficiencies, mucosal-cutaneous barrier or metabolic dysfunction, neutropenia, aging, excessive use of antibiotics, cytotoxic therapy and transplantation, as predispose risk factors and conditions, significantly influence the higher incidence of IFIs [4]. Moreover, the incidence and prevalence of nosocomial IFI are still high and Candida species represent the third most common blood isolate from hospitalized patients. Out of all IFI, 45% of invasive candidosis are from intensive care units (ICU). Candida bloodstream infection is prolonging stay in the ICU with mortality ranges from 40 to 71%. Also, it can cause complications during the clinical course of primary disease and increases treatment costs [5].
As opposed to IFI, superficial fungal infections (SFI) of the keratin rich structures are rare systemic and serious. These infections are caused commonly by dermatophytes, yeasts or non-dermatophytic molds and represent one of the dominant infections worldwide with global prevalence ranges from 22–25% [6], [7]. The lack of rapid diagnostic methods for SFI consequently influence that only 3% of physicians in general practice and 40% dermatologists require mycological analyses before prescription of empiric treatment with systemic antifungals [8]. Additionally, these infections can affect the patient's quality of life as a potential cause of social, professional and emotional problems [9]. Thus, fungal infections of the skin, nail or hair could not be considered as a cosmetic problem of relatively minor significance. Given the fact that deep infections and dissemination of pathogen can be expected in immunodeficiency, some genetic abnormalities such as CARD9 mutation, and leukemia [10], [11], the prompt diagnosis and treatment of SFIs could prevent complications and unwanted clinical outcomes.
So far, in the group of SFI, mucosal (oropharyngeal, vulvovaginal, intestinal) fungal infections have also been included. Today, the prevalence of oropharyngeal and vulvovaginal mucosal candidosis and Candida-overgrowth in intestines significantly rises up and the treatment of chronic/recurrent form is a big challenge for physicians. Also, frequent changes of local therapy by systemic, with the goal to improve the treatment, probably influences the emergence of resistant Candida species to azoles [12], [13], [14], [15], [16]. New, highly specific and sensitive rapid tests will disable misdiagnosis and unnecessary proscribing of antifungals, which will be in accordance with recently initiate appeal that systemic antifungals have to be kept for IFI [17].
Historically, it is very often highlighted that traditional/conventional methods for diagnosis of SFI and IFI are time consuming and low sensitive. The propagating/cultivating the causative agent is method with only deficiency in required time for isolation, but when we can conduct it, it is the unique analysis that provide antigen (Ag) detection, molecular, biochemical identification of etiological agents and antimicrobial susceptibility testing. On the other hand, histopathological examination of the tissue samples that is considered as a “gold standard” for IFI has a major shortcoming that lays in the complexity of the invasive biopsy procedure [18], [19], [20].
This review discusses currently available data on the advantages and disadvantages of some rapid assays for fungal detection, identification and the diagnosis of SFIs and IFIs.
2. Rapid assays for diagnosis of superficial fungal infections (SFIs)
2.1. Conventional microscopy examination (ME)
Conventional microscopic examination (ME) is cheap, easy-to use and fast method for detection of fungi in a patient's sample. Although staining procedures offer additional information concerning morphology of microorganisms, ME in a form of wet mount technique provides rapid detection of fungal blastoconidia-yeast and pseudohyphal-hyphal forms in patient's material (Fig. 1 ) [21]. Wet mount with chlorlactophenol or KOH as reagents is still irreplaceable technique for screening of patients with suspected SFI of the skin, hair and nail. Additionally, diagnosis of vaginal discharge by wet mount microscopy is useful for the diagnosis of Candida vaginitis - one of the most prevalent genital infections in women [22], [23]. Nowadays, specific staining with chemicals or fluorescent dyes (acridin-orange, calcofluor wight, blankoflor) can improve visualization and detection of fungi [18], [24], [25].
Fig. 1.
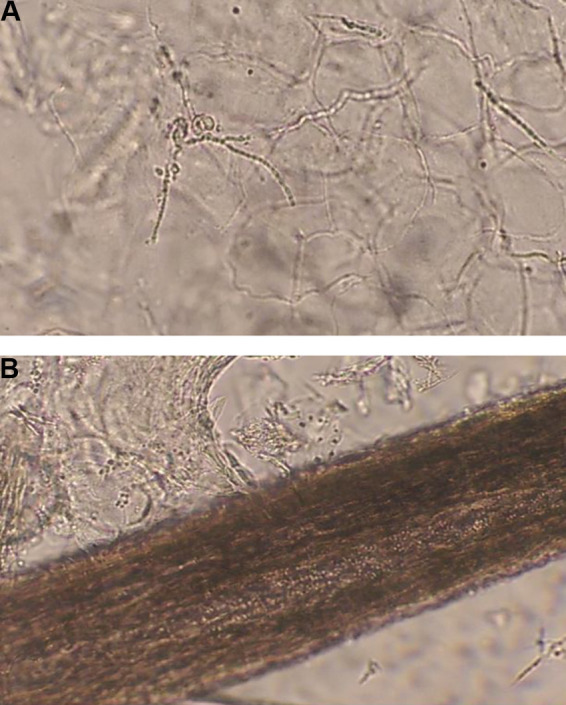
A. Fungal mycelia in patients material. B. Multiple fungal spores in hair.
2.2. Immunochromatographic assays
Currently, there are efforts for establishing the imunochromatographic (IC) assays for the determination of dermatophyte [26] and Candida Ag [27], which will represent significant progress in the diagnosis of SFIs and genital candidosis, respectively. Fast, easy to perform and easy to interpret, these assays can be a satisfactory replacement of ME.
So far, there have been reports of IC assays in which Trichophyton antibodies (Ab) have been used with colloidal gold particles to capture Ag of different dermatophyte species such as T. rubrum, T. mentagrophytes, T. violaceum, T. tonsurans, Microsporum canis, M gypseum, Epidermophyton floccosum. Comparing the diagnostic performances of IC for dermatophytes detection in skin and nail samples by ME, Higashi et al. reported that IC sensitivity and specificity were 83.5% and 66.7% for all specimens, 81.1% and 76.2% for scarified skin and 85.4% and 58.3% for samples taken from the nail plate, respectively [26]. Based on these results, authors recommended IC assay, as easy to use and rapid tool, for screening of dermatophyte infections, but for definitive diagnosis, ME and culture are still recommended methods. In the study conducted by Noriki and Ishida, in-house developed IC assays showed very good diagnostic performances for dermatophyte detection in nail samples [28].
Similarly, another IC assay which used the colloidal gold-anti-mannan IgG conjugate for Candida detection in vaginal swabs was reported recently. Advantage of this newly developed IC assay is the possibility of use in clinics and laboratories and detection of Candida spp. in the concentration of 104 CFU/mL of patients sample in 30 min without expensive equipments. However, in comparison with culture, IC Candida assay in vaginal swabs showed the high specificity 99.3%, positive predictive value 98.0, followed by lower sensitivity 80.3%, and negative predictive value 92% for C. albicans. For the second most prevalent species C. glabrata (causing more-intensive and chronic form of genital candidosis), concentration has to be more then 105 CFU/mL in patients sample [27].
Despite the few data in reference literature and the lack of commercial IC assays for SFI, further research, design and establishment of these assays for SFI will be of great significance for supporting conventional methods and facilitating the diagnosis of dermatophytosis and genital candidosis in women.
2.3. Molecular assays
During the last forty years, a number of molecular techniques have been developed for the identification of dermatophytes at species or strain level. These include fingerprinting methods, conventional PCR (employing selected genetic markers e.g. internal transcribed spacers (ITS), chitin synthase 1 gene (chs1), topoisomerase II gene, small and large ribosomal RNA subunit and non-transcribed spacer/NTS region), ITS restriction fragment length polymorphism (RFLP) and total mitochondrial DNA (mtDNA) analyses which change the view to the taxonomy of dermatophytes [29], [30], [31], [32]. Molecular diagnosis provides species-level identification of dermatophytes more accurately regarding their development in human, soil or animals. Recent studies showed that the conventional PCR of ITS and chs1 regions might be helpful tool for a better understanding of dermatophyte identification, taxonomy, ecology and epidemiology [30]. These findings revolutionized the diagnosis of SFI caused by dermatophytes and the use of “in house” or rare commercial molecular tools enable their rapid determination directly in patients sample within 48 h. As for “in house” molecular tools, various types of PCR techniques have been developed in the last decade. These assays had evolution from conventional PCR, nested PCR, pan-nested PCR which was recommended as gold standard with higher sensitivity compared to conventional methods [33], followed by multiplex real-time PCR which provides detection of T. rubrum, T. interdigitale, T. violaceum and M. audouinii [34] and later developed single-tube dermatophyte qPCR tool based on ITS1 sequences which design allows determination of 11 species of three dermatophyte genera (Trichophyton, Microsporum and Epidermophyton) directly in patients sample [35]. Besides, reported multiplex PCR for detection of T. rubrum and T. mentagrophytes in nail samples, which is based on chitin synthase I and ITS region, was proved as molecular tool with excellent diagnostic performance (95% sensitivity and 100% specificity) [36]. Few commercial assays have also been designed and some of them with high diagnostic performance such as duplex PCR which combines pan-dermatophyte PCR with a Trichophyton rubrum-specific PCR. This assay provides rapid detection and identification of Trichophyton rubrum in nail specimens and has shown specificity and sensitivity of 94% and 85%, respectively, in comparison with conventional methods [37].
On the other hand, some multiplex PCR methods for dermathophytes or dermatophytes/Candida species in clinical samples were also developed and clinical evaluation is still in progress [38]. Recently, published results [39] showed that real-time PCR with specific pan-dermatophyte primer for detection of most agents from this group of fungi in clinical samples detected more infected patients compared with ME. However, with unsatisfactory sensitivity of 87.5% and positive predicative value of 66.5%, this method cannot be suggested yet as sufficient replacement of conventional diagnosis.
In addition, new data of tinea capitis in Sub-Saharan Africa highlighted the importance of early detection of causative agents. The use of the PCR-ELISA and rapid determination of antropophylic species (T. violaceum, M. audouinii, T. soudanense, T. rubrum) allow the effective and appropriate therapy, monitoring of possible source of infection and implementation of preventive measures with the goal of preventing the spread of causative agents and infection [40].
By all means, the development, standardization, as well as the commercialization of molecular tools are of great importance for the perspective in the rapid diagnostics of SFI.
3. Rapid assays for diagnosis of invasive fungal infections (IFI)
Prolong survival of high-risk patients (transplantation, immunodeficiency, immunomodulatory regimens, malignancy, longer stay in ICU, etc.) with IFI primarily depends on timely, accurate diagnosis and antifungal treatment [41]. Rapid advances in the field of diagnostic methods for prompt diagnosis of IFI caused by Candida spp., Aspergillus spp., Pneumocystis jirovecii (P. jirovecii) have been achieved in recent years [42]. However, the standardization of currently available immunological, PCR and fluorescence in situ hybridization assays is still ongoing worldwide. Besides, the development and utilization of highly specific and sensitive diagnostic tests for detection of IFI caused by other fungal species is also required [41]. Conventional methods and designed rapid assays for the diagnosis of IFI were summarized in the Table 1 .
Table 1.
Summary of rapid assays for fungi identification and invasive fungal infections (IFIS) diagnosis.
| Invasive fungal infections and non-culture based assays in laboratory mycology | ||||
|---|---|---|---|---|
| Suspected fungi and patients samples | Conventional microscopic examination | Immuno assays | Molecular assays | |
| Candida spp. or other fungal insolates | Positive blood culture or isolates from primarly sterile regions | PNA FISH – QuickFISH (20 min) MALDI-TOF MS (20–60 min) |
||
| Candida spp. infection | Serum | Positive blood culture/Bactec + Gram staining (24–48 h) |
Screening – Ag detection by ICT (20 min) Candida MN Ag/anti Candida MN Ab-biomarkers by ELISA (3–4 h) 1,3 β-D-glucan – chromogenic, quantitative enzyme immunoassay (EIA) (40 min) |
Standard, nested, real-time or multiplex PCR (1 h directly from positive blood cultures – 3 h for sera samples) |
| Cryptococcus spp.infection | Cerebrospinal fluid/ Sputum |
Indian ink preparation of CSF wet mounts (1 h) Standard CSF, sputum or skin culture (48–72 h) |
Screening – Ag detection by ICT (≤ 10 min) Latex agglutination tests |
Multiplex PCR analysis of CSF (1 h) Quantitative PCR (q-PCR) (within 3 h) |
| Aspergillus spp. infection | BAL fluid/ Sputum/ Blood Tissues |
Standard BAL fluid or sputum culture (4–7 days) | Screening – Ag detectionby ICT (20 min) | Standard, nested, real-time, quantitative or multiplex PCR (4–8 h) |
|
Pneumocystis jirovecii infection |
BAL fluid/ Induced sputum/ Serum/ Oropharyngeal wash fluid |
Standard Wright-Giemsa and modified Giemsa stain methods of BAL fluid or induced sputum (up to 60 min) |
Direct immunofluorescence assay 1,3 β-D-glucan – chromogenic, quantitative enzyme immunoassay (EIA) (40 min) |
Standard, nested, real-time, quantitative or multiplex PCR (2.5–4 h) |
ICT: immunochromatographic test; Ag: antigen; MALDI-TOF MS: Matrix-assisted laser desorption ionization-time of flight mass spectrometry; PNA FISH: peptide nucleic acid fluorescent in situ hybridization.
3.1. Conventional ME for IFIs
Conventional ME can be used in diagnosis of IFI but require a high level of expert knowledge, which represents the main disadvantage of this procedure [21], [22], [23], [24], [25]. Detection of invasive Aspergillus hyphae by direct microscopic examination of samples obtained from primarily sterile regions is equated as proven invasive fungal disease [43]. On the other hand, this infection can be “probable” in patients with positive microscopic finding of sputum or broncho alveolar lavage (BAL) fluid in combination with the appropriate clinical criteria and predisposing factors. Also, cryptococal meningitis can be rapidly diagnosed if encapsulated Cryptococcus neoformans (Cry. neoformans) yeast cells are visible by ME of cerebrospinal fluid (CSF) [44]. Adding of Indian ink in wet mounts of CSF sediment can improve Cryptococcus detection and the diagnostic efficacy.
Specific staining of patients samples with Giemsa, Wright, Wright-Giemsa, Fontana-Masson, Grocott-methenamins silver (GMS) or fluorescent dyes (acridine orange, calcofluor whight or blankofor) can improve visualization of fungi and their detection by ME [18], [24], [25]. Specific staining methods are most commonly used for the detection of P. jiroveci (carinii) in lower respiratory tract specimens [45] or to identify the presence of intracellular forms of Histoplasma capsulatum in BAL, bone marrow, peripheral blood smears or touch preparations of lymph nodes and other tissues [46]. Acridine orange allows the detection of P. jiroveci in purulent specimens [47] . These fluorescent stains can bind to the cellulose and chitin in the fungal cell wall [21] and have become very useful tool in laboratory mycology. Also, any fungi can be detected by this methods but their identification is difficult or impossible in many cases.
3.2. Fungal Ag assays
3.2.1. -IC assays for fungal Ag-
The most important advantage of immunoassay is the possibility of performing directly on a clinical sample obtained from patients. IC assays for fungal Ags are low-cost, easy to use, and applicable at the patient's bedside, and can be performed easy to a number of samples without special equipment (even without electricity). However, currently, there are only few commercially available immunochromatographic assays that enable the detection of surface or soluble yeasts or Aspergillus Ag in clinical samples. Their detailed characteristics are presented in Table 2 .
Table 2.
Characteristics of immunochromatographic tests for the detection of invasive fungal infections.
| Species | Type of sample | Detection | Time to results | Sp/Sn of evaluated tests (%) | NPV and PPV of evaluated tests (%) | Manufacturer | Approximative cost | References |
|---|---|---|---|---|---|---|---|---|
| Aspergillus sp. | Bronchoalveolar lavage | Ag | 10–15 min | 81/80 | 96/44 | OLM Diagnostics, Newcastle upon Tyne | £10 | [48] |
| Cryptococcus sp. | Cerebrospinal fluid | Ag | ≤ 10 min | 99.1–100/99.3–100 | 98.7/99.5 | Immy Inc, Norman, Oklahoma, USA | $2 per strip in low-income countries/$5 in high-income countries | [49] |
| Candida albicans | Whole blood and serum | Ag | 20 min | Data not available | Data not available | Chembio Diagnostic Systems, Inc., NY | Data not available | [50] |
Ag: antigen; Sn: sensitivity; Sp: specificity; NPV: negative predictive value; PPV: positive predictive value.
3.2.2. Detection of cryptococcal glucuroxilomannan (GXM), Candida mannan (MN), Aspergillus galactomannan (GM) and 1,3 β-D-glucan (BDG) in patients samples
One of the most significant achievements in mycology is the development, evaluation and manufacture of immunoassays for diagnosis of IFI. With the introduction of immunoassays for Ag detection in patients biological fluids such as serum, BAL or CSF, the diagnosis of IFIs, previously diagnosed only by histopathological examination of tissue samples after biopsy, have become more rapid and non invasive.
Latex agglutination tests for cryptococcal GXM are chip, sensitive, specific and easy to perform. The polysaccharide capsular GXM that is produced in large quantities by the Crytococcus can be detected in both CSF and blood samples [51]. Also, detection of Candida MN, Aspergillus GM, 1,3 β-D-glucan (BDG) in serum, BAL or other fluids significantly improved the diagnosis of IFI [3], [52].
In the diagnosis of candidemia, MN and BDG are currently available. MN is a major component of Candida cell wall, accounting for up to 7% of total dry cell weight, which is released in bloodstream during Candida replication during infection. The Platelia Candida Ag test (Bio-Rad Laboratories, Marnes-la-Coquette, France) enables the detection of MN in blood (serum) samples in ELISA format [53]. Several retrospective and prospective studies have evaluated the utility of MN detection for the diagnosis of invasive candidosis in haematological and ICU patients with an overall sensitivity (Sn) of 58% and specificity (Sp) of 93% [53], [54], [55], [56]. The Sn of the test seems to vary with the infecting Candida species, being the highest in the case of C. albicans [55]. A positive MN test occurs several days before radiological detection of hepatosplenic candidosis or positive blood cultures (BC) [55]. In high-risk patients, the test should be performed two to three times per week since the presence of MN Ag in blood is short-lived due to rapid clearance during each episode of candidemia with the concomitant appearance of anti-MN Ab [3].
Another Ag-based test detects the presence of BDG, the important component of the cell wall of most fungi, in sera samples and is approved by Food and Drug Administration of the United States of America [57], [58]. The test has been evaluated, mostly in ICU patients, with an overall Sn of 77% and Sp of 85% for subjects with proven or probable IFI [56], [59]. However, BDG is not species specific; therefore it is a pan fungal assay. Results of many large studies have shown that with higher values of Sn (81.4%) and Sp (78.1%), extremely low value of diagnostic odds ratio (15.5%) have been obtained and BDG concentration vary in IFI due to different fungal species. White et al. proved that BDG concentration for proven/probable IFI (209 pg/mL) is significantly higher then in possible/suspeced cases and control population (73 pg/mL) and it is different regarding the cause of IFI. BDG is useful for diagnosis of invasive aspergillosis, invasive candidosis and P. jirovecii pneumonia, but BDG concentration vary based on causative agents [60]. Additionally, positive BDG in the cases of P. jirovecii pneumonia cannot distinguish infection from colonization, to promote the higher sensitivity BDG concentration of 45 pg/mL has to be considered as positive, but only concentration threshold of 300 pg/mL increased specificity. The interpretation could be only with additional clinical, radiologic examination, immuno- or molecular assays [61], [62]. The test should be applied twice weekly and a single positive test is indicative of infection. True positive cases usually show falling BDG titter that eventually becomes negative in patients responding to antifungal treatment, a trend that cannot be observed in patients who does not respond to therapy.
False positive BDG results may occur due to several reasons such as haemodialysis, abdominal surgery, treatment with β-lactam antibiotics and concomitant presence of lipopolysaccharide originationg from Gram-negative bacteremia, which makes its application rather difficult in clinical settings [56], [59]. However, based on excellent negative predictive value of this test (nearly 100%), lack of BDG detection is most useful for exclusion of IFI [56], [59]. Also, the colonization of individuals with Candida sp. has no apparent effect on the BDG test outcome [54], [56], [58].
As for the use of GM in diagnosis of invasive aspergilosis, recently published data suggest that detection of this Aspergillus Ag in blood and parallel PCR diagnostics provide very high sensitivity of 99% with specificity of 64% which influence 100% negative predictive value in high risk patients and enable the consideration of no-existing invasive aspergillosis in these patients and no need for antifungal therapy. Contrary, positivity of both tests has positive predictive value of 88% that suggest probably occurrence of invasive aspergillosis [63]. However, the cross-reactivity of Fusarium spp. in the Aspergillus GM ELISA Assay has been observed and needs to be taken into consideration in some patients, contraries or settings.
3.3. Molecular assays
The most significant progress in the diagnosis of infectious diseases, fungal infections included, has been made with the introduction of molecular biology. Molecular detection and identification as well as nucleic acid testing (NAT) have a great importance in the diagnosis of infectious diseases, especially in the presence of a suggestive patient history and clinical manifestations of the infection. These methods can be used as a sensitive and specific tool for the detection and identification of numerous microorganisms in patient specimens. The application of NAT can enhance the speed of diagnosis. Some of these tests can be completed in a few hours and have high Sn and Sp [64], [65], [66]. However, the disadvantages include the high cost of reagents and instruments, appropriately trained staff and well-equipped laboratories that are able to fulfil the highest standards. Additionally, possibility of samples or reagents contamination could be also the lack of these assays [64], [65], [67], [68].
3.4. Peptide nucleic acid fluorescent in situ hybridization (PNA-FISH) assay
Peptide nucleic acid fluorescent in situ hybridization (PNA-FISH) is a brand new, rapid, molecular diagnostic platform developed by AdvanDx (Woburn, Massachusetts) based on proprietary of PNA probe technology. The PNA-FISH uses a variety of currently approved probes, including ones for Staphylococcus aureus, coagulase-negative staphylococci (CoNS), Enterococcus faecalis and other enterococci, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, C. albicans, C. glabrata, C. parapsilosis, C. krusei, and C. tropicalis (AdvanDx) [69], [70], [71], [72], [73]. The PNA-FISH uses fluorescent-labelled probes to target species-specific rRNA sequences in a high Sn and Sp fluorescence in situ hybridization assay. The probes bind to the target RNA tightly, without electrostatic repulsion from the charged RNA backbone and since rRNA is amplified in viable cells of growing cultures, there is no need for its further amplification [70], [74], [75]. Cell lyses, usually used for isolation of genetic material, are also not necessary since the PNA probes hybridize to rRNA inside the bacteria or yeast enabling the whole cell analysis. The assay requires only limited sample preparation as cells do not need to be lysed to isolate genetic material, allowing a simple test procedure with visual results that match Gram-stain morphology [70], [73].
The major characteristics of PNA-FISH in a routine monitoring of analyses of sepsis are safety, Sp, Sn, the potential for high-throughput testing and economical feasibility. The PNA-FISH is easy to perform in clinical laboratory and does not require a significant capital equipment costs unlike microarrays or matrix-assisted laser desorption ionization-time of flight mass spectroscopy (MALDI-TOF) [71], [73], [75]. The PNA-FISH method requires only microscope equipped with a fluorescent lamp and dual band filters for results interpretation. The accuracy and Sp of PNA-FISH can significantly affect antibiotic and antifungal utilization, allowing more targeted therapy, reduction of treatment duration, with an overall reduction in healthcare costs and increased benefit for patients [70], [74]. Recently, this methodology is additionally improved by introducing new QuickFISH, developed by AdvanDx, that enables very fast (20 minutes) identification of bacteria or yeasts from positive blood samples.
3.5. Molecular diagnosis of P. jirovecii pneumonia
P. jirovecii is an opportunistic unicellular fungal pathogen that causes an acute and life-threatening Pneumocystis pneumonia in immunocompromised hosts [76], [77], such as HIV-infected patients (especially those who do not know that they are HIV positive, do not comply with or respond to antiretroviral therapy or Pneumocystis prophylaxis) [78], solid organ transplant and haematopoietic stem cell transplant recipients, patients with malignant diseases, non-HIV-infected patients receiving immunosuppressive medications (e.g. corticosteroids, monoclonal antibodies or cytokine inhibitors) for autoimmune or inflammatory diseases and subjects with congenital immunodeficiencies [79], [80], [81]. The clinical course of infection is more acute and severe in HIV-negative immunocompromised patients than in HIV-infected hosts with significantly higher rates of mortality (35–55% vs. 10–20%) [82]. In some cases, however, patients may become colonized with P. jirovecii without signs or symptoms of acute disease [78]. According to the results of the conducted studies, it has been shown that even 15–44% of HIV patients and 14–24% of non-HIV immunosuppressed patients are asymptomatic carriers of P. jirovecii [80].
In patients with Pneumocystis pneumonia, rapid and accurate establishment of the diagnosis and prompt treatment initiation are critical determinants for favorable clinical outcomes [83]. Considering the fact that P. jirovecii cannot be cultured in vitro, microscopic visualization of P. jirovecii cysts and/or trophozoites in lower respiratory samples such as induced sputum and BAL fluids still remains the gold standard for the diagnosis of Pneumocystis pneumonia [84]. However, this method is largely subjective, nonspecific, low sensitive, and depends on the skill and experience of the observer, and the type of sample [85]. Despite all disadvantages, ME is still frequently used in resource-limited laboratories for the detection of P. jirovecii in clinical specimens. This applies in particular to immunofluorescent assay (IFA) which is, compared with Calcofluor white, Grocott-Gomori methenamine silver, and Diff-Quik staining method, significantly more sensitive (90.8% vs. 73.8%, 79.4% and 49.2%) but less specific (81.9% vs. 99.6%, 99.2% and 99.6%) [86], [87].
In recent two decades, introduction of molecular detection methods into the clinical laboratory practice has led to significant progress in the diagnosis of Pneumocystis pneumonia [88]. Nowadays, these methods have greatly expanded their scope of application and are also successfully used for investigation of potential outbreak scenarios, determination of epidemiology and transmission of Pneumocystis pneumonia, and, as opposed to conventional microscopy, offer a high degree of objectivity [81], [83]. PCR, as one of the most popular molecular diagnostic techniques in clinical microbiology at this moment, has been reported as a valuable tool for detection of P. jirovecii in different non-invasively and invasively collected clinical samples from the respiratory tract [79]. In addition, a newly developed, improved version of the PCR assay – real-time PCR has contributed to a significant reduction in turnaround time (results are available after 2.5 h including DNA extraction) and an overall increase in diagnostic sensitivity compared to IFA [83]. Moreover, as opposed to nested PCR, such one-step assays are easier to perform and decrease the risk of carry-over contamination [76].
Until today, various samples have been used for PCR detection of P. jirovecii pneumonia. PCR on BAL sample has higher diagnostic sensitivity (97.1–100%) [79], [81] in comparison with non-invasive samples such as sputum (91.4%) [79] or oropharyngeal wash fluid (76%)(81), but is difficult to obtain in some critically ill patients due to the increased risk of respiratory deterioration [80]. Also, in patients with non-productive cough suspected of having P. jirovecii pneumonia, induced sputum is not easy to obtain [79]. It is important to emphasize that, although highly sensitive, negative PCR results of BAL fluid do not provide reliable exclusion of P. jirovecii pneumonia given that the false negative results can occur when mutation at position 210 (C210T) of the mitochondrial large subunit ribosomal RNA (mtLSUrRNA) gene of P. jirovecii is present [89]. On the other hand, studies have shown that PCR analysis of oropharyngeal wash fluid is more specific (93%) than BAL (86.1–87%) and sputum (86.1%), which indicates that the PCR detection of P. jirovecii in the upper part of respiratory tract is a good indicator of P. jirovecii pneumonia [79], [81].
In order to overcome insufficient specificity of PCR on BAL fluid and sputum, a relatively new concept in the diagnosis of P. jirovecii pneumonia has been developed. This method is characterized by minimal invasiveness and includes PCR detection of cell-free DNA, fragments of DNA which are present extra cellular in different body fluids, in serum samples [90]. The pioneering experimental studies have shown that diagnostic sensitivity of PCR detection of P. jirovecii DNA in sera may vary from 0–100% [90], [91]. However, the most recent study has shown that, when compared with GMS staining as the gold standard, the sensitivity of cell-free DNA PCR is the same as BAL fluid PCR and sputum PCR, while the specificity is much higher [79]. When above mentioned studies are analyzed together, it can be concluded that diagnostic performances of PCR on serum DNA depend largely on the amount of sample used for DNA extraction, DNA extraction method (magnetic bead methods are more efficient in serum cell free-DNA extraction than Proteinase K-phenol-chloroform) and the type of primers used for detection of Pneumocystis [Pneumocystis heat shock protein 70 (HSP70) gene has higher sensitivity and specificity in Pneumocystis detection than Pneumocystis mt LSU rRNA gene] [79], [90], [91].
3.6. Molecular diagnosis of invasive aspergillosis
Invasive pulmonary aspergillosis is a devastating opportunistic infection caused by molds belonging to the genus Aspergillus. This infection mainly affects immunocompromized hosts, such as patients with cancer or hematologic malignancies, allogeneic hematopoietic stem cell and solid organ transplant recipients (especially lung transplant recipients), patients with advanced AIDS, and those presenting with chronic granulomatous disease [92], [93]. In recent years, increase in incidence of invasive pulmonary aspergillosis has been noted in patients with lympho proliferative syndromes and in medical ICU patients, particularly in those treated with corticosteroids. Nowadays, despite the advances in understanding the disease and development of highly effective antifungals such as voriconazole and posaconazole, global mortality and morbidity rates are still high [94].
Timely and accurate diagnosis and early initiation of appropriate antifungal treatment significantly improve disease prognosis [93]. Diagnosis of invasive pulmonary aspergillosis is traditionally based on the isolation of fungi of the genus Aspergillus in culture in combination with CME or radiographic findings [95]. However, the sensitivity of culture is low and may take a long time to results. Besides, cultures are incapable of differentiating between infection, colonization, or contamination. On the other hand, ME, as inexpensive and easy-to-perform complementary diagnostic method, improves positive predictive value by confirming positive culture results and provides information about the infecting cell morphology, the state of infected tissues, and biofilm formation, but is not organism specific [41]. Immuno assays which are based on the detection of fungal cell wall biomarkers, such as BDG or Aspergillus GM, have significantly reduced turn-around times compared with ME, but it has been shown that they can be low sensitive and specific in certain patient groups [95].
Molecular techniques represent a novel approach to the diagnosis of invasive aspergillosis. Currently, PCR is the most widely applied molecular diagnostic test, which enables amplification and detection of extremely low concentrations (from 1 to 10 pg) of Aspergillus DNA in various clinical specimens such as BAL fluid, serum, plasma, whole blood and biopsy material [94], [96]. Also, PCR assays can quickly identify species within the genus Aspergillus from cultures and non-cultured samples which is of great significance for epidemiologic studies and clinical practice given the fact that newly described species such as Aspergillus lentulus and Aspergillus calidoustus have been shown to have elevated minimum inhibitory concentrations (MICs) to several antifungal drugs, including azoles [95]. Due to the lack of standardization and potential false-positive results, PCR was in 2002 excluded from European Organization for the Research and Treatment of Cancer/Mycoses Study Group (EORTC/MSG) definitions [97]. In 2015, a group of experts conducted an investigation in which Aspergillus PCR was compared with GM and BDG and, based on obtained results; they concluded that “PCR is now mature enough for inclusion in the EORTC/MSG definitions” [98].
Studies have shown that performance of molecular tests varies depending on the specimen type and patient population [95], [96]. PCR analysis of blood fractions is the most desired testing modality given that it requires minimally invasive sampling procedure [95]. Also, it is important to emphasize that this method is less exposed to accidental contamination and, in comparison with PCR on BAL samples, offers higher diagnostic sensitivity when screening [94].
Moreover, it has been noted that the diagnosis of invasive aspergillosis can be excluded if consistently negative results are obtained during the testing of multiple blood samples [98]. Conducted meta-analyses have shown that Aspergillus PCR on blood samples achieves Sn ranging from 77 to 88% and Sp from 75 to 94.1% [63], [93], [99]. When different fractions of blood are compared, meta-analysis showed that PCR assays of whole blood are more Sn (86% vs. 76%), but less Sp (73% vs. 85%) than those of sera. However, this difference is not statistically significant [93]. On the other hand, another study which included hematology patients at risk of invasive aspergillosis reported the highest diagnostic Sn (91%) of Aspergillus PCR testing of plasma in comparison with PCR testing of serum (80%) and whole blood (55%) [100]. This disagreement in the results of two above mentioned studies can be due to the differences in sample processing and prolonged specimen storage methods for the whole-blood specimens used in the second study [95]. In high-risk patients, at least two positive whole-blood PCR specimens per patient may dramatically increase Sp to 95% with positive predictive value of 90%, which is very indicative for invasive aspergillosis. However, this approach significantly decreases Sn to 64%, which indicates that this diagnostic strategy is more valuable as a confirmatory tool than as a screening tool [93]. Another factor that may also affect the Sn of PCR testing is current immunological status of the patient. In the study, which examined diagnostic performance of PCR analysis of sera obtained from neutropenic and non-neutropenic patients, it was shown that this testing modality has higher Sn in neutropenic patients (82.1%) than in non-neutropenic patients (62.5%), but without statistical significance. Likewise, the same study noted that the initial level of Aspergillus DNA is highly predictive of the 90-day mortality rate (below 150 copies/mL–low probability; above 150 copies/mL–high probability), which is of great significance for identification of patients who may benefit from more intensive care [96]. Finally, based on all of the above facts, it can be concluded that PCR analysis of serum or plasma seems to be diagnostic modality with acceptable Sn and Sp. Also, these sample types, as opposed to whole blood, do not require preanalytical cell lysis steps and allow other biomarker diagnostic tests, such as GM, to be run [93], [95].
BAL fluid represents another commonly used sample for the diagnosis of invasive aspergillosis. Compared with blood samples, the Sp of Aspergillus PCR BAL testing is significantly higher [98]. Based on the results of conducted meta-analysis, the pooled Sp of PCR on BAL fluid specimens was even 94% for proven/probable invasive aspergillosis, while Sn was 79% [101]. However, one of the major limitations of PCR testing of respiratory samples is the inability to differentiate airway colonization from invasive disease. Also, the risk of sample contamination at the patient's bedside or in the laboratory by airborne spores is high which may lead to the occurrence of false positive results. Furthermore, it is estimated that even 25% of BAL samples from healthy subjects would be falsely positive after molecular testing which is significantly higher than in other diagnostic modalities (e.g. detection of GM) [94]. Quantitative PCR (qPCR) detection of high fungal burdens may be indicative of invasive aspergillosis, but this diagnostic modality has still many limitations [95].
In recent years, resistance to azoles in A. fumigatus has emerged as a global health problem [102]. Based on published results of conducted studies, the resistance occurs as a result of TR34/L98H and TR46/Y121F/T289A mutations in the cyp51A gene and its promoter region [103]. These mutations were first found in the Netherlands in 1998 (TR34/L98H) and 2009 (TR46/Y121F/T289A) [102] and have since been reported in multiple European countries, Middle East, Asia, Africa, Australia and the United States [104]. Azole-resistant invasive aspergillosis is difficult to diagnose using traditionally culture-based methods given that Aspergillus cultures are negative in the majority of patients. On the other hand, immunoassays, which provide detection of fungal cell wall biomarkers, are unable to identify patients with azole-resistant disease [102]. Consequently, two commercial multiplex real-time PCR assays (MycoGENIE® (Ademtech, Pessac, France) and AsperGenius (PathoNostics, Maastricht, the Netherlands)) have been developed to enable the detection of both A. fumigatus DNA and the most prevalent cyp51A mutations responsible for azole resistance in clinical samples. These assays have been evaluated using BAL fluid and serum samples. The AsperGenius® multiplex real-time PCR test includes two PCRs – the species PCR and the resistance PCR. In respiratory samples, the species PCR generated theSn and Sp of 88.9% and 89.3%, respectively, in patients with hematological disorders, while the resistance PCR was able to detect resistance-associated mutations in a culture-negative patient with invasive aspergillosis. As for the serum samples, species assay showed the Sn and Sp of 78.6% and 100%, respectively in patients in whom fungal disease status had been previously defined using the revised EORTC criteria [105], [106], [107]. On the other hand, MycoGENIE® showed theSn of 92.9% and Sp of 92.9% for detection of Aspergillus DNA in respiratory samples, while in serum samples, the Sn and Sp were 100% and 84.6%, respectively. It is important to emphasize that all isolates harboring the TR34/L98H mutations were accurately detected [108].
3.7. Multiplex PCR (mPCR) panels
Recently, multiplex PCR and real-time mPCR molecular tests have been developed for detection and identification of various closely related pathogens causing respiratory, gastrointestinal and sexually transmitted infections as well as meningitis and sepsis. Because of similar clinical symptoms, signs and possible simultaneous presence of several different agents, the differential diagnosis of these infections includes a large number of potential agents, which are clinically indistinguishable. Use of molecular panels allows timely detection and discrimination of the infecting pathogens, which is important to optimize medication, prevent secondary spread of infection and unnecessary antibiotic use in viral infections, and may help in decisions regarding hospitalization and infection control measures. Also, these panels include internal control for monitoring of every procedure step [109], [110], [111], [112], [113], [114], [115], [116]. In medical mycology, the use of multiplex PCR for pathogens detection implies panels for detection of yeasts in blood of patients with sepsis and panels for Cry. neoformans detection in CSF of patients with meningitis/encephalitis. Characteristics of these molecular panels are shown in the Table 3 .
Table 3.
Performances of molecular multipathogen panels for detection of fungi.
| Molecular panels performances |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pathogens | Time for results obtaining/hands-on time | Sn/Sp of evaluated tests (%) | Trade Name | Manufacturer | Preextraction required | Principle | Detection methodology | Reference | |
| Blood multipathogen detection panels | C. albicans; C. glabrata; C. krusei; C. parapsilosis; C. tropicalis; Staphylococcus spp.; S. aureus; Streptococcus spp.; S. agalactiae; S. pyogenes; S. pneumonia; Enterococcus spp.; L. monocytogenes; Enterobacteriaceae; E. coli; Enterobacter cloacae complex; Klebsiella. Oxytoca; K. pneumoniae; Serratia marcescens; Proteus spp.; A. baumannii; H. influenzae; N. meningitidis; P. aeruginosa; antibiotic resistance markers mecA, vanA/vanB, KPC (27 pathogens) | 65 min/5 min | 99.5%/91.7% for fungi 98.5%/100% for Gram – bacteria 96.7%/93.7% for Gram+ bacteria |
FilmArray® BCID | BioFire Diagnostics, Salt Lake City, UT | No | Nested multiplex PCR | Endpoint melting curve analysis | [117] |
| C. albicans; C. tropicalis; C. parapsilosis; C. krusei; C. glabrata; A. fumigatus; E. coli; Klebsiella pneumoniae/oxytoca; Serratia marcescens; Enterobacter cloacae/aerogenes; P. mirabilis; P. aeruginosa; A. baumannii; S. maltophilia; S. aureus; CoNS; Str. pneumoniae; Streptococcus spp.; E. faecium; E. faecalis; (25 pathogens) | 6–8 h/3 h | 69–90.5%/65–87%for newborns 75%/92% for adults |
LightCycler® SeptiFast Test MGRADE | Roche Diagnostics GmbH, Manheim, Germany | Yes | Multiplex Real Time PCR | Endpoint melting curve analysis | [118], [119] | |
| 13 fungal pathogens, over 60 bacterial pathogens, and three resistance genes (mecA, vanA, and vanB) in positive blood cultures | 3–3.5 h/90 min | 94.7%/98.8% 99%/98% for fungal targets |
Prove-it® Sepsis | Mobidiag, Helsinki, Finland | Yes | Multiplex PCR | Microarray Colorimetric Read-out |
[120], [121] | |
| 345 fungi and bacteria | 8 h/2.5 h | 87.0%/85.8% | SepsiTest® | Molzym, Bremen, Germany | Yes | Broad-range PCR | Sequencing | [122] | |
| 6 fungi, 73 Gram (+) bacteria, 3 Drug resistance markers, 12 Gram (−) bacteria (more than 90 pathogens), 27 pathogens can be identified to the species level | 5–7 h/No data available | 64%/96% | Magicplex™ Sepsis Real-time Test | Seegene, Seoul, Korea | Yes | Multiplex Real Time PCR based on READ™ technology | Multiple fluorophore detection | [122], [123] | |
|
A. fumigatus; C. albicans; C. dubliniensis; C. glabrata; C. tropicalis; C. krusei; C. parapsilosis; Gram-positive bacteria: S. pneumoniae; S. pyogenes; S. sanguinis; S. agalactiae; S. dysgalactiae; S. mutans; E. faecium; E. faecalis; C. perfringens; S. aureus; S. epidermidis; S. saprophyticus; S. haemolyticus; S. hominis Gram-negative bacteria: E. coli; P. aeruginosa; K. pneumonia; K. oxytoca; E. cloacae; E. aerogenes; N. meningitides; M. morganii; P. mirabilis; A. baumannii; B. fragilis; S. marcescens; B. cepacia; S. maltophilia; P. buccae; P. intermedia; P. melaninogenica; Five most common resistance markers: methicillin mecA, vancomycin vanA, vancomycin vanB, β-lactamase blaSHV, β-lactamase blaCTX-M (34 bacterial and 7 fungal pathogens) |
6–8 h/70 min | 60%/75% | Vyoo® | SIRS-Lab GmbH, Jena, Germany | Yes | Multiplex PCR | Gel-electrophoresis | [111], [124] | |
| Cerebrospinal fluid multipathogen detection panels |
C. neoformans/gattii; E. coli K1; H. influenzae; L. monocytogenes; N. meningitidis; S. agalactiae; S. pneumoniae; CMV; EnV; HSV-1/2/3; HHV−6; hPEV; VZV (14 pathogens) | 1 h/2 min | 100% for 9 of 14 analytes/99.2% | FilmArray® ME Panel | BioFire Diagnostics, Salt Lake City, UT | No | Nested multiplex PCR | Endpoint melting curve analysis | [113], [125] |
| Respiratory multipathogen detection panels |
Inf A; Inf B; Inf C; hRV; hCoV (NL63, 229E, OC43, HKU1); PIV 1–4; hMPV A/B; hBoV; hRSV A/B; AdV; EnV; EVs; hPEV; C. pneumonia; S. aureus; S. pneumonia; Haemophilus influenzae spp.; CMV; P. jirovecii; H. influenzae B; Bordetella spp. (except B. parapertussis); M. pneumoniae; M. catarrhalis; K. pneumoniae; Legionella spp.; Salmonella spp. (33 pathogens) | 4 h/No data available | 77.89–92.2%/94.37–99.5% |
FTD® Respiratory Pathogens 33 | Fast Track Diagnostics, Luxembourg | Yes | Multiplex Real-Time PCR based on TaqMan® technology | Multiple fluorophore detection | [126], [127] |
| P. jirovecii; H. capsulatum; C. neoformans/C. gattii (4 fungal pathogens) | No data available | 90.7–100%/100% | Not registered | In-house | Yes | Multiplex Real-Time PCR | Multiple fluorophore detection | [128] | |
Sn: sensitivity; Sp: specificity; Acinetobacter baumannii: A. baumannii; Aspergillus fumigatus: A. fumigatus; Escherichia coli: E. coli; Klebsiella pneumoniae: K. pneumoniae; Klebsiella oxytoca: K. oxytoca; Serratia marcescens: S. marcescens; Proteus mirabilis: P. mirabilis; Pseudomonas aeruginosa: P. aeruginosa; Stenotrophomonas maltophilia: S. maltophilia; Enterococcus faecium: E. faecium; Enterococcus faecalis: E. faecalis; Candida albicans: C. albicans; Candida tropicalis: C. tropicalis; Candida parapsilosis: C. parapsilosis; Candida krusei: C. krusei; Candida glabrata: C. glabrata; Candida dubliniensis: C. dubliniensis; Clostridum perfrigens: C. perfrigens; Cryptococcus neoformans/Cryptococcus gattii: C. neoformans/gattii; Enterococcus faecalis: E. faecalis; Enterococcus faecium: E. faecium; Escherichia coli: E. coli; Histoplasma capsulatum: H. capsulatum; Haemophilus influenzae: H. influenzae; Neisseria meningitidis: N. meningitidis; Pseudomonas aeruginosa: P. aeruginosa; Listeria monocytogenes: L. monocytogenes; Morganella morganii: M. morganii; Pneumocystis jiroveci: P. jirovecii; Prevotella buccae: P. buccae; Prevotella intermedia: P. intermedia; Prevotella melaninogenica: P. melaninogenica; Staphylococcus aureus: S. aureus; CoNS (Coagulase negative Staphylococci); Staphylococcus aureus: S. aureus; Staphylococcus epidermidis: S. epidermidis; Staphylococcus haemoliticus: S. haemoliticus; Staphylococcus hominis: S. hominis; Staphylococcus saprophyticus: S. saprophyticus; Streptococcus agalactiae: S. agalactiae; Streptococcus pyogenes: S. pyogenes; Streptococcus pneumoniae: S. pneumoniae; Streptococcus bovis: S. bovis; Streptococcus dysgalactiae: S. dysgalactiae; Streptococcus mutans: S. mutans; Streptococcus sanguinis: S. sanguinis; Cytomegalovirus: CMV; enterovirus: EnV; herpes simplex virus: HSV; human herpesvirus: HHV; human parechovirus: hPEV; varicella-zoster virus: VZV; Inf: influenza virus; hRV: human rhinovirus; hCoV: human coronavirus; PIV: parainfluenzavirus; hMPV: human metapneumovirus; hBoV: human bocavirus; hRSV: human respiratory syncytial virus; AdV: adenovirus; EVs: echovirus; hPEV: human parechovirus
3.8. Fungal blood multipathogen molecular panels
In recent years, different rapid molecular methods have been developed to speed up the identification of the microorganisms that grow in blood-culture (BC) bottles. On the first place, MALDI-TOF MS Sepsityper Kit™ can be used to directly analyze positive BC in real time and to provide definitive species identification within 20 to 60 minutes, with the identification rate of 85.5% [129]. The MALDI-TOF has the advantage of rapid turn-around time and ability to identify a large number of microorganisms directly. Disadvantages include the high start-up costs associated with the purchase of mass-spectrometry equipment and training of personnel. As an alternative to MALDI-TOF, Film Array Blood Culture Identification (BCID) Panel (Bio Fire, USA), a multiplexed PCR based diagnostic test, can be used for positive BC. The BCID can detect 19 bacteria, five Candida spp. and three antimicrobial resistance determinants (mec A, van A/B, bla-KPC) in 1 h from the time of BC positivity [69]. This method has a several advantages: the large number of targets can be evaluated in a single test and all steps of the assay, from nucleic acid extraction to interpretation of amplification data, are performed in closed system and using a single pouch on a minimally processed clinical sample. The required laboratory procedures are not technologically complex and can be performed by persons who do not have training in molecular techniques [117]. Other methods such as Nano particle Probe Technology (Nucleic Acid Extraction and PCR Amplification) and Nano sphere's Verigene can be used for identification. The platform consists of two assay panels: the BC-GP assay and the BC-GN assay. Also it can detect drug resistance markers such as: mec A, van A/B, KPC, NDM, CTX-M, VIM, IMP and OXA genes [69].
3.9. Fungal meningitis/encephalitis multipathogen molecular panels
With a single PCR test, Film Array Meningitis/Encephalitis panel (Bio Fire Diagnostics, LLC, Salt Lake City, UT, USA), the detection of several common pathogens (bacteria, viruses and fungi) that cause meningitis, including Cry. neoformans, is possible. In recently published study, authors compared the performance of the standard application of BioFire Film Array Meningitis/Encephalitis panel in 48 patients. Based on obtained results, it was shown that Film Array Meningitis/Encephalitis panel detected organisms that can be missed by the conventional laboratory analyses. However, it cannot completely replace conventional laboratory methods, because it does not detect all microorganisms responsible for meningitis and encephalitis. The Film Array ME panel offers comprehensive, standardized and rapid testing of CSF with minimal volume usage in 1 hour [130].
4. Future perspectives
T2 magnetic resonance (MR) assay for the rapid diagnosis of candidemia in whole blood is one of tests in the new era of molecular diagnostics which can provide fast and species – specific (C. albicans, C. tropicalis, C. glabrata, C. krusei, C. parapsilosis) detection directly in the clinical samples. Regarding the fact that T2 MR has 91% sensitivity and > 98% specificity, does not require cultivation as well as sample purification or preparation, it can be highlighted that this test represents example of point-of-care diagnostic test which can ensure the prompt and accurate results in 4.2 ± 0.9 hours with the possibility of treatment modification in cases of candidemia. This novel technology has 99.4% negative predictive value in population of patients with Candida invasive infection, disease with estimated prevalence of 6%, which will decrease the number of patients on empiric therapy and resistant strains occurrence [131].
Regarding the candidemia caused by C. glabrata, so far, it was considered that fluconazole (FLC) resistance is common in C. glabrata and echinocandins are often used as first-line therapy. One of the major advantages of T2 MR is a possibility of rapid detection of C. glabrata in blood of patients with higher sensitivity (to > 84%) than culture (Se = 60%) [131] and its determination on low level of detection of 2 CFU/mL. This is very important since that early initiation of appropriate treatment can improve prognosis and significantly decreases mortality. However, further improvement of diagnostic performances of T2 MR is required given that nearly 12% of patients had indeterminate T2 Dx results (n = 245/2046). Also, the price of this method is high and does not enable day-to-day monitoring, contrarily to BC. In addition, only 5 species of Candida are detected by this assay [131], [132], [133].
However, the resistance to echinocandin therapy associated with FKS1 and FKS2 gene alterations of C. glabrata has been noticed in recent years. These findings influence the establishment of a novel (allele-specific molecular beacon and DNA melt analysis followed by asymmetric PCR) and highly accurate diagnostic assay for rapid identification of FKS mutations associated with echinocandin resistance in this Candida species. This platform can be diagnostic method for rapid detection of infections caused by C. glabrata resistant strains to echinocandin and will timely provide the choice of efficient treatment [132], [133].
Currently, there are efforts for development of mass spectrometryy (MS) procedure for fungal disaccharide (DS) detection in patient's sera for rapid diagnosis of IFI. Applaing of MALDI-TOF MS for fungal molecules detection have shown very good diagnostic efficacy for early diagnosis of IC, IA and more important invasive Mucor infection (IMI). The first researches in this field have pointed out the fact that the sensitivity of MS-DS (with application of the higher cutoff) is higher than that of MN or BDG detection. In addition, neither neutropenia nor bacteriemia have been shown to influence the detection of fungal disaccharide DD in patients with IC. Also, this method has provided earlier positive results than other tests on the first serum samples in the cases of IA. Moreover, results of evaluation of this method suggest the possibility of panfungal analyses and diagnosis of IMI for which currently there are no available commercial serological tests that are efficient for the diagnosis. However, it has to be highlighted that this biological test requires further evaluation and validation in multicenter studies [134].
5. Conclusion
Delays in the diagnosis of IFI and appropriate therapy application clearly affect the infectious diseases outcome in a negative fashion. On the other hand, non-established accurate identification of fungal pathogens that cause SFI can influence failure in implementation of appropriate anti-epidemic prevention measures. In the era of large number of commercial rapid tests, clinicians must decide which of them should be introduced to the routine work, based on an individual experience, consideration of the disease incidence in the population, the cost-effectiveness and finally adequate assessment and selection of patients who would have the most benefit from rapid treatment.
Formatting of funding sources
The study was supported by the Serbian Ministry of Education and Science [the grant No 175034 and grant No III 41007].
Disclosure of interest
The authors declare that they have no competing interest.
References
- 1.Montagna M.T., Coretti C., Lovero G., De Giglio O., Montagna O., Laforgia N. Diagnostic performance of 1→3-β-d-glucan in neonatal and pediatric patients with Candidemia. Int J Mol Sci. 2011;12:5871–5877. doi: 10.3390/ijms12095871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arsić Arsenijević V., Otašević S., Janić D., Minić P., Matijašević J., Medić D. Candida bloodstream infections in Serbia: first multicentre report of a national prospective observational survey in intensive care units. Mycoses. 2017 doi: 10.1111/myc.12700. [In press] [DOI] [PubMed] [Google Scholar]
- 3.Arsić-Arsenijević V., Milenković M., Otašević S., Pavlica D. Medical mycology and parasitology, 1st Ed. Serb Soc Med Mycol. 2012 [Google Scholar]
- 4.Pfaller M.A., Diekema D.J. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sydnor E.R., Perl T.M. Hospital epidemiology and infection control in acute-care settings. Clin Microbiol Rev. 2011;24:141–173. doi: 10.1128/CMR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otašević S., Barac A., Pekmezovic M., Tasic S., Ignjatović A., Momčilović S. The prevalence of Candida onychomycosis in Southeastern Serbia from 2011 to 2015. Mycoses. 2016;59:167–172. doi: 10.1111/myc.12448. [DOI] [PubMed] [Google Scholar]
- 7.Tasic Otašević S.A., Miladinovic-Tasic N., Djordjevic J., Ignjatović A., Stojanović P., Zdravković D. Superficial mycoses in the Niš region, Southeast-Serbia. Central Eur J Med. 2011;6:665–671. [Google Scholar]
- 8.Hayette M.P., Sacheli R. Dermatophytosis, trends in epidemiology and diagnostic approach. Cur Fungal Infect Rep. 2015;9:164–179. [Google Scholar]
- 9.Kalokasidis K., Onder M., Trakatelli M.G., Richert B., Fritz K. The effect of Q-switched Nd:YAG 1064 nm/532 nm laser in the treatment of onychomycosis in vivo. Dermatol Res Pract. 2013;2013:379725. doi: 10.1155/2013/379725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farmakiotis D., Ciurea A.M., Cahuayme-Zuniga L., Kontoyiannis D.P. The diagnostic yield of skin biopsy in patients with leukemia and suspected infection. J Infect. 2013;67:265–272. doi: 10.1016/j.jinf.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Lanternier F., Pathan S., Vincent Q.B., Liu L., Cypowyj S., Prando C. Deep dermatophytosis and inherited CARD9 deficiency. N Engl J Med. 2013;369:1704–1714. doi: 10.1056/NEJMoa1208487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sobel J.D. Vulvovaginal candidosis. Lancet. 2007;369:1961–1971. doi: 10.1016/S0140-6736(07)60917-9. [DOI] [PubMed] [Google Scholar]
- 13.Otašević S., Miladinović-Tasić N., Pešić S. Recurrent genital candidosis-diagnostic and epidemiological aspects. Proceeding of 47th days of preventive medicine; 2013 September 24–27; Niš, Serbia. Niš: University of Niš, Fuculty of Medicine; 2013. [Google Scholar]
- 14.Otasevic S., Momčilović S., Trajkovic A., Arsic-Arsenijevic V. Modelling of antifungal treatment with azoles and essential oils for non-albicans Candida spp. causing vulvo-vaginal infections. 27th European Congress of Clinical Microbiology and Infectious Diseases; Vienna, Austria; 2017. [Google Scholar]
- 15.Qiu W.Z., Ke L.R., Xia W.X., Yang J., Yu Y.H., Liang H. A retrospective study of 606 cases of nasopharyngeal carcinoma with or without oropharyngeal candidiasis during radiotherapy. PLoS One. 2017;12:e0182963. doi: 10.1371/journal.pone.0182963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrović M., Kostić M., Kostić M., Krunić N., Igić M., Pešić Z. Therapeutic alternatives of natural compounds in treatment of Candida-associated denture stomatitis. Acta Med Median. 2014;53:73–80. [Google Scholar]
- 17.Verweij P., Lyon S. Optimizing antifungal strategies to improve patient survival. Future Microbiol. 11, 2016:1211–1215. doi: 10.2217/fmb-2016-0153. 1211-1215 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Winn W.C., Jr., Allen S., Janda W., Koneman E., Procop G., Schreckenberger P. 6th Ed. Lippincott Williams and Wilkins; Philadelphia: 2006. Koneman's colour atlas and textbook of diagnostic microbiology. [Google Scholar]
- 19.Goff D.A., Jankowski C., Tenover F.C. Using rapid diagnostic tests to optimize antimicrobial selection in antimicrobial stewardship programs. Pharmacotherapy. 2012;32:677–687. doi: 10.1002/j.1875-9114.2012.01137.x. [DOI] [PubMed] [Google Scholar]
- 20.Hook E.W., 3rd, Spitters C., Reichart C.A., Neumann T.M., Quinn T.C. Use of cell culture and a rapid diagnostic assay for Chlamydia trachomatis screening. JAMA. 1994;272:867–870. [PubMed] [Google Scholar]
- 21.Brooks G.F., Carroll K.C., Butel J.S., Morse S.A., Mietzner T.A. 26th Ed. Lange Medical Books, McGraw Hill; New York: 2013. Jawetz, Melnick & Adelberg's medical microbiology. [Google Scholar]
- 22.Wilkison B.D., Sperling L.C., Spillane A.P., Meyerle J.H. How to teach the potassium hydroxide preparation: a disappearing clinical art form. Cutis. 2015;96:109–112. [PubMed] [Google Scholar]
- 23.Mylonas I., Bergauer F. Diagnosis of vaginal discharge by wet mount microscopy: a simple and underrated method. Obstet Gynecol Surv. 2011;66:359–368. doi: 10.1097/OGX.0b013e31822bdf31. [DOI] [PubMed] [Google Scholar]
- 24.Uehara Y., Yagoshi M., Tanimichi Y., Yamada H., Shimoguchi K., Yamamoto S. Impact of reporting gram stain results from blood culture bottles on the selection of antimicrobial agents. Am J Clin Pathol. 2009;132:18–25. doi: 10.1309/AJCP0H2DAMBXZUSS. [DOI] [PubMed] [Google Scholar]
- 25.Kunimatsu J., Ohmagari N., Yoshizawa A. Is Gram staining a diagnostic tool or a guide for optimal empirical therapy? Int J Infect Dis. 2013;17:e136. doi: 10.1016/j.ijid.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Higashi Y., Miyoshi H., Takeda K., Saruwatari H., Kubo H., Sakaguchi I. Evaluation of a newly-developed immunochromatography strip test for diagnosing dermatophytosis. Int J Dermatol. 2012;51:406–409. doi: 10.1111/j.1365-4632.2011.05046.x. [DOI] [PubMed] [Google Scholar]
- 27.Matsui H., Hanaki H., Takahashi K., Yokoyama A., Nakae T., Sunakawa K. Rapid detection of vaginal Candida species by newly developed immunochromatography. Clin Vaccine Immunol. 2009;16:1366–1368. doi: 10.1128/CVI.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noriki S., Ishida H. Production of an anti-dermatophyte monoclonal antibody and its application: immunochromatographic detection of dermatophytes. Med Mycol. 2016;54:808–815. doi: 10.1093/mmy/myw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cafarchia C., Otranto D., Weigl S., Campbell B.E., Parisi A., Cantacessi C. Molecular characterization of selected dermatophytes and their identification by electrophoretic mutation scanning. Electrophoresis. 2009;30:3555–3564. doi: 10.1002/elps.200900313. [DOI] [PubMed] [Google Scholar]
- 30.Cafarchia C., Iatta R., Latrofa M.S., Gräser Y., Otranto D. Molecular epidemiology, phylogeny and evolution of dermatophytes. Infect Genet Evol. 2013;20:336–351. doi: 10.1016/j.meegid.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 32.Symoens F., Jousson O., Planard C., Fratti M., Staib P., Mignon B. Molecular analysis and mating behaviour of the Trichophyton mentagrophytes species complex. Int J Med Microbiol. 2011;301:260–266. doi: 10.1016/j.ijmm.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Garg J., Tilak R., Garg A., Prakash P., Gulati A.K., Nath G. Rapid detection of dermatophytes from skin and hair. BMC Res Notes. 2009;2:60. doi: 10.1186/1756-0500-2-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arabatzis M., Bruijnesteijn van Coppenraet L.E., Kuijper E.J., de Hoog G.S., Lavrijsen A.P., Templeton K. Diagnosis of common dermatophyte infections by a novel multiplex real-time polymerase chain reaction detection/identification scheme. Br J Dermatol. 2007;157:681–689. doi: 10.1111/j.1365-2133.2007.08100.x. [DOI] [PubMed] [Google Scholar]
- 35.Bergmans A.M., van der Ent M., Klaassen A., Böhm N., Andriesse G.I., Wintermans R.G. Evaluation of a single-tube real-time PCR for detection and identification of 11 dermatophyte species in clinical material. Clin Microbiol Infect. 2010;16:704–710. doi: 10.1111/j.1469-0691.2009.02991.x. [DOI] [PubMed] [Google Scholar]
- 36.Dhib I., Fathallah A., Yaacoub A., Hadj Slama F., Said M.B., Zemni R. Multiplex PCR assay for the detection of common dermatophyte nail infections. Mycoses. 2014;57:19–26. doi: 10.1111/myc.12096. [DOI] [PubMed] [Google Scholar]
- 37.Kondori N., Abrahamsson A.L., Ataollahy N., Wennerås C. Comparison of a new commercial test, dermatophyte-PCR kit, with conventional methods for rapid detection and identification of Trichophyton rubrum in nail specimens. Med Mycol. 2010;48:1005–1008. doi: 10.3109/13693781003743130. [DOI] [PubMed] [Google Scholar]
- 38.Dingemans G., van den Bosch M., Hayette M.P., Goethel S., Rusu V., Sacheli R. Development of a new commercial qPCR assay to detect and differentiate dermatophyte infections of the skin, nails and hair. Presented at the European Congress of Clinical Microbiology and Infectious Diseases, 10–13 May 2014; Barcelona; 2014. [Google Scholar]
- 39.Motamedi M., Mirhendi H., Zomorodian K., Khodadadi H., Kharazi M., Ghasemi Z. Clinical evaluation of β-tubulin real-time PCR for rapid diagnosis of dermatophytosis, a comparison with mycological methods. Mycoses. 2017;60:692–696. doi: 10.1111/myc.12648. [DOI] [PubMed] [Google Scholar]
- 40.Wiegand C., Mugisha P., Mulyowa G.K., Elsner P., Hipler U.C., Gräser Y. Identification of the causative dermatophyte of tinea capitis in children attending Mbarara Regional Referral Hospital in Uganda by PCR-ELISA and comparison with conventional mycological diagnostic methods. Med Mycol. 2017;55:660–668. doi: 10.1093/mmy/myw112. [DOI] [PubMed] [Google Scholar]
- 41.Arvanitis M., Anagnostou T., Fuchs B.B., Caliendo A.M., Mylonakis E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin Microbiol Rev. 2014;27:490–526. doi: 10.1128/CMR.00091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lass-Flörl C., Mutschlechner W., Aigner M., Grif K., Marth C., Girschikofsky M. Utility of PCR in diagnosis of invasive fungal infections: real-life data from a multicenter study. J Clin Microbiol. 2013;51:863–868. doi: 10.1128/JCM.02965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheesbrough M. 2nd Ed. Cambridge University Press; Cambridge (UK): 2006. District laboratory practice in tropical countries part 2. [Google Scholar]
- 44.Cheesbrough M. 2nd Ed. Cambridge University Press; Cambridge (UK): 2006. District laboratory practice in tropical countries part 1. [Google Scholar]
- 45.Woods G.L., Walker D.H. Detection of infection or infectious agents by use of cytologic and histologic stains. Clin Microbiol Rev. 1996;9:382–404. doi: 10.1128/cmr.9.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Root R.K. Oxford University Press; New York: 1999. Clinical infectious diseases: a practical approach. [Google Scholar]
- 47.Goldman E., Green L.H. 2nd Ed. CRC Press; Boca Raton: 2008. Practical handbook of microbiology. [Google Scholar]
- 48.Eigl S., Prattes J., Lackner M., Willinger B., Spiess B., Reinwald M. Multicenter evaluation of a lateral-flow device test for diagnosing invasive pulmonary aspergillosis in ICU patients. Crit Care. 2015;19:178. doi: 10.1186/s13054-015-0905-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bahr N.C., Boulware D.R. Methods of rapid diagnosis for the etiology of meningitis in adults. Biomark Med. 2014;8:1085–1103. doi: 10.2217/bmm.14.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gunasekera M., Narine M., Ashton M., Esfandiari J. Development of a Dual Path Platform (DPP®) immunoassay for rapid detection of Candida albicans in human whole blood and serum. J Immunol Methods. 2015;424:7–13. doi: 10.1016/j.jim.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 51.Binnicker M.J., Jespersen D.J., Bestrom J.E., Rollins L.O. Comparison of four assays for the detection of cryptococcal antigen. Clin Vaccine Immunol. 2012;19:1988–1990. doi: 10.1128/CVI.00446-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pesic Z., Otasevic S., Mihailovic D., Petrovic S., Arsic-Arsenijevic V., Stojanov D. Alternaria-associated fungus ball of orbit nose and paranasal sinuses: case report of a rare clinical entity. Mycopathologia. 2015;180:99–103. doi: 10.1007/s11046-015-9881-6. [DOI] [PubMed] [Google Scholar]
- 53.Sendid B., Tabouret M., Poirot J.L., Mathieu D., Fruit J., Poulain D. New enzyme immunoassays for sensitive detection of circulating Candida albicans mannan and antimannan antibodies: useful combined test for diagnosis of systemic candidiasis. J Clin Microbiol. 1999;37:1510–1517. doi: 10.1128/jcm.37.5.1510-1517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alam F.F., Mustafa A.S., Khan Z.U. Comparative evaluation of (1, 3)-β-D-glucan, mannan and anti-mannan antibodies, and Candida species-specific snPCR in patients with candidaemia. BMC Infect Dis. 2007;7:103. doi: 10.1186/1471-2334-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prella M., Bille J., Pugnale M., Duvoisin B., Cavassini M., Calandra T. Early diagnosis of invasive candidiasis with mannan antigenemia and antimannan antibodies. Diagn Microbiol Infect Dis. 2005;51:95–101. doi: 10.1016/j.diagmicrobio.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 56.Mokaddas E., Khan Z.U., Ahmad S., Nampoory M.R., Burhamah M. Value of (1-3)-β-d-glucan, Candida mannan and Candida DNA detection in the diagnosis of candidaemia. Clin Microbiol Infect. 2011;17:1549–1553. doi: 10.1111/j.1469-0691.2011.03608.x. [DOI] [PubMed] [Google Scholar]
- 57.De Pauw B., Walsh T.J., Donnelly J.P., Stevens D.A., Edwards J.E., Calandra T. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008;46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Odabasi Z., Mattiuzzi G., Estey E., Kantarjian H., Saeki F., Ridge R.J. β-D-glucan as a diagnostic adjunct for invasive fungal infections: validation, cutoff development and performance in patients with acute myelogenous leukemia and myelodysplastic syndrome. Clin Infect Dis. 2004;39:199–205. doi: 10.1086/421944. [DOI] [PubMed] [Google Scholar]
- 59.Koo S., Bryar J.M., Page J.H., Baden L.R., Marty F.M. Diagnostic performance of the (1-- > 3)-β-D-glucan assay for invasive fungal disease. Clin Infect Dis. 2009;49:1650–1659. doi: 10.1086/647942. [DOI] [PubMed] [Google Scholar]
- 60.White P.L., Price J.S., Posso R.B., Barnes R.A. An evaluation of the performance of the Dynamiker® Fungus (1-3)-β-D-Glucan Assay to assist in the diagnosis of invasive aspergillosis, invasive candidosis and Pneumocystis pneumonia. Med Mycol. 2017;55:843–850. doi: 10.1093/mmy/myx004. [DOI] [PubMed] [Google Scholar]
- 61.White P.L., Posso R.B., Gorton R.L., Price J.S., Wey E., Barnes R.A. An evaluation of the performance of the Dynamiker® Fungus (1-3)-β-D-Glucan Assay to assist in the diagnosis of Pneumocystis pneumonia. Med Mycol. 2017 doi: 10.1093/mmy/myx097. [In press] [DOI] [PubMed] [Google Scholar]
- 62.Skvarc M., Skvarc P.R. Proceeding from 4. Simpozijum-Dijagnoza i terapija gljivičnih infekcija. Društvo medicinskih mikologa Srbije; Beograd: 2016. Diagnostic value of (1-3)-ß-D-glucan guided real-time PCR diagnostics of Pneumocystis jirovecii pneumonia (PCP) for non-HIV immune-compromised patients. [Google Scholar]
- 63.Arvanitis M., Anagnostou T., Mylonakis E. Galactomannan and polymerase chain reaction-based screening for invasive aspergillosis among high-risk hematology patients: a diagnostic meta-analysis. Clin Infect Dis. 2015;61:1263–1272. doi: 10.1093/cid/civ555. [DOI] [PubMed] [Google Scholar]
- 64.Wisplinghoff H., Bischoff T., Tallent S.M., Seifert H., Wenzel R.P., Edmond M.B. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Inf Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 65.Cobo F. Application of molecular diagnostic techniques for viral testing. Open Virol J. 2012;6:104–114. doi: 10.2174/1874357901206010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lorincz A. CRC Press; Boca Raton: 2006. Nucleic acid testing for human disease. [Google Scholar]
- 67.Healy M., Huong J., Bittner T., Lising M., Frye S., Raza S. Microbial DNA typing by automated repetitive-sequence-based PCR. J Clin Microbiol. 2005;43:199–207. doi: 10.1128/JCM.43.1.199-207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Espy M.J., Uhl J.R., Sloan L.M., Buckwalter S.P., Jones M.F., Vetter E.A. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bauer K.A., Perez K.K., Forrest G.N., Goff D.A. Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin Infect Dis. 2014;59:S134–S145. doi: 10.1093/cid/ciu547. [DOI] [PubMed] [Google Scholar]
- 70.Reller M.E., Mallonee A.B., Kwiatkowski N.P., Merz W.G. Use of peptide nucleic acid-fluorescence in situ hybridization for definitive, rapid identification of five common Candida species. J Clin Microbiol. 2007;45:3802–3803. doi: 10.1128/JCM.01127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morgan M., Marlowe E., Della-Latta P., Salimnia H., Novak-Weekley S., Wu F. Multicenter evaluation of a new shortened peptide nucleic acid fluorescence in situ hybridization procedure for species identification of select Gram-negative bacilli from blood cultures. J Clin Microbiol. 2010;48:2268–2270. doi: 10.1128/JCM.00166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forrest G.N., Roghmann M.C., Toombs L.S., Johnson J.K., Weekes E., Lincalis D.P. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob Agents Chemother. 2008;52:3558–3563. doi: 10.1128/AAC.00283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forrest G.N., Mehta S., Weekes E., Lincalis D.P., Johnson J.K., Venezia R.A. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J Antimicrob Chemother. 2006;58:154–158. doi: 10.1093/jac/dkl146. [DOI] [PubMed] [Google Scholar]
- 74.Sogaard M., Hansen D.S., Fiandaca M.J., Stender H., Schonheyder H.C. Peptide nucleic acid fluorescence in situ hybridization for rapid detection of Klebsiella pneumoniae from positive blood cultures. J Med Microbiol. 2007;56:914–917. doi: 10.1099/jmm.0.46829-0. [DOI] [PubMed] [Google Scholar]
- 75.Hall L., Le Febre K.M., Deml S.M., Wohlfiel S.L., Wengenack N.L. Evaluation of the yeast traffic light PNA FISH probes for identification of Candida species from positive blood cultures. J Clin Microbiol. 2012;50:1446–1448. doi: 10.1128/JCM.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sokulska M., Kicia M., Wesołowska M., Hendrich A.B. Pneumocystis jirovecii – from a commensal to pathogen: clinical and diagnostic review. Parasitol Res. 2015;114:3577–3585. doi: 10.1007/s00436-015-4678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fauchier T., Hasseine L., Gari-Toussaint M., Casanova V., Marty P.M., Pomares C. Detection of Pneumocystis jirovecii by quantitative PCR To differentiate colonization and pneumonia in immunocompromised HIV-positive and HIV-negative patients. J Clin Microbiol. 2016;54:1487–1495. doi: 10.1128/JCM.03174-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morris A., Norris K.A. Colonization by Pneumocystis jirovecii and its role in disease. Clin Microbiol Rev. 2012;25:297–317. doi: 10.1128/CMR.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang D., Hu Y., Li T., Rong H.-M., Tong Z.-H. Diagnosis of Pneumocystis jirovecii pneumonia with serum cell-free DNA in non-HIV-infected immunocompromised patients. Oncotarget. 2017;8:71946–71953. doi: 10.18632/oncotarget.18037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hviid C.J., Lund M., Sørensen A., Ellermann-Eriksen S., Jespersen B., Dam M.Y. Detection of Pneumocystis jirovecii in oral wash from immunosuppressed patients as a diagnostic tool. PLoS One. 2017;12:e0174012. doi: 10.1371/journal.pone.0174012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.White P.L., Backx M., Barnes R.A. Diagnosis and management of Pneumocystis jirovecii infection. Expert Rev Anti Infect Ther. 2017;15:435–447. doi: 10.1080/14787210.2017.1305887. [DOI] [PubMed] [Google Scholar]
- 82.Alanio A., Desoubeaux G., Sarfati C., Hamane S., Bergeron A., Azoulay E. Real-time PCR assay-based strategy for differentiation between active Pneumocystis jirovecii pneumonia and colonization in immunocompromised patients. Clin Microbiol Infect. 2011;17:1531–1537. doi: 10.1111/j.1469-0691.2010.03400.x. [DOI] [PubMed] [Google Scholar]
- 83.Church D.L., Ambasta A., Wilmer A., Williscroft H., Ritchie G., Pillai D.R. Development and validation of a Pneumocystis jirovecii real-time polymerase chain reaction assay for diagnosis of Pneumocystis pneumonia. Can J Infect Dis Med Microbiol. 2015;26:263–267. doi: 10.1155/2015/138787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mori S., Sugimoto M. Pneumocystis jirovecii pneumonia in rheumatoid arthritis patients: risks and prophylaxis recommendations. Clin Med Insights Circ Respir Pulm Med. 2015;9:29–40. doi: 10.4137/CCRPM.S23286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harris J.R., Marston B.J., Sangrujee N., DuPlessis D., Park B. Cost-Effectiveness analysis of diagnostic options for Pneumocystis pneumonia (PCP) PLoS One. 2011;6:e23158. doi: 10.1371/journal.pone.0023158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moodley B., Tempia S., Frean J.A. Comparison of quantitative real-time PCR and direct immunofluorescence for the detection of Pneumocystis jirovecii. PLoS One. 2017;12:e0180589. doi: 10.1371/journal.pone.0180589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Procop G.W., Haddad S., Quinn J., Wilson M.L., Henshaw N.G., Reller L.B. Detection of Pneumocystis jiroveci in respiratory specimens by four staining methods. J Clin Microbiol. 2004;42:3333–3335. doi: 10.1128/JCM.42.7.3333-3335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caliendo A.M., Hewitt P.L., Allega J.M., Keen A., Ruoff K.L., Ferraro M.J. Performance of a PCR assay for detection of Pneumocystis carinii from respiratory specimens. J Clin Microbiol. 1998;36:979–982. doi: 10.1128/jcm.36.4.979-982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Le Gal S., Robert-Gangneux F., Pépino Y., Belaz S., Damiani C., Guéguen P. A mis-leading false negative result of Pneumocystis real-time PCR assay due to a rare punctual mutation: a French multicenter study. Med Mycol. 2017;55:180–184. doi: 10.1093/mmy/myw051. [DOI] [PubMed] [Google Scholar]
- 90.Fréalle E., Gantois N., Aliouat-Denis C.M., Leroy S., Zawadzki C., Perkhofer S. Comparison of different blood compartments for the detection of circulating DNA using a rat model of Pneumocystis pneumonia. Med Mycol. 2015;53:754–759. doi: 10.1093/mmy/myv050. [DOI] [PubMed] [Google Scholar]
- 91.van Halsema C., Johnson L., Baxter J., Douthwaite S., Clowes Y., Guiver M. Short communication: diagnosis of Pneumocystis jirovecii Pneumonia by detection of DNA in blood and oropharyngeal wash, compared with sputum. AIDS Res Hum Retroviruses. 2016;32:463–466. doi: 10.1089/AID.2015.0213. [DOI] [PubMed] [Google Scholar]
- 92.Sherif R., Segal B.H. Pulmonary aspergillosis: clinical presentation, diagnostic tests, management and complications. Curr Opin Pulm Med. 2010;16:242–250. doi: 10.1097/MCP.0b013e328337d6de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arvanitis M., Ziakas P.D., Zacharioudakis I.M., Zervou F.N., Caliendo A.M., Mylonakis E. PCR in diagnosis of invasive aspergillosis: a meta-analysis of diagnostic performance. J Clin Microbiol. 2014;52:3731–3742. doi: 10.1128/JCM.01365-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Desoubeaux G., Bailly É., Chandenier J. Diagnosis of invasive pulmonary aspergillosis: updates and recommendations. Med Mal Infect. 2014;44:89–101. doi: 10.1016/j.medmal.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 95.Powers-Fletcher M.V., Hanson K.E. Molecular diagnostic testing for Aspergillus. J Clin Microbiol. 2016;54:2655–2660. doi: 10.1128/JCM.00818-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Imbert S., Gauthier L., Joly I., Brossas J.Y., Uzunov M., Touafek F. Aspergillus PCR in serum for the diagnosis, follow-up and prognosis of invasive aspergillosis in neutropenic and nonneutropenic patients. Clin Microbiol Infect. 2016;22:562. doi: 10.1016/j.cmi.2016.01.027. [e1-e8] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ascioglu S., Rex J.H., de Pauw B., Bennett J.E., Bille J., Crokaert F. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin Infect Dis. 2002;34:7–14. doi: 10.1086/323335. [DOI] [PubMed] [Google Scholar]
- 98.White P.L., Wingard J.R., Bretagne S., Löffler J., Patterson T.F., Slavin M.A. Aspergillus polymerase chain reaction: systematic review of evidence for clinical use in comparison with antigen testing. Clin Infect Dis. 2015;61:1293–1303. doi: 10.1093/cid/civ507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mengoli C., Cruciani M., Barnes R.A., Loeffler J., Donnelly J.P. Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect Dis. 2009;9:89–96. doi: 10.1016/S1473-3099(09)70019-2. [DOI] [PubMed] [Google Scholar]
- 100.Springer J., White P.L., Hamilton S., Michel D., Barnes R.A., Einsele H. Comparison of performance characteristics of Aspergillus PCR in testing a range of blood-based samples in accordance with international methodological recommendations. J Clin Microbiol. 2016;54:705–711. doi: 10.1128/JCM.02814-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tuon F.F. A systematic literature review on the diagnosis of invasive aspergillosis using polymerase chain reaction (PCR) from bronchoalveolar lavage clinical samples. Rev Iberoam Micol. 2007;24:89–94. [PubMed] [Google Scholar]
- 102.Verweij P.E., Chowdhary A., Melchers W.J.G., Meis J.F. Azole resistance in Aspergillus fumigatus: can we retain the clinical use of mold-active antifungal azoles? Clin Infect Dis. 2016;62:362–368. doi: 10.1093/cid/civ885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alvarez-Moreno C., Lavergne R.-A., Hagen F., Morio F., Meis J.F., Le Pape P. Azole-resistant Aspergillus fumigatus harboring TR34/L98H, TR46/Y121F/T289A and TR53 mutations related to flower fields in Colombia. Sci Rep. 2017;7:45631. doi: 10.1038/srep45631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wiederhold N.P., Gil V.G., Gutierrez F., Lindner J.R., Albataineh M.T., McCarthy D.I. First detection of TR34 L98H and TR46 Y121F T289A Cyp51 mutations in Aspergillus fumigatus isolates in the United States. J Clin Microbiol. 2016;54:168–171. doi: 10.1128/JCM.02478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.White P.L., Posso R.B., Barnes R.A. Analytical and clinical evaluation of the pathonostics aspergenius assay for detection of invasive aspergillosis and resistance to azole antifungal drugs directly from plasma samples. J Clin Microbiol. 2017;55:2356–2366. doi: 10.1128/JCM.00411-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chong G.M., van der Beek M.T., von dem Borne P.A., Boelens J., Steel E., Kampinga G.A. PCR-based detection of Aspergillus fumigatus Cyp51A mutations on bronchoalveolar lavage: a multicentre validation of the AsperGenius assay® in 201 patients with haematological disease suspected for invasive aspergillosis. J Antimicrob Chemother. 2016;71:3528–3535. doi: 10.1093/jac/dkw323. [DOI] [PubMed] [Google Scholar]
- 107.Chong G.L., van de Sande W.W., Dingemans G.J., Gaajetaan G.R., Vonk A.G., Hayette M.P. Validation of a new Aspergillus real-time PCR assay for direct detection of Aspergillus and azole resistance of Aspergillus fumigatus on bronchoalveolar lavage fluid. J Clin Microbiol. 2015;53:868–874. doi: 10.1128/JCM.03216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dannaoui E., Gabriel F., Gaboyard M., Lagardere G., Audebert L., Quesne G. Molecular diagnosis of invasive aspergillosis and detection of azole resistance by a newly commercialized PCR Kit. J Clin Microbiol. 2017;55:3210–3218. doi: 10.1128/JCM.01032-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Wesenbeeck L., Meeuws H., Van Immerseel A., Ispas G., Schmidt K., Houspie L. Comparison of the FilmArray RP, Verigene RV+, and Prodesse ProFLU+/FAST+ multiplex platforms for detection of influenza viruses in clinical samples from the 2011–2012 influenza season in Belgium. J Clin Microbiol. 2013;51:2977–2985. doi: 10.1128/JCM.00911-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gray J., Coupland L.J. The increasing application of multiplex nucleic acid detection tests to the diagnosis of syndromic infections. Epidemiol Infect. 2014;142:1–11. doi: 10.1017/S0950268813002367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lebovitz E.E., Burbelo P.D. Commercial multiplex technologies for the microbiological diagnosis of sepsis. Mol Diagn Ther. 2013;17:221–231. doi: 10.1007/s40291-013-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shin S.Y., Kwon K.C., Park J.W., Kim J.M., Shin S.Y., Koo S.H. Evaluation of the Seeplex® meningitis ACE detection kit for the detection of 12 common bacterial and viral pathogens of acute meningitis. Ann Lab Med. 2012;32:44–49. doi: 10.3343/alm.2012.32.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rhein J., Bahr N.C., Hemmert A.C., Cloud J.L., Bellamkonda S., Oswald C. Diagnostic performance of a multiplex PCR assay for meningitis in an HIV-infected population in Uganda. Diagn Microbiol Infect Dis. 2016;84:268–273. doi: 10.1016/j.diagmicrobio.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Samra Z., Rosenberg S., Madar-Shapiro L. Direct simultaneous detection of 6 sexually transmitted pathogens from clinical specimens by multiplex polymerase chain reaction and auto-capillary electrophoresis. Diagn Microbiol Infect Dis. 2011;70:17–21. doi: 10.1016/j.diagmicrobio.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 115.Choe H.S., Lee D.S., Lee S.J., Hong S.H., Park D.C., Lee M.K. Performance of Anyplex™ II multiplex real-time PCR for the diagnosis of seven sexually transmitted infections: comparison with currently available methods. Int J Infect Dis. 2013;17:e1134–e1140. doi: 10.1016/j.ijid.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 116.Poritz M.A., Blaschke A.J., Byington C.L., Meyers L., Nilsson K., Jones D.E. FilmArray, an automated nested multiplex PCR system for multipathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6:e26047. doi: 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Altun O., Almuhayawi M., Ullberg M., Ozenci V. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol. 2013;51:4130–4136. doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chang S.S., Hsieh W.H., Liu T.S., Lee S.H., Wang C.H., Chou H.C. system for rapid detection of pathogens in patients with presumed sepsis – a systemic review and meta-analysis. PLoS One. 2013;8:e62323. doi: 10.1371/journal.pone.0062323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ibarra O.J., Valdez T.P., Mendez V.E., Rojas L.A., Flores L.G., Bocanegra C.A. Evaluation of the Light-Cycler® SeptiFast test in newborns with suspicion of nosocomial sepsis. Iran J Pediatr. 2015;25:e253. doi: 10.5812/ijp.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tissari P., Zumla A., Tarkka E., Mero S., Savolainen L., Vaara M. Accurate and rapid identification of bacterial species from positive blood cultures with a DNA-based microarray platform: an observational study. Lancet. 2010;375:224–230. doi: 10.1016/S0140-6736(09)61569-5. [DOI] [PubMed] [Google Scholar]
- 121.Aittakorpi A., Kuusela P., Koukila-Kähkölä P., Vaara M., Petrou M., Gant V. Accurate and rapid identification of Candida spp. frequently associated with fungemia by using PCR and the microarray-based prove-it sepsis assay. J Clin Microbiol. 2012;50:3635–3640. doi: 10.1128/JCM.01461-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Loonen A.J., de Jager C.P., Tosserams J., Kusters R., Hilbink M., Wever P.C. Biomarkers and molecular analysis to improve bloodstream infection diagnostics in an emergency care unit. PLoS One. 2014;9:e87315. doi: 10.1371/journal.pone.0087315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ljungström L., Enroth H., Claesson B.E., Ovemyr I., Karlsson J., Fröberg B. Clinical evaluation of commercial nucleic acid amplification tests in patients with suspected sepsis. BMC Infect Dis. 2015;15:199. doi: 10.1186/s12879-015-0938-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fitting C., Parlato M., Adib-Conquy M., Memain N., Philippart F., Misset B. DNAemia detection by multiplex PCR and biomarkers for infection in systemic inflammatory response syndrome patients. PLoS One. 2012;7:e38916. doi: 10.1371/journal.pone.0038916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Leber A.L., Everhart K., Balada-Llasat J.M., Cullison J., Daly J., Holt S. Multicenter evaluation of biofire filmarray meningitis/encephalitis panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol. 2016;54:2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Salez N., Vabret A., Leruez-Ville M., Andreoletti L., Carrat F., Renois F. Evaluation of four commercial multiplex molecular tests for the diagnosis of acute respiratory infections. PLoS One. 2015;10:e0130378. doi: 10.1371/journal.pone.0130378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bierbaum S., Forster J., Berner R., Rücker G., Rohde G., Neumann-Haefelin D. Detection of respiratory viruses using a multiplex real-time PCR assay in Germany, 2009/10. Arch Virol. 2014;159:669–676. doi: 10.1007/s00705-013-1876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gago S., Esteban C., Valero C., Zaragoza Ó., Puig de la Bellacasa J., Buitrago M.J. A Multiplex real-time PCR assay for identification of Pneumocystis jirovecii, Histoplasma capsulatum and Cryptococcus neoformans/Cryptococcus gattii in samples from AIDS patients with opportunistic pneumonia. J Clin Microbiol. 2014;52:1168–1176. doi: 10.1128/JCM.02895-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Buchana B.W., Riebe K.M., Ledeboera N.A. Comparison of the MALDI biotyper system using sepsityper specimen processing to routine microbiological methods for identification of bacteria from positive blood culture bottles. JCM. 2012;50:346–352. doi: 10.1128/JCM.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wootton S.H., Aguilera E., Salazar L., Hemmert A.C., Hasbun R. Enhancing pathogen identification in patients with meningitis and a negative Gram stain using the BioFire FilmArray® meningitis/encephalitis panel. Ann Clin Microbiol Antimicrob. 2016;15:26. doi: 10.1186/s12941-016-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mylonakis E., Clancy C.J., Ostrosky-Zeichner L., Garey K.W., Alangaden G.J., Vazquez J.A. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis. 2015;60:892–899. doi: 10.1093/cid/ciu959. [DOI] [PubMed] [Google Scholar]
- 132.Zhao Y., Nagasaki Y., Kordalewska M., Press E.G., Shields R.K., Nguyen M.H. Rapid detection of FKS-associated echinocandin resistance in Candida glabrata. Antimicrob Agents Chemother. 2016;60:6573–6577. doi: 10.1128/AAC.01574-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Alexander B.D., Johnson M.D., Pfeiffer C.D., Jiménez-Ortigosa C., Catania J., Booker R. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis. 2013;56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mery A., Sendid B., François N., Cornu M., Poissy J., Guerardel Y. Application of mass spectrometry technology to early diagnosis of invasive fungal infections. J Clin Microbiol. 2016;54:2786–2797. doi: 10.1128/JCM.01655-16. [DOI] [PMC free article] [PubMed] [Google Scholar]


