Abstract
Background
Acute respiratory infections are the leading cause of mortality in children worldwide, especially in developing countries. Pneumonia accounts for 16% of all deaths of children under 5 years of age and was the cause of death of 935 000 children in 2015. Despite its frequency and severity, information regarding its etiology is limited. The aim of this study was to identify respiratory viruses associated with community-acquired pneumonia (CAP) in children younger than 5 years old.
Methods
One thousand four hundred and four children younger than 5 years of age with a clinical and/or radiological diagnosis of CAP in 11 hospitals in Mexico were included. Nasal washes were collected, placed in viral medium, and frozen at −70 °C until processing. The first 832 samples were processed using the multiplex Bio‐Plex/Luminex system and the remaining 572 samples using the Anyplex multiplex RT-PCR. Clinical data regarding diagnosis, clinical signs and symptoms, radiographic pattern, and risk factors were obtained and recorded.
Results
Of the samples tested, 81.6% were positive for viruses. Respiratory syncytial virus (types A and B) was found in 23.7%, human enterovirus/rhinovirus in 16.6%, metapneumovirus in 5.7%, parainfluenza virus (types 1–4) in 5.5%, influenza virus (types A and B) in 3.6%, adenovirus in 2.2%, coronavirus (NL63, OC43, 229E, and HKU1) in 2.2%, and bocavirus in 0.4%. Co-infection with two or more viruses was present in 22.1%; 18.4% of the samples were negative. Using biomass for cooking, daycare attendance, absence of breastfeeding, and co-infections were found to be statistically significant risk factors for the presence of severe pneumonia.
Conclusions
Respiratory syncytial virus (types A and B), human enterovirus/rhinovirus, and metapneumovirus were the respiratory viruses identified most frequently in children younger than 5 years old with CAP. Co-infection was present in an important proportion of the children.
Keywords: Respiratory viruses, Pneumonia, Children
Introduction
Globally, pneumonia and diarrhea are the leading causes of infection in children under 5 years of age. The World Health Organization (WHO) has estimated that 150 million cases of pneumonia occur each year in children in this age group. Of these cases, 20 million will require hospitalization, with approximately 935 000 deaths (health TtftGtco, 2013). In 2013, these infections accounted for 15% and 9%, respectively, of the 6.3 million deaths in this age group (Leung et al., 2016).
It is estimated that in up to 70% of cases of pneumonia in children, the etiology is viral (Jain, 2017). Until recently, only cultures, serology, immunofluorescence tests, and ELISA were available for the diagnosis of viral pneumonia, and the seven viruses cited as the main causes of respiratory infections were respiratory syncytial virus (RSV), influenza A and B viruses, parainfluenza virus (PIV) types 1–3, and adenovirus (Pavia, 2011).
In the past decade, molecular diagnostic methods have allowed the identification of new pathogens as causes of respiratory diseases. Multiplex PCR is a highly sensitive and specific tool for the rapid identification of respiratory viruses, such as human metapneumovirus, severe acute respiratory syndrome (SARS), NL63, and HKU1 coronaviruses, PIV type 4, bocavirus, and rhinovirus types C and D (Jartti et al., 2012), especially in settings where cultures are not available.
A study published in 2015 by the US Centers for Disease Control and Prevention (CDC) on the etiology of pneumonia in 2254 children from 1 day to 17 years of age yielded positive identification of pathogens in 81% of the cases. Among these, 66% were viruses and 8% were bacteria, and both bacteria and viruses were detected in 7% of cases. The most frequent organisms found in this study were RSV (28%), human rhinovirus (27%), metapneumovirus (13%), adenovirus (11%), Mycoplasma pneumoniae (8%), PIV (7%), influenza virus (7%), coronavirus (5%), Streptococcus pneumoniae (4%), Staphylococcus aureus (1%), and Streptococcus pyogenes (1%) (Jain et al., 2015).
Although pneumonia is the leading cause of morbidity and mortality in children (health TtftGtco, 2013), especially in those under 3 years old, there have been few studies on the etiology of viral pneumonia in Mexico. Taking advantage of the availability of new molecular diagnostic technologies, a multicenter nationwide study was conducted in Mexico using multiplex PCR diagnostic kits to better understand the etiology and epidemiology of viral community-acquired pneumonia (CAP) in children under 5 years old.
Methods
Study population and study design
From March 2010 to August 2013, a cross-sectional, prospective study was conducted using a convenience sample of patients aged 1 month to 5 years with a clinical and/or radiological diagnosis of pneumonia. The study was performed in 11 hospitals in various regions of Mexico (Nuevo Hospital Civil de Guadalajara, Hospital Regional Universitario de los Servicios de Salud del Estado de Colima, Hospital Pediátrico de Coyoacán, Hospital General de Durango, Hospital General de Mexicali, Hospital Central “Dr. Ignacio Morones Prieto” San Luis Potosí, Hospital para el Niño de Toluca, Hospital General de México “Dr. Eduardo Liceaga”, Hospital de Pediatría del CMNO, Instituto Mexicano del Seguro Social (IMSS), Guadalajara, Hospital de la Niñez Oaxaqueña, and Hospital Infantil de Tlaxcala). The study was approved by the ethics and research committees of all participating institutions. The parents or guardians of the children with pneumonia were invited to have their children participate in the study. An informed consent form was signed before performing any procedure.
The case definition of pneumonia was the presence of respiratory symptoms, including respiratory distress, tachypnea, cyanosis, or cough, with or without fever of less than 1-week duration and/or an X-ray showing the presence of pulmonary infiltrates. The radiographic pattern was classified as consolidation, alveolar, interstitial, mixed, or pleural effusion. Information regarding the date of diagnosis, clinical signs and symptoms, radiographic pattern, and risk factors for pneumonia was collected from each patient in a format designed for this study.
Sample collection
Patients who attended the emergency room or who were hospitalized at the participating institutions and met the case definition and inclusion criteria were invited to participate. A questionnaire was answered by parents or guardians and a nasal wash was obtained. Nasal washes were performed by introducing 0.5–1 ml of sterile saline solution through a sterile feeding tube connected to a syringe and aspirating the solution from each nostril. The sample was placed in viral medium and frozen at −70 °C. Frozen samples were sent to the participating laboratories (Unidad de Investigación en Medicina Experimental, Facultad de Medicina, UNAM, Mexico City; CIBO IMSS Guadalajara, Facultad de Medicina, UASLP, San Luis Potosí) and then to the Unidad de Investigación en Medicina Experimental, Facultad de Medicina, UNAM, where all the samples from the participating institutions were stored until processing.
RT-PCR technique
The samples were processed using two RT-PCR multiplex assays: 832 samples were processed in the Instituto de Diagnóstico y Referencia Epidemiológicos (InDRE) using the semi-automated extraction equipment QIAcube (Qiagen Instruments AG, Switzerland) with the MiniElute kit (Qiagen, Switzerland) to isolate viral genetic material (RNA/DNA). Respiratory viruses were detected through the x-TAG Respiratory Viral Panel (RVP) with the Bio‐Plex/Luminex platform (Luminex Corporation, USA). The nucleic acid is amplified by a multiplex RT-PCR reaction using an internal control (phage MS-2). The viral regions of interest were amplified with generic primers for 19 viruses: influenza A virus (H1, H3, and H5 subtypes), influenza B virus, RSV type A, RSV type B, coronavirus (SARS, NL63, 229E, OC43, and HKU1), PIV types 1–4, metapneumovirus, adenovirus, and human enterovirus/rhinovirus.
The remaining 572 samples were processed at the Unidad de Investigación en Medicina Experimental, Facultad de Medicina, UNAM using the Anyplex II RV16 kit (Seegene, Seoul, South Korea). The nucleic acid extraction was performed manually using the Ribo_spinGeneAll vRD kit (GeneAll Biotechnology, Seoul, South Korea); before extraction, an internal control of the Anyplex kit was added to each sample. The cDNA was synthesized from extracted RNA with the cDNA Synthesis Kit Premix (Seegene). Subsequently, real-time PCR was carried out in the CFX96 equipment (Bio-Rad) through the Anyplex II RV16 Detection kit, which uses TOCE technology. Amplified respiratory viruses were visualized using Seegene Viewer software. The Anyplex RV16 kit has the capacity to detect 14 RNA viruses and two DNA viruses: RSV types A and B, influenza virus types A and B, PIV types 1–4, adenovirus, metapneumovirus, coronavirus OC43, 229E, and NL63, rhinovirus types A, B, and C, enterovirus, and bocavirus.
Statistical analysis
Frequencies and proportions of clinical and demographic variables were determined and displayed by descriptive statistics. The Kruskal–Wallis test and analysis of variance (ANOVA) were used to compare qualitative and quantitative variables, respectively. A multivariate analysis using logistic and multinomial regression was performed to detect associations between clinical features, risk factors, hospitalization, and viral detection. A p-value of <0.05 was considered statistically significant.
Results
Demographic and clinical characteristics
A total of 1404 children with a clinical and/or radiological diagnosis of pneumonia were included in the study. Seventy percent of the samples were from hospitals located in the central region of Mexico, 12.8% from Mexico City, 15.8% from the northern region, and 0.6% from the southern region of the country. No deaths were reported at any site. The mean (±standard error (SE)) age of participants was 14.3 ± 0.3 months. Some differences in the age distribution were observed between hospitals: the population of children in Mexicali and Toluca tended to be older than those in the other cities, while children admitted to the Hospital Civil de Guadalajara were younger. Thirty-eight percent of the subjects were female. The mean (±SE) height and weight of the children were 68.1 ± 4 cm and 7.85 ± 0.01 kg, respectively. Forty-one percent of the patients presented fever. The mean (±SE) respiratory rate was 50.7 ± 0.3, and 93.8% of patients had a cough. Mild respiratory distress was observed in 56.3%, moderate respiratory distress in 29.9%, and severe respiratory distress in 2.4% of the study population; 11.4% of patients had no respiratory distress. Pneumonia was classified according to the WHO definitions. Using these criteria, it was possible to diagnose pneumonia in 41.4%, severe pneumonia in 55.1%, and very severe disease in 3.4% of the cases.
Radiographic features and risk factors
Most of the patients had a radiographic interstitial pattern (45.4%); however a significant proportion of patients had alveolar infiltrates (36.6%), consolidation (9.3%), and mixed infiltrates (8.6%). Only 0.1% had evidence of pleural effusion. According to the risk factors associated with pneumonia described in the literature, the following were found in the present study population: an incomplete vaccination schedule in 55.7%, exposure to household tobacco smoke in 43.3%, absence of breastfeeding in 43.1%, use of biomass for cooking in 19.2%, daycare attendance in 7.1%, and immunosuppression in 7%. A total of 30% had comorbidities. Logistic regression and multinomial regression were performed to assess whether the risk factors associated with pneumonia reported in the literature were associated with severe pneumonia. Using biomass for cooking, daycare attendance, absence of breastfeeding, and co-infections were found to be statistically significant risk factors for the presence of severe pneumonia. In contrast, other factors that have been reported to increase the risk of pneumonia, such as exposure to household tobacco smoke, immunosuppression, incomplete vaccination schedule, lack of influenza vaccine, and comorbidities did not show a statistically significant association with severe pneumonia (Table 1 ).
Table 1.
Logistic and multinomial regression for severe pneumonia and risk factors.
| Risk factors | OR | 95% CI | p-Value |
|---|---|---|---|
| Domestic smoking | 0.68 | 0.51–0.91 | 0.02 |
| Biomass use | 1.70 | 1.31–2.27 | <0.001 |
| Immunocompromise | 1.05 | 0.87–1.26 | 0.63 |
| Daycare attendance | 1.26 | 1.01–1.58 | 0.01 |
| Incomplete vaccination schedule | 0.98 | 0.67–1.22 | 0.19 |
| Absence of influenza vaccine | 1.23 | 0.90–1.70 | 0.12 |
| Absence of breastfeeding | 1.34 | 1.05–1.72 | 0.01 |
| Viral co-infection | 1.41 | 1.0–1.98 | 0.04 |
OR, odds ratio; CI, confidence interval.
Viral detection and co-infections
A viral nucleic acid was detected in 81.6% of the 1404 samples collected. A single agent was observed in 835 samples (59.5%) and a co-infection with two or more viruses in 311 samples (22.1%); 258 samples (18.4%) were negative.
Respiratory viruses detected in order of frequency were 23.2% RSV types A and B, 16.6% enterovirus/rhinovirus, 5.7% metapneumovirus, 5.5% PIV types 1–4, 3.6% influenza virus types A and B, 2.2% adenovirus, 2.2% coronavirus NL63, OC43, 229E, and HKU1, and 0.4% bocavirus. Of the 311 cases (22.1%) with co-infections, two viruses were detected in 80.7% (251 samples), three viruses in 15.8% (49 samples), four viruses in 0.3% (one sample), and five viruses in 3.2% (10 samples). The most frequent viruses associated with viral co-infection were enterovirus/rhinovirus in 52% (163 samples) and RSV type A in 54% (169 samples). The most frequent co-infections were enterovirus/rhinovirus and RSV type A in 29% (90 samples), followed by enterovirus/rhinovirus and PIV type 3 in 6.4% (20 samples), enterovirus/rhinovirus and adenovirus in 6.1% (19 samples), enterovirus/rhinovirus and metapneumovirus in 5.8% (18 samples), and RSV type A and adenovirus in 5.1% (16 samples). In the samples in which two viruses were detected, enterovirus/rhinovirus was found in co-infection with five different viruses (adenovirus, RSV type A, PIV type 3, metapneumovirus, and PIV type 1), and RSV type A was found in co-infection with nine different viruses (coronavirus OC43, adenovirus, enterovirus/rhinovirus, coronavirus 229E, coronavirus NL63, bocavirus, influenza virus type A, RSV type B, and metapneumovirus). In the cases with co-infections with three viruses, enterovirus/rhinovirus was found in seven of nine combinations; enterovirus/rhinovirus was found in all combinations of co-infections with four and five viruses. RSV was also found in six of the nine combinations of three viruses and in all of the co-infections with four and five viruses (Table 2, Table 3 ).
Table 2.
Frequencies of viral detection of single agents.
| Virus | n (%) |
|---|---|
| Respiratory syncytial virus type A (RSVA) | 320 (22.8) |
| Human enterovirus/human rhinovirus (HEV/HRV) | 233 (16.6) |
| Metapneumovirus | 80 (5.7) |
| Influenza A virus | 41 (2.9) |
| Parainfluenza virus type 3 (PIV3) | 35 (2.5) |
| Adenovirus | 31 (2.2) |
| Parainfluenza virus type 4 (PIV4) | 21 (1.5) |
| Parainfluenza virus type 2 (PIV2) | 12 (0.9) |
| Respiratory syncytial virus type B (RSVB) | 12 (0.9) |
| Human coronavirus NL63 (CoVNL63) | 11 (0.8) |
| Influenza B virus | 10 (0.7) |
| Human coronavirus OC43 (CoVOC43) | 9 (0.6) |
| Parainfluenza virus type 1 (PIV1) | 8 (0.6) |
| Bocavirus | 6 (0.4) |
| Human coronavirus 229E (CoV229E) | 4 (0.3) |
| Human coronavirus HKU1 (CoVHKU1) | 2 (0.1) |
| Negative | 259 (18.4) |
| Total | 835 (77.9) |
Table 3.
Viral co-infections with two to five viruses in children with community-acquired pneumoniaa.
| Number of cases positive for two viruses | |||||
| RSVA | HEV/HRV | Bocavirus | PIV3 | CoV229E | |
| HEV/HRV | 90 | ||||
| Adenovirus | 16 | 19 | 3 | 3 | |
| Metapneumovirus | 6 | 18 | 4 | ||
| Bocavirus | 11 | 14 | |||
| PIV3 | 20 | ||||
| RSVB | 17 | ||||
| Influenza A virus | 9 | ||||
| CoVNL63 | 7 | 2 | |||
| PIV1 | 6 | ||||
| CoVOC43 | 3 | ||||
| CoV229E | 1 | 2 | |||
| Number of cases positive for three viruses | |||||
| RSVA | HEV/HRV | Bocavirus | 11 | ||
| RSVA | HEV/HRV | Adenovirus | 5 | ||
| RSVA | HEV/HRV | Metapneumovirus | 4 | ||
| RSVA | HEV/HRV | PIV4 | 3 | ||
| RSVB | HEV/HRV | PIV 2 | 5 | ||
| RSVA | Adenovirus | PIV1 | 4 | ||
| HEV/HRV | CoVNL63 | PIV3 | 6 | ||
| HEV/HRV | Bocavirus | PIV1 | 2 | ||
| HRV | Bocavirus | CoVOC43 | 9 | ||
| Number of cases positive for four viruses | |||||
| RSVA | HEV/HRV | Adenovirus | Influenza B virus | 1 | |
| Number of cases positive for five viruses | |||||
| RSVA | HEV/HRV | Adenovirus | Influenza A virus | Bocavirus | 5 |
| RSVA | HEV/HRV | RSVB | PIV3 | CoVOC43 | 5 |
RSVA, respiratory syncytial virus type A; HEV/HRV, human enterovirus/human rhinovirus; PIV3, parainfluenza virus type 3, CoV229E, human coronavirus 229E; CoVNL63, human coronavirus NL63; PIV1, parainfluenza virus type 1; RSVB, respiratory syncytial virus type B; CoVOC43, human coronavirus OC43; PIV2, parainfluenza virus type 2; PIV4, parainfluenza virus type 4.
The total numbers of co-infections for two, three, four, and five respiratory viruses are reported.
Seasonality
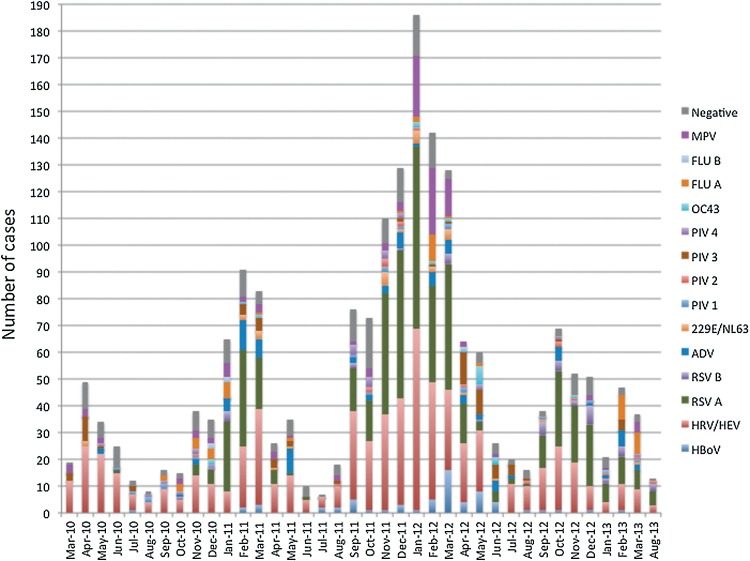
Children with CAP requiring hospital admission were admitted most frequently in winter (41.1% of cases) and autumn (30.6%), and less frequently during the spring (19.1%) and summer (9.1%). Rhinovirus, bocavirus, PIV, and adenovirus did not present a seasonal pattern; cases were observed throughout the year. In contrast, influenza A and B viruses were present in the winter and spring, RSV was present in the autumn and winter, and metapneumovirus was observed in autumn, winter, and spring, but not in summer (Figure 1 ).
Figure 1.
Frequency of respiratory viruses in children with community-acquired pneumonia.
MPV, metapneumovirus; FLU B, influenza B virus; FLU A, influenza A virus; OC43, human coronavirus OC43; PIV4, parainfluenza virus type 4; PIV3, parainfluenza virus type 3; PIV2, parainfluenza virus type 2; PIV1, parainfluenza virus type 1; 229E/NL63, coronavirus 229E and NL63; ADV, adenovirus; RSV B, respiratory syncytial virus type B; RSV A, respiratory syncytial virus type A; HRV/HEV, human rhinovirus/human enterovirus; HBoV, human bocavirus.
Discussion
Of the 1404 samples collected from children under 5 years of age with CAP, positive viral detection was identified in 81.6%, a higher prevalence than that reported in studies conducted in previous decades when molecular diagnostic methods were not available. In hospitalized children with pneumonia admitted to tertiary referral centers, a respiratory virus was detected in 47% of patients in a hospital in San Luis Potosí (Noyola et al., 2004) and 14% at the Hospital Infantil de Mexico “Federico Gómez” (Wong-Chew et al., 2010). Other studies that have included hospitalized patients in secondary care hospitals have reported a positivity rate between 39% and 60% (Noyola et al., 2004). In addition, studies in the community setting have described up to 65% of viral detection (Manjarrez et al., 2003, Noyola et al., 2005).
The overall positivity rate observed in this study is slightly higher than the frequency reported by the CDC in a study conducted in pediatric patients with pneumonia, in which 66% detection of viruses and 7% of co-infections with viruses and bacteria were found (Jain et al., 2015). The present study results also showed a higher percentage of respiratory viruses compared to the study reported by the present authors’ group in collaboration with the Biotechnology Institute (IBT) UNAM, which was conducted in the state of Veracruz. In that study, 525 children aged 1 month to 15 years with upper respiratory tract infections, assessed in private pediatric practices, were evaluated clinically and diagnostically with the use of multiplex molecular techniques to search for respiratory viruses; a virus was detected in 71.6% of these children (Wong-Chew et al., 2015).
The viruses detected most frequently in the group of children under 5 years of age with lower respiratory infections in the present study were RSV types A and B (23.7%), enterovirus/rhinovirus (16.5%), metapneumovirus (5.7%), PIV types 1–4 (5.5%), and influenza virus types A and B (3.6%). These results are similar to those reported by Jain et al. in a study involving children younger than 18 years of age with CAP requiring hospitalization in three hospitals in Memphis, Nashville, and Salt Lake City; the most commonly detected viruses were RSV (28%), human rhinovirus (27%), and metapneumovirus (13%) (Jain et al., 2015). RSV has been reported previously as the most common respiratory pathogen in hospitalized children with lower respiratory infections (Noyola et al., 2004, Noyola et al., 2007), as well as in children in the community (Manjarrez et al., 2003). In the aforementioned study, performed in the state of Veracruz in outpatients with respiratory infections, the two most common pathogens were RSV (detected in 19.6% of patients) and rhinovirus (detected in 17.5%), followed by PIV (10.5%), influenza (10.4%), coronavirus (8.7%), adenovirus (7.2%), enterovirus (5.9%), metapneumovirus (5.3%), and bocavirus (2.5%) (Wong-Chew et al., 2015).
In a recent study of children hospitalized with respiratory tract infections and respiratory distress at the Instituto Nacional de Enfermedades Respiratorias (Mexico City), viral infections were detected in 59.7% of 432 enrolled patients, with human rhinovirus being the most frequently detected (76, 26.6%). Other viruses included influenza A virus in 27, RSV in 51, PIV in seven, adenovirus in six, and metapneumovirus in 13 (Moreno-Valencia et al., 2015). One limitation of the present study is that the xTAG multiplex RT-PCR method does not differentiate between rhinovirus and enterovirus, so the potential role of rhinoviruses could be overestimated with the enterovirus detections added.
In the present prospective study in which patients with severe lower respiratory tract infections were included, viral co-infections were found in 22.1% of patients, which is a higher percentage than that found in the study of upper respiratory infections in Veracruz (14.1%) (Wong-Chew et al., 2015). Although some characteristics of the study populations are different (such as the recruitment time and age distribution), the higher percentage of co-infections in patients with pneumonia compared with upper respiratory infections in the Veracruz study, might suggest that co-infections cause more severe disease, suggesting complementary interaction among these viruses. This is also suggested by the multivariate analysis, in which it was found that co-infections are a risk factor for severe pneumonia. These observations are consistent with those of other authors who have reported RSV co-infections to be associated with a more severe disease than infections caused by a single virus (Harada et al., 2013). However, whether the viral detection in nasal washes reflects the pathogen responsible for the lower respiratory disease or only reflects a nasopharyngeal commensal or asymptomatic infection is not known with certainty, particularly when highly sensitive molecular methods are used (Jansen et al., 2011).
The seasonal prevalence of RSV in autumn and winter, of influenza in winter and spring, and of metapneumovirus from September to May observed in this study is consistent with other reports. The year-round detection of other viruses, such as rhinovirus, enterovirus, adenovirus, and PIV, has also been reported previously (WHO, 2014, Mahony, 2008, Fisman, 2012).
In the present study, 45.4% of the cases presented in winter, and although 90.3% required hospitalization, only 55.1% had severe pneumonia; 56.3% had mild respiratory distress, 93.8% had a cough, and 41% presented an interstitial radiographic pattern on admission, characteristics that are consistent with previous reports.
One of the advantages of current molecular techniques is the high sensitivity to detect multiple pathogens. On the other hand, interpretation of the results provided by these assays requires further examination. Because of the increasing detection of some viruses, particularly rhinoviruses, coronaviruses, and bocavirus in patients with pneumonia, their specific contribution to lower respiratory tract infections needs to be evaluated. A recent study compared the frequency of detection of respiratory viruses in patients with CAP to the frequency of detection in asymptomatic subjects (Self et al., 2016). RSV, human metapneumovirus, and influenza virus were notably more frequent in children with CAP (26.6%, 15.1%, and 3.4%, respectively) than in asymptomatic children (1.9%, 1.5%, and 0%, respectively) (Self et al., 2016). In contrast, there were less striking differences in the detection of rhinoviruses (21.9% vs. 17.3% in patients with pneumonia and asymptomatic subjects, respectively) and coronaviruses (4.5% and 1.5% in patients with pneumonia and asymptomatic subjects, respectively). In fact, after multivariate regression analysis, rhinoviruses were found not to be significantly associated with the presence of pneumonia (Self et al., 2016). Similar results have been reported by others, who have detected respiratory viruses in a large proportion of children without pneumonia (Zar et al., 2016, Rhedin et al., 2015).
Viruses that are detected consistently more frequently in patients with pneumonia include RSV and influenza viruses, while rhinovirus, enterovirus, and coronaviruses have been reported not to be associated with pneumonia, when compared to a control group (Zar et al., 2016, Rhedin et al., 2015, Spichak et al., 2016). In these studies, the association between pneumonia and the presence of adenovirus or metapneumovirus has been inconsistent. Also, a recent meta-analysis that compared the frequency of detection of respiratory viruses in children with pneumonia and subjects without pneumonia found that RSV, influenza virus, metapneumovirus, and rhinovirus were associated with the presence of pneumonia, while adenovirus, coronavirus, and bocavirus were as frequent in patients as in controls (Shi et al., 2015).
Overall, these studies indicate that, although rhinovirus, enterovirus, coronavirus, and bocavirus may be detected frequently in patients with pneumonia, the impact of these viruses as a cause of lower respiratory tract infection is not clear. Moreover, the viral load was not measured in the present study, but some authors have mentioned that it could determine disease severity, while others state that the viral load is not important. This is controversial and needs further studies.
Many factors have previously been associated with a higher risk of lower respiratory tract infection in children (Fonseca et al., 1996, Koch et al., 2003, Suzuki et al., 2009). Some of these have been shown consistently to increase the risk of severe infection, while others have shown inconsistent results among different studies. In this study, in addition to the presence of co-infection, an association was found between other factors and the risk of severe pneumonia: lack of breastfeeding, use of biomass for cooking, and daycare attendance. Lack of breastfeeding and indoor air pollution have consistently been associated with severe lower respiratory tract infection and were considered as definite risk factors in a meta-analysis published in 2013 (Jackson et al., 2013). In that report, parental smoking showed inconsistent results among studies and was considered as a likely risk factor for severe infection. In the present study it showed a protective effect, although there is no apparent explanation for this finding. Finally, daycare attendance was sporadically reported in association with severe infections and was considered a possible risk factor. Inadequate breastfeeding, indoor pollution, and second-hand smoke exposure have also been found to be risk factors for mortality in children with acute lower respiratory infections (Sonego et al., 2015).
The results of the present study provide additional information that strengthens the association between patient characteristics and the risk of developing severe pneumonia. The high frequency of some of these factors in the children included in this study (exposure to tobacco smoke in 43.3%, lack of breastfeeding in 43.1%, and using biomass for cooking in 19.2%) indicates areas that still require interventions in Mexico. Current campaigns to encourage breastfeeding should be strengthened, and new strategies to reduce exposure to indoor pollution need to be developed.
A limitation of this study is that it did not include a random sample of patients from all of Mexico; however, this is the first prospective study to assess patients from a large number of hospitals in the north and central regions and to a lesser extent from the southern region of the country. Another limitation of the study is that some participating hospitals included a significant number of patients, while other centers only recruited from three to nine patients (Tlaxcala, Mexicali, and Oaxaca). Most previously reported studies from Mexico have included data from a single institution. In contrast, this is the first multicenter active surveillance study of viral agents associated with CAP in children under 5 years of age in Mexico, in an era where the advancement of molecular biology technology has allowed the detection of multiple pathogens in a single sample. This study also provides updated information on the prevalence of known viral pathogens and others that are of more recent discovery, such as human metapneumovirus, SARS, NL63, and HKU1 coronaviruses, PIV type 4, bocavirus, and rhinovirus type C, as well as the role of each pathogen in disease compared to other pathogens.
Author contributions
Wong-Chew Rosa María: study design, data analysis, data interpretation, writing the manuscript, study coordinator, wrote grants for the study. García-León Miguel L.: study design, data interpretation, coordinator, sample analysis, review of the manuscript. González-Rodríguez Alejandra Pamela, López-Martínez Irma, Hernández-Andrade Teresa, Alpuche-Aranda Celia M.: nasal specimens, molecular biology processing, review of the manuscript. Noyola Daniel E., Pérez-González Luis F., Gaitán-Meza Jesús, Villaseñor-Sierra Alberto, Martínez-Aguilar Gerardo, Rivera-Nuñez Victor Hugo, Newton-Sánchez Oscar A., Firo-Reyes Verónica, Del Río-Almendarez Carlos N., Ortiz-García Enrique R., Navarrete-Navarro Susana, Soria-Rodríguez Carmen, Carrasco-Castillo Adoniram, Sánchez-Medina Eneida: patient recruitment, sample collection, review of the manuscript. Santos-Preciado José I.: study design, obtained grants for the study, review of the manuscript.
Ethical approval
This study was approved by the Institutional Review Board of the School of Medicine, Universidad Nacional Autónoma de México and the institutional review boards of all participating institutions. Written consent was obtained from parents or guardians.
Funding
This work was supported by grant 69852 (to Santos-Preciado JI) and grant 87691 (to Santos- Preciado JI) from the Consejo Nacional de Ciencia y Tecnologia (CONACYT).
Conflict of interest
Daniel Noyola reports personal fees as a speaker for Abbvie; Eneida Sánchez reports personal fees as a speaker for Sanofi Pasteur; Rosa M. Wong-Chew reports personal fees from Sanofi Pasteur and GSK for advisory boards, outside the submitted work.
Corresponding Editor: Eskild Petersen, Aarhus, Denmark
Footnotes
This work was presented in part as abstracts at ID Week 2013, San Francisco, CA, USA in October 2013; ID week 2014, Philadelphia, PA, USA in October 2014; the annual meeting of the Asociación Mexicana de Infectología y Microbiología Clínica, in Puebla, Puebla in May 2011, in León, Guanajuato in May 2012, in Guadalajara, Jalisco in May 2013, and in Acapulco, Guerrero in May 2014. The full work was presented at the Virology Meeting, Puente de Ixtla, Morelos in November 2015.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ijid.2017.06.020.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Fisman D. Seasonality of viral infections: mechanisms and unknowns. Clin Microbiol Infect. 2012;18(10):946–954. doi: 10.1111/j.1469-0691.2012.03968.x. [DOI] [PubMed] [Google Scholar]
- Fonseca W., Kirkwood B.R., Victora C.G., Fuchs S.R., Flores J.A., Misago C. Risk factors for childhood pneumonia among the urban poor in Fortaleza, Brazil: a case–control study. Bulletin of the World Health Organization. 1996;74(2):199–208. [PMC free article] [PubMed] [Google Scholar]
- Harada Y., Kinoshita F., Yoshida L.M., Minh le N., Suzuki M., Morimoto K. Does respiratory virus coinfection increases the clinical severity of acute respiratory infection among children infected with respiratory syncytial virus? Pediatr Infect Dis J. 2013;32(5):441–445. doi: 10.1097/INF.0b013e31828ba08c. [DOI] [PubMed] [Google Scholar]
- health TtftGtco . 2013. The World We Want. Health in the post-2015 agenda: report of the Global Thematic Consultation on Health. http://www.saluteinternazionale.info/wp-content/uploads/2013/07/Health-in-the-post-2015-agenda-Report-on-the-Global-Thematic-Consultation-on-Health-April-2013.pdf. (accessed January, 2016.) [Google Scholar]
- Jackson S., Mathews K.H., Pulanic D., Falconer R., Rudan I., Campbell H. Risk factors for severe acute lower respiratory infections in children: a systematic review and meta-analysis. Croatian medical journal. 2013;54(2):110–121. doi: 10.3325/cmj.2013.54.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Williams D.J., Arnold S.R., Ampofo K., Bramley A.M., Reed C. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. 2015;372(9):835–845. doi: 10.1056/NEJMoa1405870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S. Epidemiology of Viral Pneumonia. Clin Chest Med. 2017;38(1):1–9. doi: 10.1016/j.ccm.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.R., Wieringa J., Koekkoek S.M., Visser C.E., Pajkrt D., Molenkamp R. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol. 2011;49(7):2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jartti T., Jartti L., Ruuskanen O., Soderlund-Venermo M. New respiratory viral infections. Curr Opin Pulm Med. 2012;18(3):271–278. doi: 10.1097/MCP.0b013e328351f8d4. [DOI] [PubMed] [Google Scholar]
- Koch A., Mølbak K., Homøe P., Sørensen P., Hjuler T., Olesen M.E. Risk factors for acute respiratory tract infections in young Greenlandic children. American journal of epidemiology. 2003;158(4):374–384. doi: 10.1093/aje/kwg143. [DOI] [PubMed] [Google Scholar]
- Leung D.T., Chisti M.J., Pavia A.T. Prevention and Control of Childhood Pneumonia and Diarrhea. Pediatr Clin North Am. 2016;63(1):67–79. doi: 10.1016/j.pcl.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J.B. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21(4):716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjarrez M.E., Rosete D.P., Rincon M., Villalba J., Cravioto A., Cabrera R. Comparative viral frequency in Mexican children under 5 years of age with and without upper respiratory symptoms. J Med Microbiol. 2003;52(Pt 7):579–583. doi: 10.1099/jmm.0.05007-0. [DOI] [PubMed] [Google Scholar]
- Moreno-Valencia Y., Hernandez-Hernandez V.A., Romero-Espinoza J.A., Coronel-Tellez R.H., Castillejos-Lopez M., Hernandez A. Detection and Characterization of respiratory viruses causing Acute Respiratory Illness and Asthma Exacerbation in children during Three Different Season (2011-2014) in Mexico City. Influenza Other Respir Viruses. 2015 doi: 10.1111/irv.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyola D.E., Rodriguez-Moreno G., Sanchez-Alvarado J., Martinez-Wagner R., Ochoa-Zavala J.R. Viral etiology of lower respiratory tract infections in hospitalized children in Mexico. Pediatr Infect Dis J. 2004;23(2):118–123. doi: 10.1097/01.inf.0000110269.46528.a5. [DOI] [PubMed] [Google Scholar]
- Noyola D.E., Alpuche-Solis A.G., Herrera-Diaz A., Soria-Guerra R.E., Sanchez-Alvarado J., Lopez-Revilla R. Human metapneumovirus infections in Mexico: epidemiological and clinical characteristics. J Med Microbiol. 2005;54(Pt 10):969–974. doi: 10.1099/jmm.0.46052-0. [DOI] [PubMed] [Google Scholar]
- Noyola D.E., Zuviri-Gonzalez A., Castro-Garcia J.A., Ochoa-Zavala J.R. Impact of respiratory syncytial virus on hospital admissions in children younger than 3 years of age. J Infect. 2007;54(2):180–184. doi: 10.1016/j.jinf.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Pavia A.T. Viral infections of the lower respiratory tract: old viruses, new viruses, and the role of diagnosis. Clin Infect Dis. 2011;52(Suppl 4):S284–9. doi: 10.1093/cid/cir043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhedin S., Lindstrand A., Hjelmgren A., Ryd-Rinder M., Öhrmalm L., Tolfvenstam T. Respiratory viruses associated with community-acquired pneumonia in children: matched case-control study. Thorax. 2015;70(9):847–853. doi: 10.1136/thoraxjnl-2015-206933. [DOI] [PubMed] [Google Scholar]
- Self W.H., Williams D.J., Zhu Y., Ampofo K., Pavia A.T., Chappell J.D. Respiratory Viral Detection in Children and Adults: Comparing Asymptomatic Controls and Patients With Community-Acquired Pneumonia. J Infect Dis. 2016;213(4):584–591. doi: 10.1093/infdis/jiv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T., McLean K., Campbell H., Nair H. Aetiological role of common respiratory viruses in acute lower respiratory infections in children under five years: A systematic review and meta-analysis. Journal of global health. 2015;5(1):010408. doi: 10.7189/jogh.05.010408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonego M., Pellegrin M.C., Becker G., Lazzerini M. Risk factors for mortality from acute lower respiratory infections (ALRI) in children under five years of age in low and middle-income countries: a systematic review and meta-analysis of observational studies. PloS one. 2015;10(1):e0116380. doi: 10.1371/journal.pone.0116380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spichak T.V., Yatsyshina S.B., capital Ka C.L.K.A.C., capital Ka C.S.S., Korppi M.O. Is the role of rhinoviruses as causative agents of pediatric community-acquired pneumonia over-estimated? Eur J Pediatr. 2016 doi: 10.1007/s00431-016-2791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Thiem V.D., Yanai H., Matsubayashi T., Yoshida L.M., Tho L.H. Association of environmental tobacco smoking exposure with an increased risk of hospital admissions for pneumonia in children under 5 years of age in Vietnam. Thorax. 2009;64(6):484–489. doi: 10.1136/thx.2008.106385. [DOI] [PubMed] [Google Scholar]
- WHO . World Health Statistics 2014. In: Data WLCiP, editor. Global health Indicators. World Health Organization; Switzerland: 2014. [Google Scholar]
- Wong-Chew R.M., Farfan-Quiroz R., Sanchez-Huerta J.L., Nava-Frias M., Casasola-Flores J., Santos-Preciado J.I. [Frequency of respiratory viruses and clinical characteristics in children attending a care center in Mexico City] Salud Publica Mex. 2010;52(6):528–532. [PubMed] [Google Scholar]
- Wong-Chew R.M., Espinoza M.A., Taboada B., Aponte F.E., Arias-Ortiz M.A., Monge-Martínez J. Prevalence of respiratory virus in symptomatic children in private physician office settings in five communities of the state of Veracruz, Mexico. BMC Res Notes. 2015;8:261. doi: 10.1186/s13104-015-1239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar H.J., Barnett W., Stadler A., Gardner-Lubbe S., Myer L., Nicol M.P. Aetiology of childhood pneumonia in a well vaccinated South African birth cohort: a nested case-control study of the Drakenstein Child Health Study. The Lancet Respiratory medicine. 2016;4(6):463–472. doi: 10.1016/S2213-2600(16)00096-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.