Abstract
All members of the herpesviridae contain within their large tegument protein a cysteine protease module that displays deubiquitinating activity. We report the crystal structure of the cysteine protease domain of murine cytomegalovirus M48 (M48USP) in a complex with a ubiquitin (Ub)-based suicide substrate. M48USP adopts a papain-like fold, with the active-site cysteine forming a thioether linkage to the suicide substrate. The Ub core participates in an extensive hydrophobic interaction with an exposed β hairpin loop of M48USP. This Ub binding mode contributes to Ub specificity and is distinct from that observed in other deubiquitinating enzymes. Both the arrangement of active-site residues and the architecture of the interface with Ub lead us to classify this domain as the founding member of a previously unknown class of deubiquitinating enzymes.
Keywords: PROTEINS
Introduction
The covalent modification of target proteins by ubiquitin (Ub), a small 76 residue protein, occurs in various linkage types and can produce many different outcomes. Processes as diverse as cell-cycle regulation, receptor trafficking, signal transduction, epigenetic regulation of gene expression, DNA repair, regulated proteolysis, and destruction of the target protein by the proteasome are under the control of Ub addition (Glickman and Ciechanover, 2002, Schnell and Hicke, 2003). Moreover, the processing of pathogen-derived proteins by the Ub/proteasome pathway is a prerequisite for their display to the immune system to initiate the host's defense (York and Rock, 1996).
Ubiquitination, like many other posttranslational modifications, is a reversible process, allowing regulation of the balance between the unmodified and modified states of the target protein. Different collections of enzymes are responsible for driving either the forward or the reverse reaction, providing dynamic control of the modification state. A significant portion of the genome is invested in the process of Ub addition and removal: there are ∼500 Ub ligases in the human genome catalyzing the forward reaction in conjunction with one Ub-activating (E1) and several Ub-conjugating enzymes (E2s), whereas ∼100 deubiquitinating enzymes (DUBs) are currently predicted through bioinformatics (Nijman et al., 2005).
All viral pathogens and many intracellular bacteria rely critically on the host synthetic machinery for their propagation. Accordingly, these pathogens have learned to exploit or subvert to their own advantage the cellular enzymatic machinery that could otherwise be deployed against them. A number of these pathogens have thus evolved an elaborate molecular armory that includes pathogen-encoded ubiquitinating and deubiquitinating activities, as well as proteins capable of manipulating/hijacking the host's Ub machinery. The known functions of these pathogen-encoded activities are quite diverse. Regulation of the cell cycle, activation or suppression of the NF-κB pathway, destruction of the class I MHC glycoprotein, and regulation of intracellular trafficking are but a few examples (Gao and Luo [2006] and references cited therein).
How can we detect Ub-related activities that relate to host-pathogen interactions? A functional proteomic approach—where active DUBs are targeted selectively in crude cell extracts with epitope-tagged, Ub-based suicide substrates, retrieved by immunoprecipitation, and identified by mass spectrometry—has proved particularly useful in identifying new deubiquitinating enzymes (Borodovsky et al., 2002, Ovaa et al., 2004). Using this approach, we identified a unique deubiquitinating activity present in herpes simplex virus-1 (HSV-1/α subfamily)-infected cells (Kattenhorn et al., 2005). The DUB activity mapped to the N-terminal portion of UL36, a large (3156 residues) and presumably multifunctional protein of the α-herpesvirus HSV-1. UL36 is a constituent of the amorphous proteinaceous layer located between the nucleocapsid and the envelope of the herpesvirus particle, referred to as tegument. All known herpesviruses sequenced to date contain homologs of this large tegument protein. The putative key catalytic residues of the protease domain found in UL36 are conserved throughout all herpesviridae subfamilies: α, β, and γ (Kattenhorn et al., 2005) (Schlieker et al., 2005). Deubiquitinating activity was confirmed for the UL36 homologs of murine cytomegalovirus (MCMV/β subfamily), human cytomegalovirus (HCMV/β subfamily), and Epstein-Barr virus (EBV/γ subfamily), M48, UL48, and BPLF1, respectively (Schlieker et al., 2005, Wang et al., 2006). Although the in vivo function of this activity is currently unknown, its strict conservation through all herpesvirus subfamilies suggests an important role in their biology. To gain insights into the structural and functional properties of this activity, we set out to determine the structure of this enzyme.
We report the crystal structure of the Ub-specific protease (USP) domain of MCMV (M48USP) in a covalent complex with a Ub-based suicide inhibitor, Ub vinylmethylester (UbVME). A comparison of the M48USP topology to eukaryotic DUBs and other proteases established that M48USP is the founding member of a structurally distinct class of deubiquitinating enzymes, which we designate herpesvirus tegument USPs (htUSPs). The presence of this family of cysteine proteases has implications for its possible role as a target to manipulate the interaction of MCMV and by inference, other herpesviruses, with their host.
Results and Discussion
Construct Design and Enzymatic Properties of M48USP
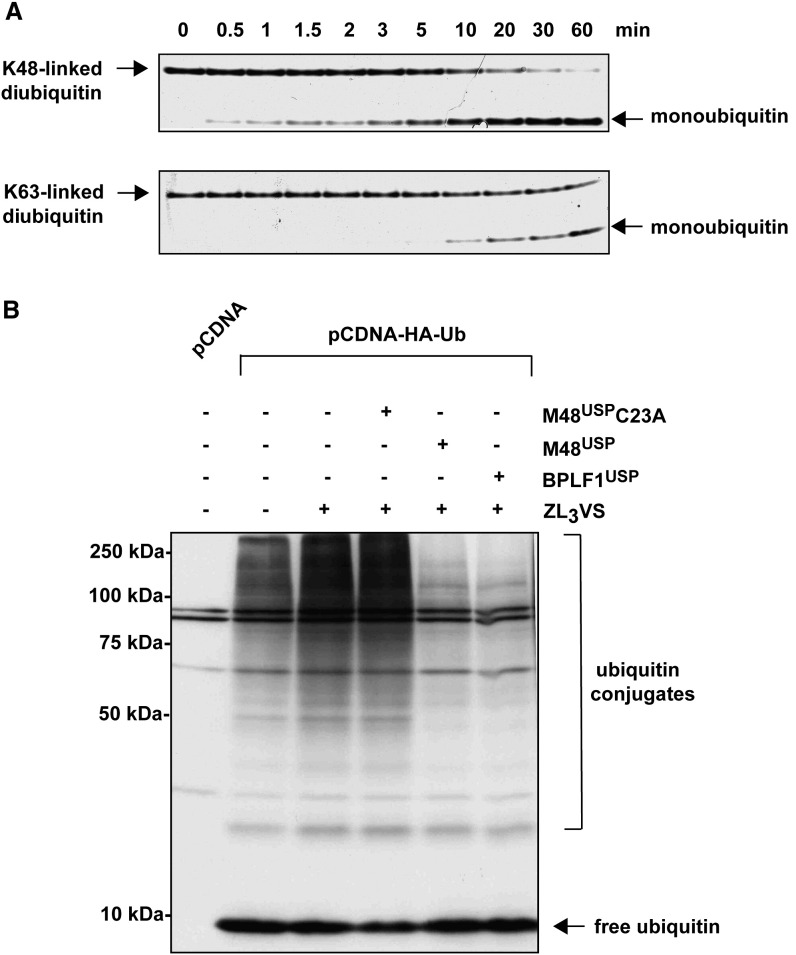
A 205 residue module in the N-terminal portion of MCMV M48 and EBV BPLF1 is necessary and sufficient for deubiquitinating activity and Ub specificity (Schlieker et al., 2005). The proposed catalytic triad and oxyanion residues of this cysteine protease module are conserved in all herpesviruses. The tegument protease domains otherwise display only moderate sequence conservation (≤12% identity between representatives of α, β, and γ subfamilies) yet share a high degree of structural similarity according to secondary structure predictions (Schlieker et al., 2005). Neither BLAST searches (Altschul et al., 1997) nor threading programs that employ sequence alignment and protein-fold recognition algorithms (McGuffin et al., 2000) uncovered homologs other than herpesvirus tegument proteins (data not shown). This suggested that htUSPs have structural properties that are distinct from known DUBs and other cysteine proteases (Kattenhorn et al., 2005, Sulea et al., 2006). Accordingly, UL36USP and its homologs represent an independent family (C76) in the CA clan of proteases in the MEROPS peptidase database (Rawlings et al., 2004). To explore the structural and mechanistic properties of htUSPs, we expressed an N-terminal fragment encompassing residues 1–236 of the MCMV M48 tegument protein (M48USP), which likely comprises the entire USP domain of M48, because it is followed by an ∼70 residue region that is likely unstructured according to secondary structure predictions. To confirm that this domain is a functional deubiquitinating enzyme, we first tested the ability of M48USP to hydrolyze the fluorogenic substrate Ub C-terminal 7-amido-4-methylcoumarin (UbAMC). UbAMC hydrolysis followed Michaelis-Menten kinetics, with kcat / KM = 9.17 × 106 M−1 s−1 (data not shown). To determine whether M48USP can cleave true isopeptide-linked Ub chains, we examined its ability to attack K48-linked or K63-linked diUb by using a 500-fold excess of substrate over enzyme. Both adducts were converted to free monomeric Ub, albeit with ∼5-fold slower kinetics for K63-linked compared to K48-linked diUb ( Figure 1A).
Figure 1.
M48USP Cleaves Ub Conjugates with High Efficacy
(A) Hydrolysis of isopeptide-linked diUb by M48USP. K48- or K63-linked diUb (5 μM) was incubated with M48USP (10 nM) at 37°C, stopped at the indicated time points by addition of 2× tris/tricine sample buffer, subjected to a tris/tricine gel, and stained by colloidal blue. A densiometric quantification for these time courses (see Experimental Procedures) yields rate constants of 0.102 and 0.021/min for K48- and K63-linked diUb, respectively.
(B) M48USP cleaves cellular Ub conjugates to yield free Ub. 293T cells were transiently transfected with HA-Ub or pcDNA3.1+ as control. To enrich for polyubiquitinated species, cells were exposed to the proteasome inhibitor ZL3VS 1 hr prior to cell lysis where indicated. Soluble cell extracts were prepared 24 hr posttransfection, incubated for 1 hr in the absence or presence of enzymes, and subjected to SDS-PAGE and immunoblotting. HA-Ub was detected by using 12CA5 (anti-HA) as primary antibody.
Can M48USP also cleave Ub conjugates found in living cells? To address this issue, 293T cells were transiently transfected with HA-tagged Ub and treated with a proteasome inhibitor, ZL3VS (Bogyo et al., 1997), to allow accumulation of (poly)-ubiquitinated species. Cell lysates were prepared and incubated in the absence and presence of M48USP or its catalytically inactive counterpart, M48USP-C23A (Schlieker et al., 2005), and subjected to SDS-PAGE and immunoblotting (Figure 1B). Both free Ub and Ub conjugates were readily detectable, the latter being more prominent in samples derived from ZL3VS-treated cells. Upon exposure to nanomolar concentrations of M48USP, the majority of cellular Ub conjugates were deubiquitinated, as judged by the near-complete elimination of high molecular weight, anti-HA-reactive material and a corresponding increase in free Ub. For comparison, we also tested the homologous EBV-derived protease BPLF1USP and observed similar deubiquitinating activity (Figure 1B), confirming that the activity exerted on poly-Ub adducts is conserved among herpesviruses.
Structure Determination and Overall Structure of M48USP
Having demonstrated the activity of M48USP, we initiated crystallization trials for M48USP and BPLF1USP, neither of which yielded diffraction-quality crystals. We crystallized M48USP in a covalent complex with UbVME, a suicide inhibitor that forms a thioether bond with the active-site cysteine of DUBs (Borodovsky et al., 2002, Misaghi et al., 2005). The structure was determined by using crystals of SeMet-labeled M48USP covalently bound to UbVME in a single-wavelength anomalous dispersion (SAD) experiment. Continuous electron density was observed for residues 1–232 of the two M48USP molecules and all residues of the two UbVME molecules in the asymmetric unit. The final model was refined to 1.8 Å resolution to an R factor of 15.8% (Rfree = 21.4%) (see Table 1 for data collection and refinement statistics). Because the Cα atoms of both complexes can be superimposed with 0.3 Å root-mean-square deviation (rmsd), we limit our discussion to a single complex of M48USP with UbVME (chains A and C).
Table 1.
Crystallographic Data and Refinement Statistics
| Data Statistics | |
|---|---|
| Space group | P1 |
| Beamline | ID 24.2 (APS) |
| Wavelength (Å) | 0.9797 (Se Peak) |
| Unit cell parameters | |
| a, b, c (Å) | 40.9, 57.3, 67.3 |
| α, β, γ (°) | 73.4, 85.4, 88.5 |
| Resolution range (Å)a | 50.0–1.80 (1.86–1.80) |
| Number of observations | 216,924 (12,199) |
| Number of unique reflections | 52,323 (5,074) |
| Completeness (%) | 97.4 (84.2) |
| Redundancy | 4.2 (2.4) |
| 〈I/σ(I)〉 | 8.3 (3.5) |
| Rsym (%)b | 9.4 (26.7) |
| Refinement Statistics | |
| Resolution range (Å) | 50.0–1.80 (1.83–1.80) |
| Reflections in work set | 51,609 (3,690) |
| Reflections in Rfree set | 1,052 (81) |
| Residues (atoms) | 712 (11,555) |
| Water molecules | 651 |
| Rwork | 15.8 (18.0) |
| Rfree | 21.4 (29.0) |
| Rmsdsc | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.44 |
Values in parentheses refer to the outer resolution shell.
Rmerge = ∑h∑i|I(h) − I(h)i| / ∑h∑iI(h)I, where h is a unique reflection index, I(h)I is the intensity of symmetry-related reflections, and I(h) is the mean intensity.
Computed with PROCHECK.
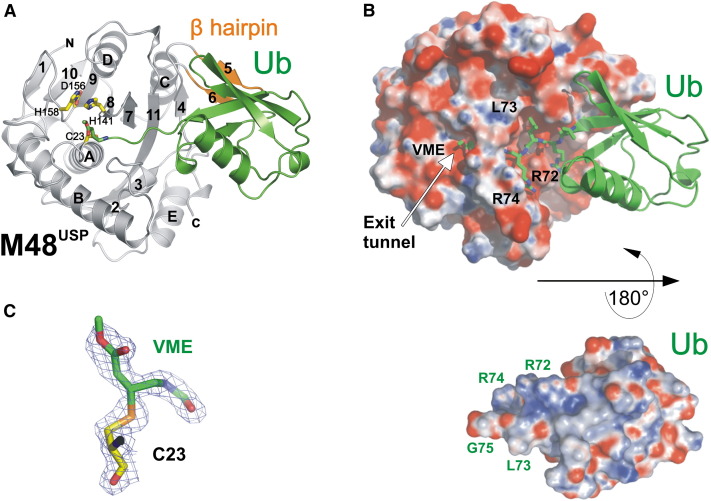
The M48USP structure is organized around a bifurcated antiparallel β sheet flanked on either side by α helices to form an α-β-α sandwich structure ( Figure 2A). The extended C terminus of UbVME lies in the deep catalytic cleft between α helix A and the β sheet of M48USP, with the active-site cysteine in thioether linkage to the former VME moiety (Figures 2B and 2C). As detailed below, the M48USP fold, although it has similarities to several cysteine proteases, is distinct from all other known DUB structures.
Figure 2.
Fold and Structure of M48USP-UbVME
(A) Ribbon representation of the M48USP structure (gray with the β hairpin in orange) in complex with UbVME (green). The secondary structure elements are labeled, and the side chains of catalytic triad residues are shown in yellow.
(B) Electrostatic surface potential representation of M48USP with bound Ub shown in a ribbon representation (top). Below, Ub, in an electrostatic surface potential representation, was rotated 180° to show the charge distribution on the face forming the interface. Note the charge complementarity between the M48USP acidic cleft and the positively charged Ub C terminus.
(C) The final 2Fo − Fc electron density map contoured at 1.3 σ indicates a covalent bond between the catalytic C23 and the Cβ atom of the former vinylmethylester moiety at the Ub C terminus.
Charge and Surface Complementarity Yield an Intimate M48USP-Ub Interface
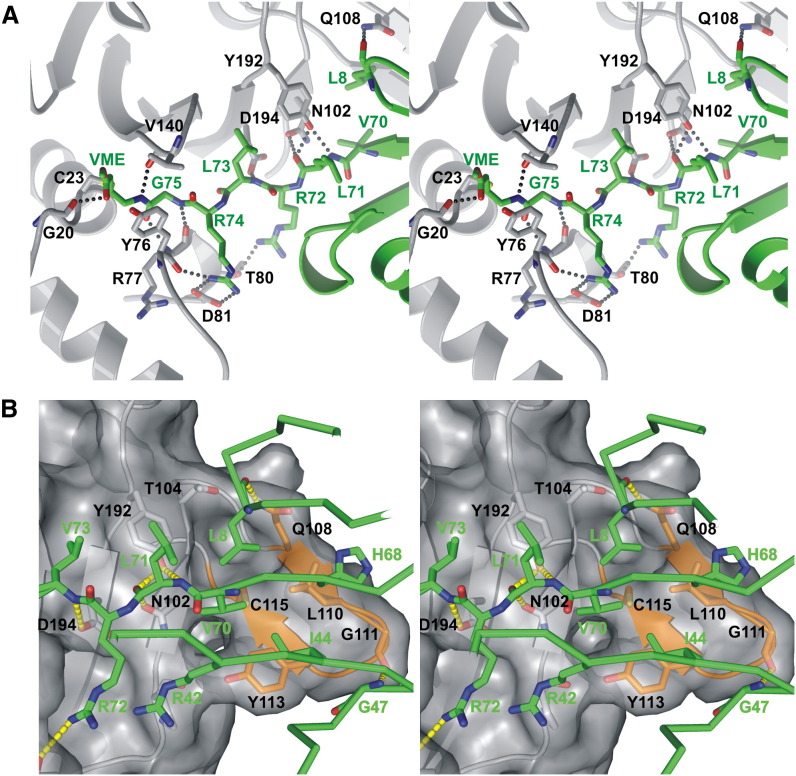
UbVME undergoes only minimal conformational changes upon binding to M48USP, as reflected by 0.33 Å rmsd over all Cα atoms when free Ub (Vijay-Kumar et al., 1987) and M48USP-bound UbVME are superimposed. Electron density indicates a 1.9 Å covalent bond between the M48USP C23 Sγ atom and the Cβ-carbon atom of the former VME moiety that replaces the C-terminal G76 of wild-type Ub (Figure 2C), reminiscent of the covalent complex between UbVME and UCH-L3 (Misaghi et al., 2005). Complex formation buries 1840 Å2 of solvent-accessible surface area, corresponding to 816 Å2 (7%) and 1024 Å2 (21%) of the total solvent-accessible surface area of free M48USP and UbVME, respectively. The size of this interface is comparable to the corresponding ones in UCH enzymes, e.g., 2300 A2 in UCH-L3 (Misaghi et al., 2005), but smaller than in HAUSP (3600 Å2) (Hu et al., 2002) and other USPs. Electrostatic surface potential calculations show a high degree of charge complementarity: positively charged surface regions of the extended Ub C terminus (residues 71–75) are deeply buried in the largely acidic cleft of M48USP (Figure 2B). The backbone of the Ub C terminus adopts an extended β conformation and is extensively coordinated by seven salt bridges and hydrogen bonds to residues lining the M48USP cleft, conferring sequence specificity for the Ub C terminus ( Figure 3A). In addition, a hydrophobic pocket lined by M48USP V140 and Y192 accommodates the Ub L73 side chain (Figures 2B and 3A). Binding of the C-terminal Ub segment in an extended conformation in the active-site cleft is a common feature of DUBs, as observed in the crystal structures of Ub derivatives in complex with UCH-L3, HAUSP, USP2, and USP14 (Hu et al., 2002, Hu et al., 2005, Misaghi et al., 2004, Renatus et al., 2006).
Figure 3.
Interactions between UbVME and M48USP
(A) Stereo view of the extended C terminus and attached VME moiety of Ub in the M48USP active-site cleft. M48USP and UbVME are in gray and green, respectively. Nitrogen atoms are shown in blue, and oxygen atoms in red. Dashed lines indicate hydrogen bonds. Note that the Ub C terminus features an extended β conformation and is extensively coordinated by several hydrogen bonds to M48USP residues. V140 and Y76 of M48USP are in van der Waals contact and form a canopy over the active site.
(B) Stereo view of the interactions between the Ub core (green) and M48USP (gray) or its β hairpin (orange). The gray transparent M48USP surface highlights the shape complementarity of the interface, which is mainly lined by hydrophobic residues.
Apart from contacts with the C-terminal Ub stretch, there are a number of additional interactions between Ub and M48USP. Most notably, an exposed β hairpin, formed by M48USP β strands 5 and 6 and located in line with the central eight-stranded β sheet, contacts the Ub core domain (Figure 2A). The hydrophobic patch formed by Ub residues L8, R42, I44, H68, and V70 contacts the β hairpin residues L110, Y113, and C115 (Figure 3B). In addition, several hydrogen bonds are formed between backbone atoms of the two proteins (Figure 3B).
Members of the UCH family like UCH-L3 and Yuh1 lack an additional Ub-interacting subdomain beyond the active-site cleft that accommodates the C-terminal Ub residues. Other important contacts are formed to two loop regions (residues 8–9 and 39–42) neighboring this C-terminal Ub stretch (Johnston et al., 1999, Misaghi et al., 2005). This binding pattern allows significant conformational freedom of Ub when bound to UCH-L3 (Misaghi et al., 2005). In contrast, extensive interactions mediated by the M48USP β hairpin likely restrain movement of Ub relative to the M48 protease domain, as suggested from the two structurally very similar M48USP-Ub complexes in the asymmetric unit (see above).
Overall, the Ub-M48USP interaction is very intimate, as indicated by shape complementarity (Sc) calculations, which yield a high Sc of 0.77 compared to an average of 0.70–0.74 for oligomeric interfaces (Lawrence and Colman, 1993). The surface complementarity also results in exclusion of water molecules from the interface, which contrasts with DUBs of the USP family such as HAUSP or USP2 (Hu et al., 2002, Renatus et al., 2006). In both USP-Ub complexes, ∼20 water molecules are coordinated in the interface between the USP finger domain and the Ub core domain, and Ub specificity is therefore dictated by several water-mediated contacts (Hu et al., 2002, Renatus et al., 2006).
Active-Site Conformation
The active site that surrounds the covalent bond between the catalytic C23 and the former VME moiety of UbVME is canopied by the Y76 and V140 side chains forming a van der Waals interaction above the active site (Figure 3A). Consequently, the entry of ubiquitinated substrates into the active site would require a small structural rearrangement around either of these two residues. This is analogous to the USP-Ub structures (Hu et al., 2002, Hu et al., 2005, Renatus et al., 2006) but stands in contrast to the UCH family members, which have a long crossover loop that covers the active site (Das et al., 2006, Johnston et al., 1997, Misaghi et al., 2005). The exit tunnel (Figure 2B) is occupied in our structure by the VME moiety and in the case of bound natural substrates would readily accommodate the isopeptide-linked lysine side chain of the target protein.
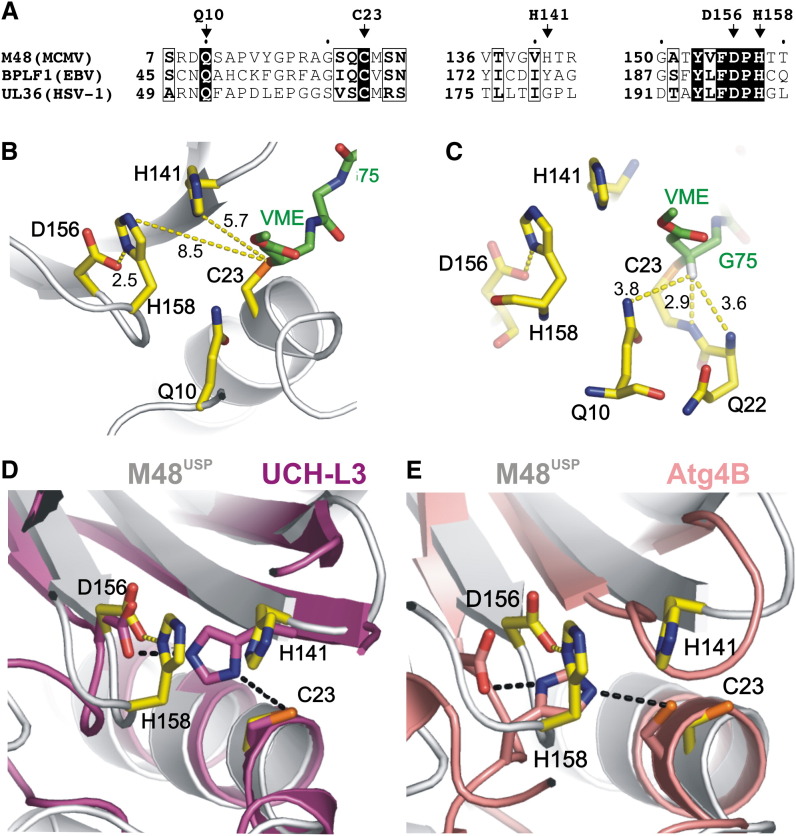
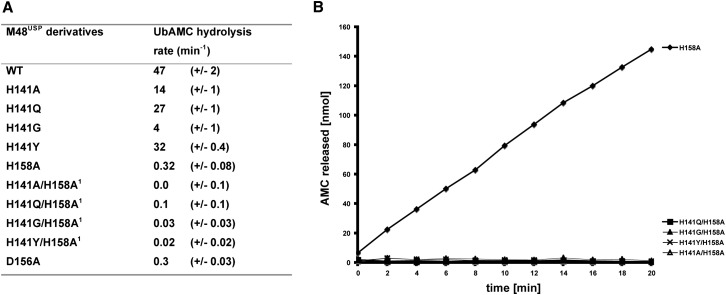
In light of the strict conservation of Q10, C23, D156, and H158 throughout all herpesviridae subfamilies ( Figure 4A and Figure S1A, in the Supplemental Data available with this article online; numbering according to M48 sequence), we proposed that these residues are involved in the formation of the oxyanion hole (Q10) and the aspartic acid-histidine-cysteine catalytic triad (Kattenhorn et al., 2005, Schlieker et al., 2005). This prediction is unambiguously confirmed for C23, which forms a thioether linkage to the UbVME suicide inhibitor (Figure 2C). The addition of C23 to the former VME moiety imposes an sp3 configuration on the VME β carbon atom, thus resembling the tetrahedral reaction intermediate during catalysis, but with the oxyanion replaced by a hydrogen atom. This hydrogen is located in a pocket that is lined by the conserved Q10 and corresponds to the oxyanion hole observed in other cysteine proteases (Figure 4C). Thus, the transition-state oxyanion is likely coordinated by the Q10 side chain and the backbone NH groups of Q22 and the catalytically active C23 (Figure 4C). Furthermore, the catalytic aspartic acid residue can be assigned to D156 due to its location and distance to the catalytic C23 (Figure 4B). Consistently, an D156A substitution results in a 180-fold reduction of catalytic activity ( Figure 5A).
Figure 4.
Two M48USP Histidines Can Participate in the Catalytic Triad
(A) Sequence alignment of herpesvirus-encoded USPs M48 (MCMV), UL36 (HSV-1), and BPLF1 (EBV). Conserved residues are boxed, and conserved catalytic triad residues (C23, D156, and H158) and Q10 of the oxyanion hole residues are indicated and shown in (B) as yellow sticks. Furthermore, H141, which is not conserved but might participate in catalysis, is also shown. The covalently bound UbVME is represented by green sticks. Note that the catalytic triad is misaligned because both histidines are >5 Å from C23.
(C) The proposed oxyanion hole is occupied by the hydrogen atom of the β carbon atom of the former vinylmethylester moiety, thus resembling a catalytic intermediate except that the oxyanion is replaced by a hydrogen. The Q10 side chain and NH groups of C23 and Q22 are <4 Å from this hydrogen atom and are thus likely to coordinate the oxyanion during catalysis.
(D) Superposition of the active sites of M48USP (yellow) and UCH-L3 (pink). H141 corresponds to the location of the UCH-L3 catalytic histidine, and a conformational change of H141 may result in a similar functional catalytic triad alignment as observed for UCH-L3.
(E) Superposition of catalytic triad residues of M48USP (yellow) and human Atg4B (salmon), the only known cysteine protease structure that features a catalytic triad arrangement that aligns its catalytic histidine with M48USP H158. A different rotamer conformation for M48USP H158 may therefore yield an aligned catalytic triad.
Figure 5.
Effect of Active-Site Residue Mutations on M48USP Catalytic Activity
(A) Catalytic activities of M48USP and mutant derivatives (100 pM) were determined with UbAMC (100 nM) as substrate at subsaturating concentration ([UbAMC] < KM). Hydrolytic activities are given as the ratio of initial velocities (nM AMC released/min) and total enzyme concentration E (nM). Errors are the standard deviations calculated from three independent experiments.
(B) The mutants indicated by “1” in (A) have no catalytic activity, within experimental error. The corresponding mutants were therefore tested at 50-fold higher enzyme concentrations (5 nM) and saturating substrate concentrations (5 μM; [UbAMC] >> KM), further demonstrating that they are completely inactive.
Surprisingly, two histidine residues, H141 and H158, are found neighboring the catalytic C23. H158 is conserved throughout the herpesviridae and forms a hydrogen bond to the conserved D156 (Figure 4B) and for these reasons is the plausible candidate to act as catalytic base. However, both histidines are >5 Å from the catalytic C23, resulting in a misaligned catalytic triad (Figure 4B). A side-chain rearrangement is required to position either histidine within hydrogen bonding distance to C23 and D156, in order to form a functional proton relay system. Possible scenarios for either rearrangement can be derived from structurally related cysteine proteases. Figure 4D shows that in superimposed structures of M48USP and UCH-L3, C23, D156, and H141 correspond to UCH-L3 catalytic triad residues. Therefore, to form a functional catalytic triad, the M48USP H141 side chain may adopt a rotamer conformation observed for the corresponding UCH-L3 histidine, as is observed in the vast majority of cysteine proteases. On the other hand, the conserved M48USP H158 could instead participate in catalysis after adopting a conformation that corresponds to the unusual histidine alignment found in the catalytic triad of the structurally related human cysteine protease Atg4B (Figure 4E).
To address which histidine serves as catalytic base, we exchanged either or both histidine residues to alanine by site-directed mutagenesis. Enzymatic activity was assessed by measuring the hydrolysis of UbAMC. Both single substitutions remained enzymatically active, with a 3.5-fold and a 146-fold reduction of hydrolytic activity compared to WT enzyme for the H141A and H158A mutants, respectively (Figure 5A). In contrast, the double-mutant protein (H141A/H158A) completely lacked catalytic activity (Figure 5A). However, activity was partially restored in a concentration-dependent manner upon addition of imidazole (data not shown). We conclude that H158 is the principal catalytic histidine as judged by the more pronounced reduction of catalytic activity of the H158A mutant, consistent with H158 being conserved throughout the htUSPs. Nevertheless, the residual activity of H158A is still substantial at saturating substrate concentrations (kcat = 1.38/min, Figure 5B). To confirm that the residual catalytic activity observed for H158A indeed involves H141, we mutated H141 to glycine or tyrosine, the corresponding residues in HSV-1 and EBV, respectively (Figure 4A), or to glutamine—a conservative replacement—into a H158A background (Figure 5A). All three double mutants were catalytically inactive, even when tested at 50-fold-higher enzyme and saturating substrate concentrations (Figure 5B). Therefore, neither glycine, tyrosine, nor glutamine can replace H141 when H158 is changed to alanine. Because the H141Q and H141Y single mutants had 58% and 68% of WT activity, respectively (Figure 5A), the inactivity of the double mutants is unlikely to result from structural distortions, albeit we cannot formally exclude this possibility.
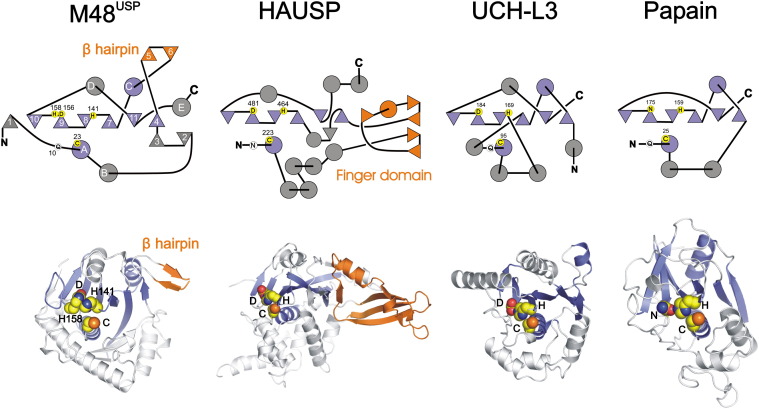
In summary, our structural and mutagenesis data suggest that the conserved H158 is the principal catalytic histidine, whereas H141, only present in M48USP and not in the other herpesvirus homologs, might participate in catalysis when H158 is absent. Interestingly, the principal catalytic triad histidine, H158, is located on a secondary structure element different from that in most cysteine proteases, whereas H141 is located on a position homologous to the catalytic triad histidine in papain ( Figure 6). The presence of two histidines in the M48USP active site suggests how the location within the tertiary structure of catalytic triad residues might change while the original fold of an enzyme is retained, providing a rationale for the catalytic-site diversity observed in cysteine proteases.
Figure 6.
Topological Comparison of the M48USP Structure to Other Cysteine Proteases and Deubiquitinating Enzymes
The topographies (top) and ribbon diagrams (bottom) of M48USP and representatives of the USP (HAUSP) and UCH family (UCH-L3) of DUBs, and papain, are shown. Circles and triangles represent α helices and β sheets, respectively. In the topology diagram of M48USP, secondary structure elements are numbered sequentially according to Figure 2A. Secondary structure elements conserved within the cysteine protease superfamily, exemplified by papain, are shown in blue. The exposed β hairpin of M48USP and the finger domain of HAUSP are orange; both interact with the Ub core. Catalytic triad residues are in yellow circles, whereas oxyanion hole residues are in white circles. Note that the sequential order of the more efficient M48USP catalytic triad (C23, D156, and H158) is different compared to other cysteine proteases and that M48USP shares more similarities to the papain topography than to the two DUB representatives.
Ub Specificity and Ubl Discrimination
A number of DUBs display promiscuity, targeting not only Ub but also some Ub-like modifiers (Ubls). For example, USP21 and UCH-L3 can cleave Ub and NEDD8 (Gong et al., 2000, Misaghi et al., 2005, Wada et al., 1998), whereas USP14 is bispecific for Ub and ISG15 (Hemelaar et al., 2004). In contrast, M48USP and its relatives in the herpesvirus family are highly specific for Ub, because NEDD8-, SUMO-, and ISG15-based suicide inhibitors fail to inhibit enzymatic activity, even when present in a 100-fold molar excess over the respective enzyme (Kattenhorn et al., 2005, Schlieker et al., 2005).
What is the structural basis for the observed specificity? In general, DUBs feature a largely acidic catalytic cleft that accommodates the complementary basic C-terminal Ub stretch (71LRLRGG76), implicating the cleft in substrate recognition and specificity (Hu et al., 2002). For instance, the C-terminal sequence of SUMO (72QEQTGG76) is clearly different and of opposite charge compared to the Ub C terminus, and consequently, SUMO cannot bind to M48USP. Other Ubls, however, terminate in a sequence similar or identical to Ub (Figure S1B). For example, the ISG15 C-terminal sequence LRLRGG is identical to that of Ub. Therefore, additional interactions outside the active-site cleft must determine substrate specificity. A superposition of the structure of free ISG15 (Narasimhan et al., 2005) on UbVME bound to M48USP indicates that the ISG15 C terminus would require only minor structural changes to enter the M48USP active site (data not shown). However, the M48USP β hairpin that tightly interacts with the Ub core domain (Figure 3A) lacks surface complementarity for the ISG15 core domain (data not shown), suggesting that several unfavorable contacts between the β hairpin of M48USP and ISG15 mainly account for the discrimination between Ub and ISG15.
As pointed out by Shen and coworkers, free NEDD8 exhibits a different C-terminal conformation compared to free Ub, due to the substitution of R72 for alanine in NEDD8 (Shen et al., 2005). This difference was suggested to be crucial for Ub discrimination of the NEDD8-specific protease NEDP1 (Shen et al., 2005) and, by analogy, could also explain why NEDD8 is not a M48USP substrate. In addition, NEDD8 A72 cannot form the hydrogen bond to M48USP T80 as observed for Ub R72 (Figure 3A). To test whether the NEDD8 A72 indeed determines M48USP specificity, we generated a NEDD8-based suicide inhibitor (NEDD8-VME) and its mutant derivative (NEDD8A72R-VME). In agreement with our previous data (Schlieker et al., 2005), we observed negligible covalent adduct formation of M48USP with NEDD8-VME. In contrast, NEDD8A72R-VME readily reacted with M48USP to yield a covalent adduct (Figure S2). This indicates that R72 is a key determinant for M48USP substrate specificity because the single substitution of A72 by arginine in NEDD8 renders it a M48USP substrate.
Taken together, we propose that M48USP has a bipartite substrate recognition mechanism: the acidic cleft recognizes the positively charged Ub C-terminal sequence through electrostatic interactions and specific contacts with side chains of the Ub C terminus, whereas the β hairpin serves to further discriminate Ubls that feature the identical C-terminal stretch, as exemplified by ISG15.
The M48USP Structure Defines a Unique Class of Deubiquitinating Enzymes
All deubiquitinating enzymes known to date are either cysteine- or metalloproteases. Cysteine protease DUBs can be divided into four classes: Ub C-terminal hydrolases (UCHs), Ub-specific proteases (USPs), ovarian tumor proteases (OTUs), and Machado-Joseph disease protein domain proteases (MJDs). According to current estimates, the human genome encodes 4 UCHs, 58 USPs, 14 OTUs, and 5 MJDs (Nijman et al., 2005).
How does the M48USP structure compare to other protease structures and to which DUB class does M48USP belong? A structural similarity search using the DALI algorithm (Holm and Sander, 1993) showed that ∼140 aa in the catalytic core of M48USP share moderate structural similarity to papain-like cysteine proteases (best Z score, 7.1) and only weak similarities to DUBs like USP6 from S. cerevisiae (1vjv; Z score, 4.2) and human UCH-L3 (1uch; Z score, 2.2) (Table S1). For comparison, UCH-L3/UCH-L1 and USP2/HAUSP pairwise comparisons yield Z scores of 33 and 27, respectively, whereas a pairwise comparison of two classes, e.g., UCH-L1/HAUSP as representatives from UCH and USP families, yields scores of ∼4. We conclude that M48USP is a member of the papain protease superfamily but is only distantly related to cellular DUBs of known structure. Structural database searches (DALI) or manual superimposition of active-site residues of M48USP and members of the OTU and MJD families did not detect any structural relationships (Table S1 and data not shown).
To investigate the structural relationship of M48USP to UCHs and USPs in detail, we compared the topology of M48USP to the closest structural relatives in these families: UCH-L3 and HAUSP (Figure 6 and Table S1). All three enzymes share secondary structure elements of the papain fold. Besides the different Ub binding mode of M48USP compared to UCHs and USPs (see above), several topology differences between these DUB representatives validate their classification in three different classes. First, the order of secondary structure elements in UCH-L3 is different from that of other cysteine protease DUBs (Figure 6). Second, UCH-L3 features a crossover loop that is disordered in the substrate-free structure and ordered above the active-site cleft when Ub is bound (Johnston et al., 1997, Misaghi et al., 2005). Third, HAUSP lacks an α helix corresponding to α helix C in M48USP and features an extensive “N-terminal” insertion dubbed the finger domain that forms mainly water-mediated contacts with the Ub (Ubl) core domain during catalysis (Hu et al., 2002). Fourth, M48USP is more closely related to the classic papain-like proteases than to other DUBs and features a unique β hairpin to recognize Ub and discriminate other Ubls (see above and Table S1). Fifth, sequence alignments (Figure 4A) and our mutational data suggest that H158 is the principal participant of the catalytic triad, yielding the primary sequence order cysteine → aspartic acid → histidine. This order has so far been observed only in Atg4B (Kumanomidou et al., 2006), whereas all other cysteine proteases of known structure, including DUBs, feature a cysteine → histidine → aspartic acid/asparagine catalytic triad in their primary sequence. In conclusion, M48USP shows distinct features in its tertiary fold and catalytic triad and a unique Ub binding β hairpin. In agreement with the listing of HSV UL36 as the founding member of a new cysteine protease family in the MEROPS database (Gao and Luo, 2006, Rawlings et al., 2004), we conclude that the M48USP structure defines a previously unknown class of cysteine protease DUBs and confirms the existence of a new cysteine protease family (C76), which we term herpesvirus tegument USPs (htUSPs).
Concluding Remarks
A number of viruses modulate the Ub system of the host, both by ubiquitination and deubiquitination. Recent examples include the paramyxovirus V protein, which hijacks the DSB1-Cul4A Ub ligase (Li et al., 2006), a deubiquitinating module of the USP family embedded in a nonstructural protein encoded by severe acute respiratory syndrome (SARS) coronavirus (Ratia et al., 2006), and an adenovirus protease endowed with deubiquitinating activity (Balakirev et al., 2002). Deubiquitinating activities were also found to be encoded by pathogenic intracellular bacteria, such as Yersinia spp. (Zhou et al., 2005) and Chlamydia trachomatis (Misaghi et al., 2006). For most of these examples, the precise biological role has yet to be defined.
We have meanwhile generated an MCMV mutant in which M48 is deprived of its USP activity via an in-frame deletion. This mutant is impaired in sustaining a productive infection in mice in vivo (L.M.K, C.S., H.L.P., unpublished data). With additional data in support of an essential role of the USP activity in vivo, htUSPs, and possibly other pathogen-encoded DUBs, deserve scrutiny from the perspective of drug design for treating pathogen infections. The respective structures will be important scaffolds to facilitate the design of specific inhibitors, all the more so if the pathogen-encoded DUBs are sufficiently different from cellular proteases, as exemplified by htUSPs.
Experimental Procedures
Kinetic Assays
The M48USP domain specifying aa 1–236 of the MCMV-encoded M48 tegument protein was expressed and purified as described previously (Schlieker et al., 2005). The corresponding domain of EBV, BPLFUSP, specifying aa 1–276, was purified accordingly. Ub C-terminal 7-amido-4-methylcoumarin (UbAMC) and diubiquitin (K63- and K48-linked) were purchased from Boston Biochem. Densiometric quantifications for diUb conversions were obtained with ImageJ and subsequently fitted to a single exponential decay in Kaleidagraph. UbAMC hydrolysis assays were performed in assay buffer (50 mM Tris/HCl, 150 mM NaCl, 2 mM EDTA, 2 mM DTT [pH 7.5]), supplemented with 1 mg/ml bovine serum albumin (Roche) at 25°C. Enzyme concentrations were determined with the Bradford reagent (Biorad) and BSA (Pierce) as standard. Measurements were performed in triplicate with 20 pM (unless stated otherwise) of the respective enzyme in a total volume of 30 μl/well in a 384-well plate (NUNC) using a Spectramax M2 plate reader (Molecular Devices), with a 368 nm/467 nm filter pair and a 455 nm cutoff. 7-amido-4-methylcoumarin (Anaspec Inc.) was used for calibration. Kinetic parameters (Vmax, KM) were calculated by fitting initial velocities via a nonlinear regression to the Michaelis-Menten equation in Kaleidagraph, using substrate concentrations ranging from 0 to 5 μM. kcat was determined from the equation kcat = Vmax / [E]; with [E] = total enzyme concentration.
Deubiquitination of Cellular Ub Conjugates
293T cells at 70%–80% confluence were transfected with 5 μg of pcDNA3.1+ (Invitrogen) encoding HA-tagged Ub (or empty pcDNA3.1+ as control) per 10 cm dish, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. To enrich for polyubiquitinated species, cells were exposed to 50 μM proteasome inhibitor ZL3VS 1 hr prior to cell lysis. Soluble cell extracts were prepared 24 hr posttransfection by resuspension in 1 ml assay buffer supplemented with 0.5% (v/v) NP-40. Cellular debris was removed by centrifugation (1200 × g), and the supernatants were adjusted to 1 mg/ml protein with assay buffer supplemented with 0.1% (v/v) NP-40. The soluble extracts were then incubated in the absence or presence of enzyme (50 nM final concentration) in a total volume of 50 μl for 1 hr at 37°C. Reactions were stopped by addition of SDS-PAGE sample buffer and subjected to SDS-PAGE and immunoblotting according to standard procedures.
Preparation of M48USP-UbVME and Crystallization
M48USP bearing an N-terminal histidine tag was purified as described previously (Schlieker et al., 2005). The histidine tag was removed by PreScission protease (GE Healthcare) treatment according to the manufacturer's instructions, followed by size exclusion chromatography (S75 HiLoad, GE Healthcare) in 50 mM Tris/HCl, 150 mM NaCl, and 2 mM DTT (pH 7.5). M48USP was reacted with a ∼1.5-fold molar excess of Ub-vinylmethylester (UbVME) (Borodovsky et al., 2002) at room temperature for 2 hr to achieve quantitative covalent modification. The covalent M48USP-UbVME adduct was subjected to size exclusion chromatography (S75 HiLoad) in 10 mM HEPES, 50 mM NaCl, and 2 mM DTT (pH 7.5) to remove excess UbVME. The sample was concentrated to 10 mg/ml by ultrafiltration (Centriprep YM10, Millipore) and used in sparse matrix crystallization screens (Hampton Research, Qiagen). Initial crystals grew from a condition containing 200 mM magnesium formate and 14% PEG 3350 to maximum dimensions of 0.2 × 0.2 × 0.1 mm and were used in microseeding of SeMet-modified M48USP-UbVME mixed with equal volumes of 100 mM imidazole (pH 6.0), 20 mM MnCl2, 200 mM magnesium formate, and 12%–14% PEG3350. Crystals were harvested with a nylon loop and quickly dipped in crystallization solution supplemented with 25% PEG400 prior to flash-freezing in liquid N2.
X-Ray Data Collection, Structure Determination, and Refinement
X-ray diffraction data were collected on crystals of SeMet-labeled M48USP-UbVME at 100 K at NE-CAT beamline ID24 at the Advanced Photon Source, Argonne, IL and processed with HKL2000 (Otwinowski and Minor, 1997). The M48USP-UbVME crystal structure was determined by using a single-wavelength anomalous dispersion experiment at the Se K edge (Table 1). All 12 expected Se sites for two M48USP chains in the asymmetric unit were determined by using the program SHELXD (Schneider and Sheldrick, 2002) and used for phasing with the program SHARP (de La Fortelle and Bricogne, 1997). Density modification and subsequent automated model building in RESOLVE (Terwilliger, 2003) provided an 80% complete model. This model was further improved to 97% completeness with ARP/wARP run in alternating mode with restrained and TLS refinement in REFMAC5 (Murshudov et al., 1997). Remaining residues and the VME moieties were built manually in O (Jones et al., 1991).
Data collection and refinement statistics are summarized in Table 1. Model quality was examined with WHAT_CHECK (Hooft et al., 1996), showing that all φ, ψ torsion angles are within the allowed areas of the Ramachandran plot. Figure 2, Figure 3, Figure 4 were created with PyMOL.
Acknowledgments
C.S. is supported by an EMBO long-term fellowship (187-2005). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract No. W-31-109-ENG-38. We thank Thomas U. Schwartz and Howard C. Hang for critically reading the manuscript and Kanagalaghatta Rajashankar for beamline assistance.
Published: March 8, 2007
Footnotes
Supplemental Data include two figures and one table and can be found with this article online at http://www.molecule.org/cgi/content/full/25/5/677/DC1/.
Accession Numbers
Atomic coordinates have been deposited with the Protein Data Bank under accession code 2J7Q.
Supplemental Data
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakirev M.Y., Jaquinod M., Haas A.L., Chroboczek J. Deubiquitinating function of adenovirus proteinase. J. Virol. 2002;76:6323–6331. doi: 10.1128/JVI.76.12.6323-6331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogyo M., McMaster J.S., Gaczynska M., Tortorella D., Goldberg A.L., Ploegh H. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl. Acad. Sci. USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L., Kessler B.M. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- Das C., Hoang Q.Q., Kreinbring C.A., Luchansky S.J., Meray R.K., Ray S.S., Lansbury P.T., Ringe D., Petsko G.A. Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1. Proc. Natl. Acad. Sci. USA. 2006;103:4675–4680. doi: 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Fortelle E., Bricogne G. Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- Gao G., Luo H. The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 2006;84:5–14. doi: 10.1139/y05-144. [DOI] [PubMed] [Google Scholar]
- Glickman M.H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Gong L., Kamitani T., Millas S., Yeh E.T. Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J. Biol. Chem. 2000;275:14212–14216. doi: 10.1074/jbc.275.19.14212. [DOI] [PubMed] [Google Scholar]
- Hemelaar J., Borodovsky A., Kessler B.M., Reverter D., Cook J., Kolli N., Gan-Erdene T., Wilkinson K.D., Gill G., Lima C.D., et al. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell. Biol. 2004;24:84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Hooft R.W., Vriend G., Sander C., Abola E.E. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Hu M., Li P., Li M., Li W., Yao T., Wu J.W., Gu W., Cohen R.E., Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Hu M., Li P., Song L., Jeffrey P.D., Chenova T.A., Wilkinson K.D., Cohen R.E., Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.C., Larsen C.N., Cook W.J., Wilkinson K.D., Hill C.P. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 Å resolution. EMBO J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.C., Riddle S.M., Cohen R.E., Hill C.P. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta. Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kattenhorn L.M., Korbel G.A., Kessler B.M., Spooner E., Ploegh H.L. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell. 2005;19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Kumanomidou T., Mizushima T., Komatsu M., Suzuki A., Tanida I., Sou Y.S., Ueno T., Kominami E., Tanaka K., Yamane T. The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J. Mol. Biol. 2006;355:612–618. doi: 10.1016/j.jmb.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Lawrence M.C., Colman P.M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Li T., Chen X., Garbutt K.C., Zhou P., Zheng N. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell. 2006;124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Misaghi S., Sun Z.Y., Stern P., Gaudet R., Wagner G., Ploegh H. Structural and functional analysis of human cytomegalovirus US3 protein. J. Virol. 2004;78:413–423. doi: 10.1128/JVI.78.1.413-423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaghi S., Galardy P.J., Meester W.J., Ovaa H., Ploegh H.L., Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J. Biol. Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- Misaghi S., Balsara Z.R., Catic A., Spooner E., Ploegh H.L., Starnbach M.N. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol. Microbiol. 2006;61:142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum- likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Narasimhan J., Wang M., Fu Z., Klein J.M., Haas A.L., Kim J.J. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J. Biol. Chem. 2005;280:27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ovaa H., Kessler B.M., Rolen U., Galardy P.J., Ploegh H.L., Masucci M.G. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. USA. 2004;101:2253–2258. doi: 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N.D., Tolle D.P., Barrett A.J. MEROPS: the peptidase database. Nucleic Acids Res. 2004;32:160–164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renatus M., Parrado S.G., D'Arcy A., Eidhoff U., Gerhartz B., Hassiepen U., Pierrat B., Riedl R., Vinzenz D., Worpenberg S., Kroemer M. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C., Korbel G.A., Kattenhorn L.M., Ploegh H.L. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 2005;79:15582–15585. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T.R., Sheldrick G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Schnell J.D., Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 2003;278:35857–35860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- Shen L.N., Liu H., Dong C., Xirodimas D., Naismith J.H., Hay R.T. Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1. EMBO J. 2005;24:1341–1351. doi: 10.1038/sj.emboj.7600628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulea T., Lindner H.A., Menard R. Structural aspects of recently discovered viral deubiquitinating activities. Biol. Chem. 2006;387:853–862. doi: 10.1515/BC.2006.108. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg C.E., Cook W.J. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- Wada H., Kito K., Caskey L.S., Yeh E.T., Kamitani T. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem. Biophys. Res. Commun. 1998;251:688–692. doi: 10.1006/bbrc.1998.9532. [DOI] [PubMed] [Google Scholar]
- Wang J., Loveland A.N., Kattenhorn L.M., Ploegh H.L., Gibson W. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 2006;80:6003–6012. doi: 10.1128/JVI.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York I.A., Rock K.L. Antigen processing and presentation by the class I major histocompatibility complex. Annu. Rev. Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- Zhou H., Monack D.M., Kayagaki N., Wertz I., Yin J., Wolf B., Dixit V.M. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-kappa B activation. J. Exp. Med. 2005;202:1327–1332. doi: 10.1084/jem.20051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.