Highlights
-
•
Histophilus somni is a respiratory pathogen of cattle.
-
•
Antimicrobial susceptibility testing was performed against commonly used antimicrobial agents.
-
•
Disc diffusion and minimum inhibitory concentration assays were mostly comparable.
-
•
Isolates from Australian cattle were almost completely susceptible bar, but one resistant isolate was identified.
-
•
Genotypic investigation detected a major cluster and clonal group of H. somni.
Keywords: Histophilus somni, Bovine respiratory disease, Antimicrobial susceptibility, Disc diffusion, Minimum inhibitory concentration
Abstract
This study investigated antimicrobial resistance traits, clonal relationships and epidemiology of Histophilus somni isolated from clinically affected cattle in Queensland and New South Wales, Australia. Isolates (n = 53) were subjected to antimicrobial susceptibility testing against six antimicrobial agents (ceftiofur, enrofloxacin, florfenicol, tetracycline, tilmicosin and tulathromycin) using disc diffusion and minimum inhibitory concentration (MIC) assays. Clonal relationships were assessed using repetitive sequence PCR and descriptive epidemiological analysis was performed. The H. somni isolates appeared to be geographically clonal, with 27/53 (47%) isolates grouping in one cluster from one Australian state. On the basis of disc diffusion, 34/53 (64%) isolates were susceptible to all antimicrobial agents tested; there was intermediate susceptibility to tulathromycin in 12 isolates, tilmicosin in seven isolates and resistance to tilmicosin in one isolate. Using MIC, all but one isolate was susceptible to all antimicrobial agents tested; the non-susceptible isolate was resistant to tetracycline, but this MIC result could not be compared to disc diffusion, since there are no interpretative guidelines for disc diffusion for H. somni against tetracycline. In this study, there was little evidence of antimicrobial resistance in H. somni isolates from Australian cattle. Disc diffusion susceptibility testing results were comparable to MIC results for most antimicrobial agents tested; however, results for isolates with intermediate susceptibility or resistance to tilmicosin and tulathromycin on disc diffusion should be interpreted with caution in the absence of MIC results.
Introduction
Histophilus somni causes bovine respiratory disease (BRD) worldwide (Sandal and Inzana, 2010). Although it is a commensal of the nasopharynx (Corbeil, 2007), H. somni can be an opportunistic pathogen of cattle, predominantly causing respiratory infections, but occasionally septicaemia, myocarditis, arthritis, abortion and other systemic infections (Sandal et al., 2007).
BRD is the most economically important disease in beef cattle (Welsh et al., 2004), costing the Australian feedlot industry approximately AUD$40 million per year (Sackett et al., 2007). Antimicrobial agents including tetracycline, tilmicosin, florfenicol, tulathromycin, ceftiofur and enrofloxacin are used routinely to prevent and/or treat BRD (Welsh et al., 2004). A reliance on these drugs creates a selection pressure that may result in the emergence of drug-resistant microorganisms (Barton et al., 2003). Resistance is emerging amongst BRD pathogens, particularly to those antimicrobial agents from first generation classes (e.g. tetracycline) (Welsh et al, 2004, Portis et al, 2012). Moreover, antimicrobial resistance patterns vary according to bacterial species and geographical location (Hendriksen et al., 2008), meaning that local knowledge of susceptibilities is critical for the effective prevention and treatment of H. somni infections.
The aim of this study was to determine the antimicrobial susceptibilities of H. somni against six antimicrobial agents commonly used to control and treat bovine bacterial respiratory pathogens via both disc diffusion and minimum inhibitory concentration (MIC) testing. Although MIC is considered to be the gold-standard test method in antimicrobial susceptibility determination (Andrews, 2001), disc diffusion is commonly used in veterinary diagnostic laboratories. An additional aim of this study was to assess associations between epidemiological factors (e.g. state of origin, production type, site of isolation), clonal relationships and antimicrobial susceptibility of H. somni cultured from Australian cattle.
Materials and methods
Isolates
Fifty-three H. somni isolates were obtained in 2012 from bovine samples that had been submitted to the Animal Disease Surveillance Laboratory, Toowoomba, Queensland or Elizabeth Macarthur Agricultural Institute, Menangle, New South Wales, Australia. Isolates were derived from cattle with clinical signs of respiratory disease (n = 51), thrombotic meningoencephalitis (n = 1) or infertility (n = 1) and H. somni was considered to be the causal or a contributing pathogen. Isolates were recovered from lung samples (37/53, 70%), nasal swabs (6/53, 11%), brain swabs (3/53, 6%) and one each from a pleural swab, preputial swab and heart blood swab; the remaining four (8%) isolates were from unspecified sites. All isolates were confirmed as H. somni by clonal morphology, Gram stain and H. somni-specific PCR (Angen et al., 1998). The quality control strain H. somni ATCC 700025 was used for all testing.
A clinical history, including location, breed, sex, age, production type and if the animal was introduced onto the property or homebred, was available for all cases, together with the results of serology or molecular testing for potential contributing pathogens, including infectious bovine rhinotracheitis virus (bovine herpesvirus type 1), bovine coronavirus and bovine pestivirus (bovine viral diarrhoea virus).
Antimicrobial disc diffusion susceptibility
Disc diffusion susceptibility testing was used to determine the antimicrobial susceptibility of H. somni isolates against ceftiofur (30 µg), enrofloxacin (5 µg), florfenicol (30 µg), tilmicosin (15 µg) and tulathromycin (30 µg) according to Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical Laboratory Standards Institute, 2013). Since guidelines for tilmicosin were not available for H. somni, interpretation was based on guidelines for Mannheimia haemolytica (Blackall et al., 2007). Disc diffusion susceptibility testing was also performed for tetracycline (30 µg), although CLSI guidelines were not available for interpretation of these results. Tulathromycin discs were obtained from Becton Dickinson, while other antimicrobial discs were obtained from Oxoid.
Minimum inhibitory concentration susceptibility testing
The MICs of ceftiofur, enrofloxacin, florfenicol, tetracycline, and tilmicosin were determined according to CLSI guidelines for agar dilution (Clinical Laboratory Standards Institute, 2013). The MICs of tulathromycin were determined for only 43 isolates using the same guidelines, since there were delays in obtaining tulathromycin antimicrobial powder and 10 isolates could not be revived for testing. Tulathromycin was obtained from Zoetis, while other antimicrobial powders were obtained from Sigma Aldrich.
The MICs were determined as the lowest concentrations of antimicrobial agent in the plate that completely inhibited colony formation. All MICs were tested in duplicate independently on separate days. If duplicate tests were within one serial dilution of each other, they were accepted, and the MIC result was reported as the highest MIC. In all cases, duplicate MIC results were identical or within one serial dilution.
Enterobacterial repetitive intergenic consensus PCR
Clonality between the H. somni isolates was determined by enterobacterial repetitive intergenic consensus (ERIC) PCR (Versalovic et al., 1991). Banding patterns were analysed using GelComparII (Applied Maths) with a Dice coefficient of 0.28% and a tolerance of 2.8%. A cluster was defined as a group of isolates that shared ≥80% similarity in their ERIC-PCR patterns. Within each cluster, isolates with a similarity of >94% were considered to be a clonal group. Isolates were considered to be outliers if they were <70% similar.
Epidemiological analysis
Epidemiological analyses were performed with Epitools. 1 The effect of state (Queensland vs. New South Wales), production type (meat/feedlot vs. non-meat/feedlot) and sample site (lung vs. non-lung) for cluster 6 (the dominant cluster including 27/53 of all isolates) compared to isolates from other clusters was determined using the Fisher's exact test. Other variables were not compared, since the total number of isolates in each category were <10.
Results
Antimicrobial susceptibility testing
Using the disc diffusion method, 35/53 (66%) isolates were susceptible to all antimicrobial agents tested (Table 1 ). All isolates were susceptible to ceftiofur, enrofloxacin and florfenicol. Intermediate susceptibility against tulathromycin was exhibited by 12/53 (23%) isolates and against tilmicosin by 7/53 (13%) isolates; 2/53 (4%) isolates had intermediate susceptibility to both tulathromycin and tilmicosin, while 1/53 (2%) isolates exhibited resistance to tilmicosin.
Table 1.
Disc diffusion distribution and susceptibility zones of 53 Histophilus somni isolates.
| Antimicrobial agents | Number of isolates (%) | Disc diffusion zone sizes (mm) | ||||
|---|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | Median | Range | CLSI breakpoints | |
| Ceftiofur | 53 (100%) | 0 (0%) | 0 (0%) | 38 | 26–48 | R ≤ 17; S ≥ 21 |
| Enrofloxacin | 53 (100%) | 0 (0%) | 0 (0%) | 32 | 24–42 | R ≤ 16; S ≥ 21 |
| Florfenicol | 53 (100%) | 0 (0%) | 0 (0%) | 40 | 30–50 | R ≤ 14; S ≥ 19 |
| Tilmicosin | 45 (85%) | 7 (13%) | 1 (2%) | 14 | 10–24 | R ≤ 10; S ≥ 14 |
| Tulathromycin | 41 (77%) | 12 (23%) | 0 (0%) | 20 | 16–28 | R ≤ 14; S ≥ 18 |
| Tetracycline | NA | NA | NA | 28 | 22–36 | NA |
S, susceptible; R, resistant; NA, not available; CLSI, Clinical and Laboratory Standards Institute.
MICs, percentages of resistance to each antimicrobial agent, and MIC50 and MIC90 values are shown in Table 2 . One of 53 (2%) isolates was resistant to tetracycline, with an MIC of 32 µg/mL, while all other isolates were susceptible to all antimicrobial agents tested.
Table 2.
Distribution of minimum inhibitory concentrations (MICs) of 53 Histophilus somni isolates.

a Isolates with an MIC result as a range have been rounded up.
b Lowest concentration of antimicrobial agent capable of inhibiting the growth of 50% of isolates.
c Lowest concentration of antimicrobial agent capable of inhibiting the growth of 90% of isolates.
d Percentage of resistance.
e MICs to the right of the solid vertical lines indicate breakpoints for resistance; MICs to the left of the dotted vertical lines indicate breakpoints for susceptibility.
f Only 43 H. somni could be revived for tulathromycin MIC testing.
There was complete agreement between the results of the disc diffusion and MIC methods for ceftiofur, enrofloxacin and florfenicol; all isolates were identified as susceptible with both methods. The isolate which exhibited tetracycline resistance in the MIC (32 µg/mL) had a corresponding disc diffusion of 22 mm (Fig. 1 ).
Fig. 1.

Comparison of disc diffusion and minimum inhibitory concentration (MIC) results of Australian isolates of Histophilus somni for (a) tetracycline, (b) tilmicosin and (c) tulathromycin. Solid line, resistant breakpoint; broken line, susceptible breakpoint. Isolates with a MIC value less than the lowest concentration tested have been given the value of the lowest concentration tested. Disc diffusion breakpoints for tetracycline are not available. Overlapping of data occurs at some points.
Using CLSI breakpoints for M. haemolytica, all H. somni isolates were susceptible to tilmicosin on MIC (Fig. 1). Seven isolates had intermediate susceptibility to tilmicosin by disc diffusion, with zone diameters of 12–13 mm (intermediate breakpoints 11–13 mm); these isolates had MIC values of 2–8 µg/mL (susceptible breakpoint ≤8 µg/mL). The one resistant isolate had a zone diameter of 10 mm (resistant breakpoint ≤10 mm) and a corresponding MIC of 8 µg/mL.
All 43 isolates tested were susceptible to tulathromycin on MIC testing (Fig. 1); 11/43 (26%) isolates had intermediate susceptibility to tulathromycin by disc diffusion, all with a zone diameter of 16 mm (intermediate breakpoints 15–17 mm). These isolates had MIC values of 4–16 µg/mL (susceptible breakpoint ≤16 µg/mL).
Clonal relationships
Using ERIC-PCR, 10 clusters were identified amongst the 53 H. somni isolates (Fig. 2 ). If four outlying clusters (clusters 1, 2, 9 and 10) were removed, the remaining isolates had a similarity level of >72% (Fig. 2). Twenty-seven of 52 (51%) isolates aligned with cluster 6; 15/27 (56%) isolates within cluster 6 belonged to clonal group 6.3. Cluster 8 included 7/53 (13%) isolates and cluster 4 included 6/53 (11%) isolates. The remaining eight isolates were distributed across three clusters, each with no more than four isolates.
Fig. 2.
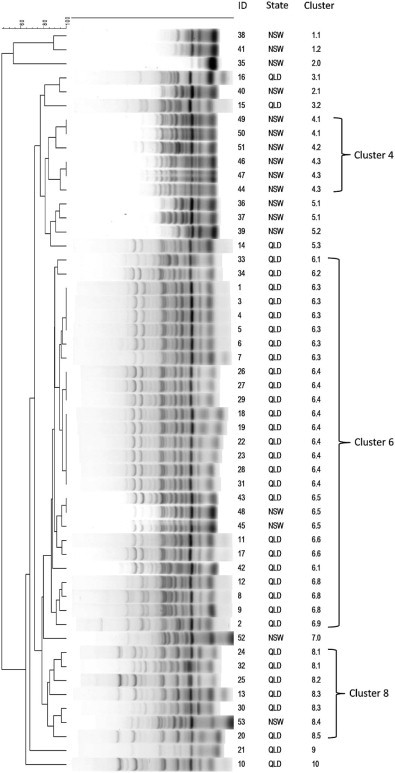
Dendrogram of enterobacterial repetitive intergenic consensus PCR fingerprint profiles of 53 Histophilus somni isolates from cattle in Australia. QLD, Queensland; NSW, New South Wales.
Epidemiology
Thirty-six H. somni isolates originated from cattle in Queensland and 17 isolates originated from cattle in New South Wales (Table 3 ). Four clusters contained isolates from both Queensland and New South Wales (clusters 3, 5, 6 and 8). Cluster 6 consisted predominately of Queensland isolates (24/27, 89%); the proportion of isolates from Queensland in cluster 6 was significantly higher than the proportion of isolates from Queensland in all the other clusters combined (P < 0.01). Isolates in cluster 6 were cultured from samples from 17 different regions; clonal group 6.3 contained only isolates from Queensland. Cluster 8 consisted mostly of Queensland isolates (6/7, 86%). Clusters 1 and 4 contained isolates exclusively from New South Wales (2 and 6 isolates, respectively). The tetracycline resistant isolate belonged to cluster 8. Most isolates (38/53, 72%) were cultured from the lungs and most isolates (41/53, 77%) were cultured from feedlot/meat cattle (Table 3). Of the 38 isolates cultured from the lungs, four of these animals were also infected with a viral respiratory pathogen; 13 samples tested negative for one or more viral pathogens, whereas 21 lung samples were not analysed. No patterns were apparent between cluster group and production type, sex, age, breed or introduction of an animal onto a property.
Table 3.
Distribution of isolates by category of epidemiological variables for all isolates, those from cluster 6 and cluster 8.
| Variable | Category | Number and percentage of isolates (n = 53) | Number cluster 6 (n = 27) | Number cluster 8 (n = 7) |
|---|---|---|---|---|
| State | Queensland | 36 (67.9%) | 24 (88.9%) | 6 (85.7%) |
| New South Wales | 17 (32.1%) | 3 (11.1%) | 1 (14.3%) | |
| Production | Meat/Feedlot | 41 (77.4%) | 23 (85.2%) | 6 (85.7%) |
| Dairy | 5 (9.4%) | 1 (3.7%) | 1 (14.3%) | |
| Unknown | 7 (13.2%) | 3 (11.1%) | 0 (0%) | |
| Sample site | Lung | 38 (71.7%) | 19 (70.4%) | 6 (85.7%) |
| Brain | 3 (5.7%) | 2 (7.4%) | 1 (14.3%) | |
| Nasal | 5 (9.4%) | 1 (3.7%) | 0 (0%) | |
| Other | 3 (5.7%) | 3 (11.1%) | 0 (0%) | |
| Unknown | 4 (7.5%) | 2 (7.4%) | 0 (0%) | |
| Year of isolation | 1989–1994 | 4 (7.5%) | 4 (14.8%) | 0 (0%) |
| 1995–2000 | 9 (17%) | 6 (22.2%) | 2 (28.6%) | |
| 2001–2005 | 9 (17%) | 7 (25.9%) | 2 (28.6%) | |
| 2006–2010 | 25 (47.2%) | 6 (22.2%) | 2 (28.6%) | |
| 2011–2012 | 2 (3.8%) | 1 (3.7%) | 1 (14.3%) | |
| Unknown | 4 (7.5%) | 3 (11.1%) | 0 (0%) | |
| Sex | Male | 13 (24.5%) | 8 (29.6%) | 1 (14.3%) |
| Female | 11 (20.8%) | 4 (14.8%) | 2 (28.6%) | |
| Unknown | 29 (54.7%) | 15 (55.6%) | 4 (42.9%) | |
| Origin | Introduced | 23 (43.4%) | 13 (48.2%) | 4 (57.1%) |
| Homebred | 7 (13.2%) | 5 (18.5%) | 0 (0%) | |
| Unknown | 23 (43.4%) | 9 (33.3%) | 3 (42.9%) | |
| Age (months) | 0–6 | 12 (22.6%) | 6 (22.2%) | 3 (42.8%) |
| 7–12 | 10 (18.9%) | 5 (18.5%) | 1 (14.3%) | |
| 13–18 | 15 (28.3%) | 7 (26%) | 2 (28.6%) | |
| 19–24 | 5 (9.4%) | 3 (11.1%) | 1 (14.3%) | |
| Unknown | 11 (20.8%) | 6 (22.2%) | 0 (0%) | |
| Other infections | IBRVa | 1 (1.8%) | 1 (3.7%) | 0 (0%) |
| Coronavirus | 2 (3.7%) | 1 (3.7%) | 0 (0%) | |
| Pestivirus | 1 (1.8%) | 0 (0%) | 1 (14.3%) | |
| Negativeb | 13 (24.5%) | 9 (33.3%) | 1 (14.3%) | |
| Not tested | 36 (67.9%) | 16 (59.3%) | 5 (71.4%) |
Infectious bovine rhinotracheitis virus.
Tested for at least one virus but all results were negative.
Discussion
Studies on BRD pathogens throughout the world, including Denmark (Aarestrup et al., 2004), Australia (Blackall et al., 2007), North America (Portis et al., 2012), Japan (Katsuda et al., 2009) and Canada (D'Amours et al., 2011), show that resistance to antimicrobial agents is increasing. The present study demonstrated that resistance against six antimicrobial agents in H. somni cultured from Australian cattle is either absent or extremely low.
This study utilised two widely accepted methods, disc diffusion and MIC, for determining antimicrobial susceptibility in H. somni isolates. The results of the two tests for tilmicosin and tulathromycin were not comparable for all isolates, since a small number of isolates had intermediate susceptibility or resistant zone sizes on disc diffusion which were determined to be susceptible by the MIC method. Caution is needed in the interpretation of tilmicosin and tulathromycin disc diffusion results for isolates displaying intermediate susceptibility or resistance in the absence of MIC results.
The finding that all isolates were susceptible to tilmicosin by MIC is supported by previous findings in another Australian study, in which all of 27 H. somni isolates tested were susceptible to tilmicosin (Blackall et al., 2007). A study in the United States investigating tilmicosin susceptibility over time (1994–2002) showed that H. somni isolates were consistently susceptible (Welsh et al., 2004). However, a later study from North America (2000–2009) identified a decrease in the susceptibility of H. somni to both tilmicosin and tulathromycin over time (Portis et al., 2012). One year prior to registration of tulathromycin in Northern America in 2004, 2–6% of BRD pathogens exhibited resistance and, by 2009, only 81% of H. somni remained susceptible (Portis et al., 2012). Therefore, continued surveillance should be a priority to detect any emergence of reduced susceptibility in H. somni.
In our study, one H. somni isolate was resistant to tetracycline by the MIC method. Resistance to tetracycline has been demonstrated in H. somni in North America by Portis et al. (2012), who observed a decrease in tetracycline susceptibility from 83% of isolates in 2000 to 47% in 2009. Tetracycline resistance has not previously been reported in Australian isolates of H. somni; however, with the detection of a highly resistant isolate in the present study (isolated in 2012), tetracycline susceptibility in H. somni should be closely monitored.
The 53 H. somni isolates formed 10 separate clusters, with the majority of isolates displaying high levels of similarity (Fig. 2). This supports previous studies suggesting there is limited genetic diversity in H. somni isolates and that the main mode of dispersal is clonal expansion (D'Amours et al., 2011). In our study, 51% of H. somni isolates belonged to cluster 6; within this cluster, clonal group 6.3 contained 56% of isolates. The isolates in cluster 6 were cultured from 1989 to 2011 and 85% were from cattle used for meat/feedlot production, but few conclusions can be drawn about the virulence potential of these isolates until further characterisation is performed.
While this study was able to demonstrate low levels of resistance in H. somni isolates tested against a panel of commercially available antimicrobial agents, there are certain limitations to the study design. The sample size (n = 53) was too small to be able to draw definitive conclusions based on epidemiological data. Data were limited to histories provided at the time of submission. Isolates were from diagnostic samples and therefore were submitted at the discretion of veterinary practitioners, and thus may not be representative of H. somni in the wider population of cattle.
Conclusions
This study demonstrated that most isolates of H. somni from cattle in Queensland and New South Wales are susceptible to antimicrobial agents that are most frequently used to treat BRD. MIC and disc diffusion data were generally comparable, with the exception of tilmicosin and tulathromycin. Identification of a H. somni isolate with tetracycline resistance from 2012 highlights the importance of continued surveillance to ensure early detection of any emerging resistance. Genotypic investigation into clonal lineages identified a major cluster (cluster 6) and a clonal group (clone 6.3) within this cluster.
Conflict of interest statement
None of the authors of this paper has a financial or personal relationship with other people or organisations that could inappropriately influence or bias the content of this paper.
Acknowledgements
The authors wish to thank the Animal Disease Surveillance Laboratory, Toowoomba, Queensland, and the Elizabeth Macarthur Agricultural Institute, Menangle, New South Wales, for data and isolates. This study was supported by grant B.FLT.0224 from Meat and Livestock Australia, with matching funds provided by the Australian Government. Preliminary results were presented at the 8th Annual Meeting of the Australian Association of Veterinary Laboratory Diagnosticians, Bundoora, Victoria, Australia, 23–23 November 2012 and at ‘Microbiology in Maleny’, Queensland Branch of the Australian Society of Microbiology, Maleny, Queensland, Australia, 24 November 2012.
Footnotes
See: http://epitools.ausvet.com.au (accessed 1 December 2014).
References
- Aarestrup F.M., Seyfarth A.M., Angen O. Antimicrobial susceptibility of Haemophilus parasuis and Histophilus somni from pigs and cattle in Denmark. Veterinary Microbiology. 2004;101:143–146. doi: 10.1016/j.vetmic.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Andrews J.M. Determination of minimum inhibitory concentrations. Journal of Antimicrobial Chemotherapy. 2001;48(Suppl. 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- Angen O., Ahrens P., Tegtmeier C. Development of a PCR test for identification of Haemophilus somnus in pure and mixed cultures. Veterinary Microbiology. 1998;63:39–48. doi: 10.1016/s0378-1135(98)00222-3. [DOI] [PubMed] [Google Scholar]
- Barton M., Pratt R., Hart W.S. Antibiotic resistance in animals. Communicable Diseases Intelligence Quarterly Report. 2003;27(Suppl.):121–126. [PubMed] [Google Scholar]
- Blackall P.J., O'Dell R., Stephens C.P. Minimal inhibitory concentration of tilmicosin against isolates of Histophilus somni from Australian cattle. Australian Veterinary Journal. 2007;85:503–504. doi: 10.1111/j.1751-0813.2007.00230.x. [DOI] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute . Fourth Ed. CLSI Document VET01-A4. Clinical Laboratory Standards Institute; Wayne, Pennsylvania, USA: 2013. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard. [Google Scholar]
- Corbeil L.B. Histophilus somni host-parasite relationships. Animal Health Research Reviews. 2007;8:151–160. doi: 10.1017/S1466252307001417. [DOI] [PubMed] [Google Scholar]
- D'Amours G.H., Ward T.I., Mulvey M.R., Read R.R., Morck D.W. Genetic diversity and tetracycline resistance genes of Histophilus somni. Veterinary Microbiology. 2011;150:362–372. doi: 10.1016/j.vetmic.2011.02.051. [DOI] [PubMed] [Google Scholar]
- Hendriksen R.S., Mevius D.J., Schroeter A., Teale C., Meunier D., Butaye P., Franco A., Utinane A., Amado A., Moreno M. Prevalence of antimicrobial resistance among bacterial pathogens isolated from cattle in different European countries: 2002–2004. Acta Veterinaria Scandinavica. 2008;50:28–38. doi: 10.1186/1751-0147-50-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuda K., Kohmoto M., Mikami O., Uchida I. Antimicrobial resistance and genetic characterization of fluoroquinolone-resistant Mannheimia haemolytica isolates from cattle with bovine pneumonia. Veterinary Microbiology. 2009;139:74–79. doi: 10.1016/j.vetmic.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Portis E., Lindeman C., Johansen L., Stoltman G. A ten-year (2000–2009) study of antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex – Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni – in the United States and Canada. Journal of Veterinary Diagnostic Investigation. 2012;24:932–944. doi: 10.1177/1040638712457559. [DOI] [PubMed] [Google Scholar]
- Sackett D., Holmes P., Abbott K., Jephcott S., Barber M. Assessing the economic cost of endemic disease on profitability of Australian beef cattle and sheep producers. 2007. http://www.mla.com.au/Research-and-development/Search-RD-reports/RD-report-details/Animal-Health-and-Biosecurity/Assessing-the-economic-cost-of-endemic-disease-on-the-profitability-of-Australian-beef-cattle-and-sheep-producers/120 Meat and Livestock Australia, Project AHW.087. accessed 7 May 2014.
- Sandal I., Inzana T.J. A genomic window into the virulence of Histophilus somni. Trends in Microbiology. 2010;18:90–99. doi: 10.1016/j.tim.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Sandal I., Hong W., Swords W.E., Inzana T.J. Characterization and comparison of biofilm development by pathogenic and commensal isolates of Histophilus somni. Journal of Bacteriology. 2007;189:8179–8185. doi: 10.1128/JB.00479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J.R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Research. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh R.D., Dye L.B., Payton M.E., Confer A.W. Isolation and antimicrobial susceptibilities of bacterial pathogens from bovine pneumonia: 1994–2002. Journal of Veterinary Diagnostic Investigation. 2004;16:426–431. doi: 10.1177/104063870401600510. [DOI] [PubMed] [Google Scholar]


