Abstract
Highly pathogenic influenza viruses continue to cause serious threat to public health due to their pandemic potential, calling for an urgent need to develop effective, safe, convenient, and universal vaccines against influenza virus infection. In this study, we constructed two recombinant protein vaccines, 2H5M2e-2H7M2e-H5FP-H7FP (hereinafter M2e-FP-1) and 2H5M2e-H5FP-2H7M2e-H7FP (hereinafter M2e-FP-2), by respectively linking highly conserved sequences of two molecules of ectodomain of M2 (M2e) and one molecule of fusion peptide (FP) epitope of hemagglutinin (HA) of H5N1 and H7N9 influenza viruses in different orders. The Escherichia coli-expressed M2e-FP-1 and M2e-FP-2 proteins induced similarly high-titer M2e-FP-specific antibodies in the immunized mice. Importantly, both proteins were able to prevent lethal challenge of heterologous H1N1 influenza virus, with significantly reduced viral titers and alleviated pathological changes in the lungs, as well as increased body weight and complete survivals, in the challenge mice. Taken together, our study demonstrates that highly conserved M2e and FP epitope of HA of H5N1 and H7N9 influenza viruses can be used as important targets for development of safe and economical universal influenza vaccines, and that the position of H7N9 M2e and H5N1 HA epitope sequences in the vaccine components has no significant effects on the immunogenicity and efficacy of M2e-FP-based subunit vaccines.
Keywords: Influenza virus, M2e, Hemagglutinin fusion peptide, Universal vaccines, Protection
Abbreviations: CPE, cytopathic effect; HA-FP, hemagglutinin fusion peptide; IPTG, isopropyl-β-d-thiogalactopyranoside; M2e, extracellular domain of M2; TCID50, 50% tissue culture infectious dose
1. Introduction
Influenza is one of the important emerging infectious diseases threatening public health worldwide. Influenza vaccines are required annually to immunize a large number of people to prevent influenza. The highly pathogenic avian influenza viruses H5N1 and H7N9, as well as 2009 pandemic H1N1 influenza virus, have infected humans and caused sporadic cases or epidemics [1], [2], [3], [4], [5], [6], [7], [8], [9], calling for continuous efforts to develop vaccines to prevent the threat of influenza. Current influenza vaccines are mainly strain-specific, targeting one or more closely related strains, and having limited efficacy to prevent divergent and new viral strains [10], [11], [12]. Thus, development of effective universal vaccines with cross-protective efficacy against variant influenza viruses has been the focus of influenza researches.
The extracellular domain of M2 (M2e) protein of influenza virus is highly conserved among all influenza A viruses, thus serving as a promising target for developing universal influenza vaccines [13], [14]. However, containing only 24 amino acids, the M2e molecule is a poor inducer of immune response, elucidating the need to improve its immunogenicity in designing influenza vaccines. A number of approaches, including conjugating M2e with other carrier proteins, such as Neisseria meningitidis outer membrane complex (OMPC) and bacterial flagellin, or increasing the copy numbers of M2e molecules, have been introduced to increase the immunogenicity and/or protective efficacy of M2e-based vaccines [15], [16], [17], [18], [19]. In addition to M2e, fusion peptide (FP) epitope in the hemagglutinin 2 (HA2) stem region of influenza virus is also highly conserved in influenza A viruses, making it as another important vaccine target [14], [20], [21]. Chimeric vaccines or synthetic peptides containing conserved HA2 sequences may induce cross-reactive and cross-protective antibodies against divergent influenza viruses [22], [23], [24], constituting a key approach to develop universal influenza vaccines.
In this study, we designed two recombinant vaccines, 2H5M2e-2H7M2e-H5FP-H7FP (hereinafter M2e-FP-1) and 2H5M2e-H5FP-2H7M2e-H7FP (hereinafter M2e-FP-2), respectively containing highly conserved sequences of two M2e molecules and one HA-FP molecule of H5N1 and H7N9 influenza viruses in two different orders, and expressed the proteins in Escherichia coli expression system. We further evaluated the immunogenicity and cross-protective efficacy of these two subunit vaccines against a heterologous strain of H1N1 influenza virus. This study demonstrates that the position of H7N9 M2e and H5N1 HA epitope sequences in the vaccine components has no significant impact on the immunogenicity and efficacy of M2e-FP-based subunit vaccines, suggesting the potential to develop these proteins as effective and economical universal influenza vaccines.
2. Materials and methods
2.1. Ethics statement
Female BALB/c mice at 6–8 weeks old were used in the study. The animal studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal protocols were approved by the Committees on the Ethics of Animal Experiments of the Beijing Institute of Microbiology and Epidemiology (Permit Number: PMB15-0012), and the New York Blood Center (Permit Number: 322.06).
2.2. Construction of recombinant vaccines
The construction of recombinant vaccines containing M2e and HA-FP of H5N1 and H7N9 influenza viruses was carried out as previously described [25]. Briefly, genes encoding the highly conserved sequences of two molecules of H5N1 M2e and one molecule of HA-FP, as well as two molecules of H7N9 M2e and one molecule of HA-FP, in two orders were synthesized, inserted into pQE30 E. coli Expression Vector, and sequenced for corrected sequences. A linker sequence encoding GGGGS was added between each molecule, and a His6 tag was added at the C-terminus for easy purification of expressed recombinant proteins.
2.3. Recombinant protein expression and purification
The recombinant M2e-FP-1 and M2e-FP-2 proteins were expressed in the E. coli expression system as previously described [25], [26] with some modifications. Briefly, the aforementioned recombinant plasmids encoding M2e-FP-1 and M2e-FP-2 proteins were transformed into E. coli (M15), cultured in LB medium overnight at 37 °C, diluted (1:100), and then continuously cultured until the OD600 value reaching 0.6–1.0. Isopropyl-β-d-thiogalactopyranoside (IPTG, Sigma) was added at the final concentration of 1 mM, and the E. coli was centrifuged to collect pellets. The sonicated E. coli supernatant containing the expressed proteins was collected, and purified using Ni-chromatography (Promega) and gel filtration chromatography. The purified recombinant proteins were concentrated, analyzed for endotoxin, and used for further studies.
2.4. SDS-PAGE and Western blot
The recombinant M2e-FP-1 and M2e-FP-2 proteins were analyzed by SDS-PAGE and Western blot as previously described [27], [28]. Briefly, the IPTG-induced proteins and non-induced controls were boiled for 10 min, separated by 10% Tris–Glycine gels, and stained by Coomassie Brilliant Blue for SDS-PAGE analysis. The same samples were also transferred to nitrocellulose membranes for Western blot analysis by blocking the blots using 5% fat-free milk in PBST overnight at 4 °C, and sequentially incubating them for 1 h at room temperature with influenza virus M2e-specific polyclonal antibodies (1:1000) (laboratory stock), and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG antibody (1:5,000, Invitrogen). The signals were then detected using ECL substrate buffer (Peirce) and Amersham Hyperfilm.
2.5. Mouse immunization and virus challenge
A/PR/8/34(H1N1) influenza virus was grown in the allantoic cavities of 9-day-old embryonated chicken eggs. Virus-containing allantoic fluid was harvested after 72 h, and stored at −80 °C until use. Mice were immunized with recombinant proteins and challenged with influenza virus using previously described protocols [27], [29] with some modifications. Briefly, BALB/c mice were intramuscularly (i.m.) immunized with M2e-FP-1 or M2e-FP-2 (10 μg/50 μl/mouse) in the presence of aluminum adjuvant, and boosted twice at a 3-week interval. Mice injected with PBS plus above adjuvant were included as a negative control. Sera were collected before each vaccination, and measured for influenza virus M2e-FP-specific IgG, IgG1, and IgG2a antibodies. One month post-last immunization, mice were challenged with A/PR/8/34(H1N1) influenza virus (103 50% tissue culture infectious dose: TCID50). The infected mice were then detected for pathological changes by H&E staining and viral titers in lung tissues at 5 days post-infection (p.i.), or observation of survivals and weight changes for 14 days p.i.
2.6. ELISA
ELISA was used to analyze influenza virus M2e-FP-specific antibody responses in the immunized mouse sera as previously described [27], [30] with some modifications. Briefly, ELISA plates were coated with recombinant M2e-FP proteins overnight at 4 °C, and blocked with 3% BSA-PBS for 2 h at 37 °C. Mouse sera at 2-serial dilutions were added to the plates, and incubated for 1 h at 37 °C. After 4 washes, the plates were incubated with HRP-conjugated anti-mouse IgG, anti-mouse IgG1, and anti-mouse IgG2a (1:5,000, Santa Cruz), respectively, for 1 h at 37 °C. The reaction was detected by 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Invitrogen), and stopped by 1 N H2SO4. The absorbance at 450 nm was measured using ELISA plate reader, and the related antibody titers were calculated.
2.7. Viral titer detection
Viral titers in lung tissues of the above challenged mice were detected as previously described [25], [31] with some modifications. Briefly, lung tissues were homogenized in minimal essential medium (MEM) to achieve 10% (w/v) suspensions. Serially diluted samples were added to monolayers of Madin–Darby canine kidney (MDCK) cells seeded in 96-well tissue culture plates, and incubated for 2 h at 37 °C. Supernatants were removed and replaced with fresh MEM, followed by incubation of MDCK cells for 72 h. Viral cytopathic effect (CPE) was observed daily, and viral titers were calculated by HA test. For the HA test, 50 μl of 0.5% turkey red blood cells (Lampire Biological Laboratories) was added to 50 μl of cell culture supernatant, and incubated at room temperature for 30 min. Wells containing HA were scored as positive. The viral titer was calculated by the Reed and Muench method, and expressed as Log10 TCID50/g of lung tissues.
2.8. Statistical analysis
The values are presented as mean with standard deviation (SD). Statistical significance was calculated by Student's t-test using GraphPad Prism statistical software. *** indicates P < 0.001.
3. Results
3.1. Construction and characterization of recombinant proteins containing highly conserved M2e and HA-FP epitope of H5N1 and H7N9 influenza viruses
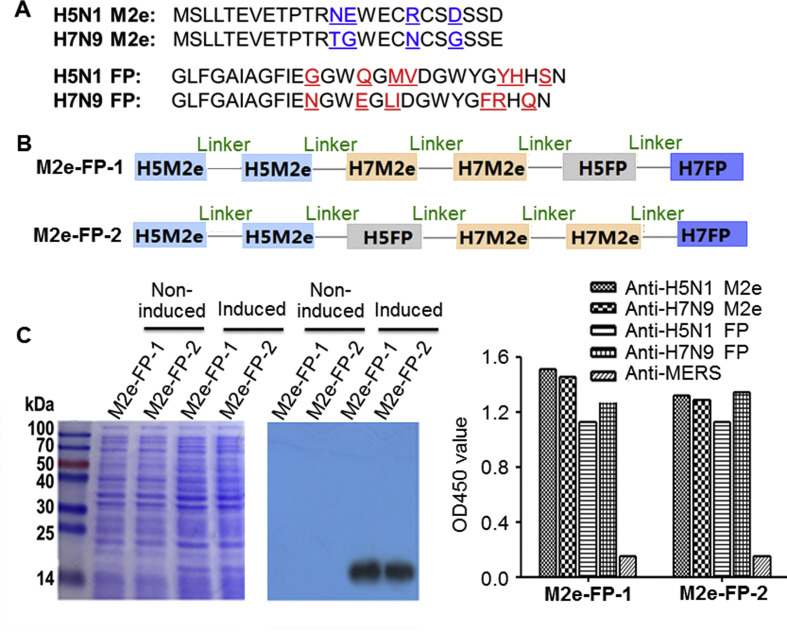
The amino acid sequences of M2e and HA-FP of H5N1, H7N9, H1N1, and H3N2 influenza viruses are highly conserved among all influenza A viruses, maintaining at least 90% homology [14]. Here, we constructed two recombinant proteins, designated M2e-FP-1 and M2e-FP-2, by respectively linking two M2e molecules and one FP epitope of HA protein of H5N1 and H7N9 influenza viruses in different orders (Fig. 1 A–B), and expressed the fusion proteins in E. coli expression system, a convenient and economical system for expressing recombinant proteins with manufacture capacity [32], [33], [34]. The proteins with the expected molecular weight size were expressed in the soluble form of the induced E. coli (Fig. 1C, left), and purified with 98.6% purity, which were recognized by M2e-specific monoclonal antibodies developed in our laboratories, while no such bands were detected in the non-induced cell controls (Fig. 1C, middle). The purified M2e-FP-1 and M2e-FP-2 proteins were used as coating antigens to detect their reactivity with sera of mice immunized with M2e or FP peptides of influenza virus H5N1 and H7N9. The results showed that these recombinant proteins reacted specifically with the respective sera, but not with the control sera (Fig. 1C, right). These data suggest that the constructed M2e and HA-FP fusion proteins were successfully expressed and maintained strong specificity to influenza virus.
Fig. 1.
Construction and expression of recombinant proteins. (A) Amino acid sequences of H5N1 and H7N9 M2e and FP of HA2 proteins. Amino acids with differences in M2e of H5N1 and H7N9 influenza viruses were shown in blue, and those in FP of H5N1 and H7N9 influenza viruses were in red. (B) Schematic structure of constructed M2e-FP-1 and M2e-FP-2 proteins. Linker sequences, GGGGS. (C) SDS-PAGE (left) and Western blot analysis (middle) of expressed M2e-FP-1 and M2e-FP-2 proteins. The protein molecular weight marker (kDa) is shown on the left. Anti-M2e antibodies (1:1000) were used for the Western blot detection. Shown on the right is the ELISA result for detection of the reactivity of M2e-FP-1 and M2e-FP-2 proteins with sera of mice immunized with M2e or FP peptides of H5N1 and H7N9 influenza viruses (1:2000). Sera of mice immunized with Middle East respiratory syndrome coronavirus (anti-MERS) were included as control.
3.2. Immunogenicity of recombinant proteins containing highly conserved M2e and HA-FP epitope of H5N1 and H7N9 influenza viruses
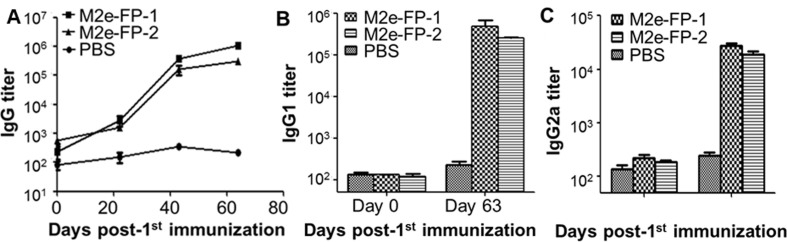
To evaluate the immunogenicity of the M2e and HA-FP fusion proteins, we immunized mice using the two proteins following the protocol described in Materials and Methods, and detected the resulting antibody responses in the mouse sera. As shown in Fig. 2 A, both proteins were able to elicit similarly levels of potent M2e-FP-specific IgG antibodies, increased to a higher titer after each boost, and maintained for at least 9 weeks during the detection period, whereas only a background level of antibodies were detected in the sera of the control mice. Further evaluation of antibody subtypes revealed that high titers of M2e-FP-specific IgG2a (Th1), particularly IgG1 (Th2), antibodies were induced in the sera of protein-immunized mice collected at 9 weeks post-immunization, but not in those of the control mice injected with PBS (Fig. 2B–C). The above results indicate that M2e-FP-1 and M2e-FP-2 maintained strong immunogenicity in inducing long-term specific antibodies, and that both fusion proteins elicited a relatively higher titer of Th2 (IgG1)-biased antibody response, thus potentially promoting the production of antibody-mediated humoral immune response.
Fig. 2.
Antibody responses in sera of mice immunized with M2e-FP-1 and M2e-FP-2 proteins. Mice were immunized with M2e-FP-1 and M2e-FP-2 proteins, or PBS as a control, and sera were collected at the indicated time points post-immunization to detect M2e-FP-specific IgG (A), IgG1 (B), and IgG2a (C) antibodies by ELISA. The antibody titers were expressed as the endpoint dilutions that remain positively detectable, and presented as mean ± SD of 5 mice in each group.
3.3. Recombinant proteins containing highly conserved M2e and HA-FP epitope of H5N1 and H7N9 influenza viruses significantly reduced viral replication and pathological changes
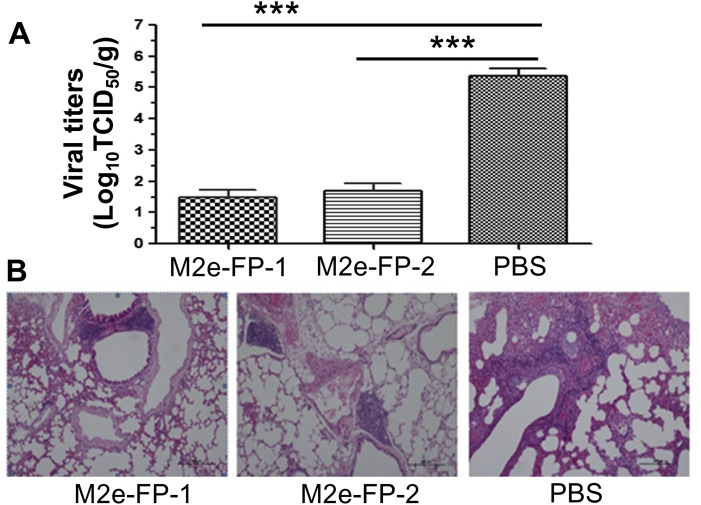
Having confirmed the immunogenicity of M2e-FP1 and M2e-FP-2 fusion proteins, next we investigated their protective immunity by challenging the immunized mice with a heterologous strain of A/PR/8/34(H1N1) influenza virus, and detected viral replication and pathological damage in the lungs at 5 days post-infection. As expected, mice immunized with the two fusion proteins had significantly lower titers of virus in their lungs than those of the control mice receiving PBS (Fig. 3 A). Analysis of the pathological changes demonstrated that both fusion proteins effectively protected mice against H1N1 infection, with obviously alleviated lung damage and the presence of BALT (bronchus-associated lymphoid tissue) surrounding the lung bronchial tube (Fig. 3B, left and middle), a characteristic helping for restoring lung tissue cells. In contrast, there was serious mesenchymal inflammation in the lung tissue of the control mice, which was represented by thickened alveolar walls, inflammatory cell infiltration, hemorrhage, and vessel inflammation (Fig. 3B, right). The above data suggest that the two fusion proteins had strong ability to inhibit viral replication and reduce virus-caused tissue damage.
Fig. 3.
Viral titers and histopathological changes in lung tissues of challenged mice. The mice immunized with M2e-FP-1 and M2e-FP-2 proteins, respectively, or PBS as a control were challenged with A/PR/8/34(H1N1) influenza virus (103 TCID50), and collected for lung tissues at 5 days p.i. to detect viral titers (A) and histopathological changes (B). The data in (A) are presented as mean ± SD of 5 mice in each group. ***P < 0.001. For (B), Representative images from lung tissues of immunized mice and control mice are shown. The tissue sections were stained by H&E straining, and observed for pathological damages under light microscopy (10× magnification).
3.4. Recombinant proteins containing highly conserved M2e and HA-FP epitope of H5N1 and H7N9 influenza viruses completely protected against heterologous challenge
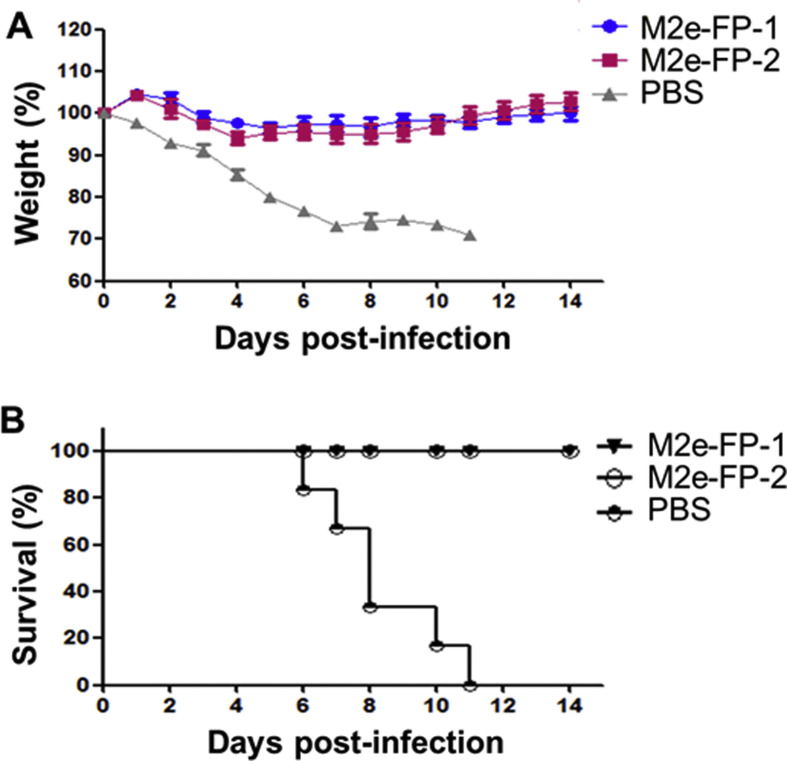
The protective efficacy of M2e-FP fusion proteins were further evaluated by challenging the immunized mice with A/PR/8/34(H1N1) influenza virus, and recording mouse survivals and weight changes for up to 14 days post-challenge. The results demonstrated that both M2e-FP-1 and M2e-FP-2 significantly prevented weight loss of the mice after viral challenge, with slightly reducing weights during 3–5 days, and then steadily increasing weights at 6 days, post-infection. By comparison, the control mice had a continuous weight loss (Fig. 4 A). In addition, all mice immunized with the two fusion proteins survived from influenza virus challenge at the end of the detection period, whereas none of the control mice injected with PBS survived from such challenge, all of which died by 11 days post-infection (Fig. 4B). These results confirm the protective efficacy of the test influenza vaccines in providing complete protection against infection of heterologous strain of influenza virus, suggesting that the position of H7N9 M2e and H5N1 HA-FP epitope sequences in the M2e-FP-based subunit vaccines had no significant impact on the protection.
Fig. 4.
Survivals and weights of immunized mice challenged with influenza virus. The mice immunized with M2e-FP-1 and M2e-FP-2 proteins, respectively, or PBS as a control were challenged with A/PR/8/34(H1N1) influenza virus (103 TCID50), observed for 14 days p.i., and calculated for percentages of weights (A) and survivals (B). The data in (A) are presented as mean ± SD of 6 mice in each group.
4. Discussion
The highly conserved influenza virus M2e and FP of HA2 can be used as effective vaccine targets to develop universal influenza vaccines with broad protective efficacy to prevent against divergent viral strains [12], [14], [35]. Indeed, we have previously indicated that a tetra-branched multiple antigenic peptide (MAP)-based vaccine (H5N1-M2e-MAP) carrying four copies of H5N1 M2e peptide conferred cross-protection against lethal infection of two clades of H5N1 and one pandemic 2009 H1N1 influenza viruses [31], [36]. We have also demonstrated the ability of a recombinant fusion protein (M2e3-ASP-1) containing three conserved H5N1 M2e molecules and a new Onchocerca volvulus activation associated protein-1 (ASP-1) adjuvant in cross-protection against two divergent strains of H5N1 influenza viruses [25]. In addition, H5N1 M2e peptide-based vaccines were developed to test the cross-protection of H5N1 M2e against infection of the new H7N9 virus, and the H5N1 M2e vaccination provided potent cross-protection against lethal challenge of H7N9 virus [37], [38]. In this study, we designed two recombinant universal influenza vaccines by respectively linking two molecules of M2e and one molecule of HA-FP sequences of H5N1 and H7N9 in two different orders, and evaluated their immunogenicity and protection against a heterologous H1N1 strain. The sequences of M2e and HA-FP used in this study are highly conserved among all influenza A virus strains, maintaining over 90% homology. As expected, both proteins linked through two different orders of M2e and FP molecules were able to induce effective antibody responses and cross-protection against lethal challenge of A/PR/8/34(H1N1) influenza virus with 100% survival rate, indicating that H5N1 and H7N9 M2e and HA-FP recombinant protein-based universal vaccines have capacity to elicit sufficient cross-protective efficacy against infection of heterologous strain of H1N1 influenza virus.
We have also found that while the two proteins, in the presence of aluminum adjuvant, elicited high titers of antibody response in the immunized mice, they failed to induce effective cellular immune response (data not shown), suggesting that M2e and HA-FP recombinant protein-induced protection might be mainly through humoral, rather than cellular, immune response-mediated immunity. Studies have shown that the mechanisms of cross-protection by influenza virus M2e-based vaccines may be through antibody-dependent cell-mediated cytotoxicity (ADCC) or complement-dependent cytolysis (CDC) [39], [40]. In addition to M2e molecules, our recombinant proteins also contain HA-FP conserved epitope sequences. Future studies are warranted to further elucidate the protective mechanisms of M2e and HA-FP-based recombinant vaccines, as well as to detect their efficacy in protecting against multiple current and future strains of influenza virus with pandemic potential.
Overall, this study suggests that simultaneous expression of M2e molecule and HA-FP peptide from several important influenza virus strains may help facilitate the development of universal vaccines with broad-spectrum cross-protective efficacy against divergent heterologous strains of influenza viruses, and that the order of M2e and HA-FP in the vaccine components has no significant effects on the immunogenicity and protective efficacy. Our data demonstrate the potential for further development of a universal influenza vaccine based on highly conserved sequences of M2e and HA-FP epitope of influenza virus.
Conflict of interest statement
The authors declared no conflict of interest.
Acknowledgments
This study was supported by Research Grant from Ministry of Health of China (AWS15J006), NIH grant (R21AI111152), and the intramural fund of New York Blood Center (NYB000414).
Contributor Information
Lanying Du, Email: ldu@nybc.org.
Yusen Zhou, Email: Yszhou@bmi.ac.cn.
References
- 1.Fineberg H.V. Pandemic preparedness and response – lessons from the H1N1 influenza of 2009. N Engl J Med. 2014;370:1335–1342. doi: 10.1056/NEJMra1208802. [DOI] [PubMed] [Google Scholar]
- 2.Rewar S., Mirdha D., Rewar P. Treatment and prevention of pandemic H1N1 influenza. Ann Glob Health. 2015;81:645–653. doi: 10.1016/j.aogh.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Zhou L., Tan Y., Kang M., Liu F., Ren R., Wang Y. Preliminary epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus, China, 2017. Emerg Infect Dis. 2017;23:1355–1359. doi: 10.3201/eid2308.170640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X., Jiang H., Wu P., Uyeki T.M., Feng L., Lai S. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis. 2017;17:822–832. doi: 10.1016/S1473-3099(17)30323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang M., Lau E.H.Y., Guan W., Yang Y., Song T., Cowling B.J. Epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus in Guangdong, 2016 to 2017. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.27.30568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paratus Business News . 22 March 2017. As deadly human bird flu cases surge and new U.S. cases are discovered, world pandemic fears grow; how to prepare.http://paratusnews.com/as-bird-flu-cases-surge-among-humans-and-new-u-s-cases-are-discovered-world-pandemic-fears-grow-how-to-prepare/ [Google Scholar]
- 7.World Health Organization . World Health Organization; Geneva, Switzerland: 19 July 2017. Human infection with avian influenza A(H7N9) virus – China.http://www.who.int/csr/don/19-july-2017-ah7n9-china/en/ [Google Scholar]
- 8.World Health Organization . World Health Organization; Geneva, Switzerland: 15 June 2017. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2017.http://www.who.int/influenza/human_animal_interface/2017_06_15_tableH5N1.pdf [Google Scholar]
- 9.Normile D. Science; 17 Feb. 2017. Bird flu strain taking a toll on humans.http://www.sciencemag.org/news/2017/02/bird-flu-strain-taking-toll-humans [Google Scholar]
- 10.Houser K., Subbarao K. Influenza vaccines: challenges and solutions. Cell Host Microbe. 2015;17:295–300. doi: 10.1016/j.chom.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krammer F., Garcia-Sastre A., Palese P. Is it possible to develop “universal” influenza virus vaccine? Toward a universal influenza virus vaccine: potential target antigens and critical aspects for vaccine development. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a028845. pii: a028845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soema P.C., Kompier R., Amorij J.P., Kersten G.F. Current and next generation influenza vaccines: formulation and production strategies. Eur J Pharm Biopharm. 2015;94:251–263. doi: 10.1016/j.ejpb.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 13.Fiers W., De Filette M., El Bakkouri K., Schepens B., Roose K., Schotsaert M. M2e-based universal influenza A vaccine. Vaccine. 2009;27:6280–6283. doi: 10.1016/j.vaccine.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Du L., Zhou Y., Jiang S. Research and development of universal influenza vaccines. Microbes Infect. 2010;12:280–286. doi: 10.1016/j.micinf.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Deng L., Kim J.R., Chang T.Z., Zhang H., Mohan T., Champion J.A. Protein nanoparticle vaccine based on flagellin carrier fused to influenza conserved epitopes confers full protection against influenza A virus challenge. Virology. 2017;509:82–89. doi: 10.1016/j.virol.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mardanova E.S., Kotlyarov R.Y., Kuprianov V.V., Stepanova L.A., Tsybalova L.M., Lomonossoff G.P. High immunogenicity of plant-produced candidate influenza vaccine based on the M2e peptide fused to flagellin. Bioengineered. 2016;7:28–32. doi: 10.1080/21655979.2015.1126017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X., Wang Y., Dong C., Hu J., Yang L. High copy numbers and N terminal insertion position of influenza A M2e fused with hepatitis B core antigen enhanced immunogenicity. Biosci Trends. 2015;9:221–227. doi: 10.5582/bst.2015.01060. [DOI] [PubMed] [Google Scholar]
- 18.Stepanova L.A., Kotlyarov R.Y., Kovaleva A.A., Potapchuk M.V., Korotkov A.V., Sergeeva M.V. Protection against multiple influenza A virus strains induced by candidate recombinant vaccine based on heterologous M2e peptides linked to flagellin. PLoS One. 2015;10:e0119520. doi: 10.1371/journal.pone.0119520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravin N.V., Blokhina E.A., Kuprianov V.V., Stepanova L.A., Shaldjan A.A., Kovaleva A.A. Development of a candidate influenza vaccine based on virus-like particles displaying influenza M2e peptide into the immunodominant loop region of hepatitis B core antigen: insertion of multiple copies of M2e increases immunogenicity and protective efficiency. Vaccine. 2015;33:3392–3397. doi: 10.1016/j.vaccine.2015.04.066. [DOI] [PubMed] [Google Scholar]
- 20.Crowe J.E., Jr. Is it possible to develop a “universal” influenza virus vaccine? Potential for a universal influenza vaccine. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a029496. pii: a029496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angeletti D., Yewdell J.W. Is it possible to develop a “universal” influenza virus vaccine? Outflanking antibody immunodominance on the road to universal influenza vaccination. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a028852. pii: a028852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong X., Yin H., Shi Y., He X., Yu Y., Guan S. Evaluation of the immunogenicity and protective effects of a trivalent chimeric norovirus P particle immunogen displaying influenza HA2 from subtypes H1, H3 and B. Emerg Microbes Infect. 2016;5:e51. doi: 10.1038/emi.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T.T., Tan G.S., Hai R., Pica N., Ngai L., Ekiert D.C. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci U S A. 2010;107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellebedy A.H., Krammer F., Li G.M., Miller M.S., Chiu C., Wrammert J. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci U S A. 2014;111:13133–13138. doi: 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao G., Du L., Xiao W., Sun S., Lin Y., Chen M. Induction of protection against divergent H5N1 influenza viruses using a recombinant fusion protein linking influenza M2e to Onchocerca volvulus activation associated protein-1 (ASP-1) adjuvant. Vaccine. 2010;28:7233–7240. doi: 10.1016/j.vaccine.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 26.Du L., Zhao G., Chan C.C., Sun S., Chen M., Liu Z. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology. 2009;393:144–150. doi: 10.1016/j.virol.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tai W., Wang Y., Fett C.A., Zhao G., Li F., Perlman S. Recombinant receptor-binding domains of multiple middle east respiratory syndrome coronaviruses (MERS-CoVs) induce cross-neutralizing antibodies against divergent human and camel MERS-CoVs and antibody escape mutants. J Virol. 2017;91 doi: 10.1128/JVI.01651-16. e01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du L., Tai W., Yang Y., Zhao G., Zhu Q., Sun S. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat Commun. 2016;7 doi: 10.1038/ncomms13473. 13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai W., Zhao G., Sun S., Guo Y., Wang Y., Tao X. A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology. 2016;499:375–382. doi: 10.1016/j.virol.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang N., Channappanavar R., Ma C., Wang L., Tang J., Garron T. Identification of an ideal adjuvant for receptor-binding domain-based subunit vaccines against middle east respiratory syndrome coronavirus. Cell Mol Immunol. 2016;13:180–190. doi: 10.1038/cmi.2015.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao G., Lin Y., Du L., Guan J., Sun S., Sui H. An M2e-based multiple antigenic peptide vaccine protects mice from lethal challenge with divergent H5N1 influenza viruses. Virol J. 2010;7:9. doi: 10.1186/1743-422X-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heikkila J. What intravenous treatment for myocardial infarction: beta-blockers, nitrates or thrombolysis? Duodecim. 1990;106:79–98. [PubMed] [Google Scholar]
- 33.Rosano G.L., Ceccarelli E.A. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia B., Jeon C.O. High-throughput recombinant protein expression in Escherichia coli: current status and future perspectives. Open Biol. 2016;6 doi: 10.1098/rsob.160196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nachbagauer R., Krammer F. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect. 2017;23:222–228. doi: 10.1016/j.cmi.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao G., Sun S., Du L., Xiao W., Ru Z., Kou Z. An H5N1 M2e-based multiple antigenic peptide vaccine confers heterosubtypic protection from lethal infection with pandemic 2009 H1N1 virus. Virol J. 2010;7:151. doi: 10.1186/1743-422X-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung H.C., Chan C.C., Poon V.K., Zhao H.J., Cheung C.Y., Fai N. An H5N1-based matrix protein 2 ectodomain tetrameric peptide vaccine provides cross-protection against lethal infection with H7N9 influenza virus. Emerg Microbes Infect. 2015;4:e22. doi: 10.1038/emi.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasan N., Ebrahimie E., Ignjatovic J., Tarigan S., Peaston A., Hemmatzadeh F. Epitope mapping of avian influenza M2e protein: different species recognise various E. PLoS One. 2016;11:e0156418. doi: 10.1371/journal.pone.0156418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y.N., Kim M.C., Lee Y.T., Kim Y.J., Kang S.M. Mechanisms of cross-protection by influenza virus M2-based vaccines. Immune Netw. 2015;15:213–221. doi: 10.4110/in.2015.15.5.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolpe A., Schepens B., Fiers W., Saelens X. M2-based influenza vaccines: recent advances and clinical potential. Expert Rev Vaccines. 2017;16:123–136. doi: 10.1080/14760584.2017.1240041. [DOI] [PubMed] [Google Scholar]