Abstract
Little information is available on persistent infection of severe acute respiratory syndrome (SARS) coronavirus (CoV). In this study, we established persistent infection of SARS-CoV in the Vero E6 cell line. Acute infection of Vero E6 with SARS-CoV produced a lytic infection with characteristic rounding cytopathic effects (CPE) and the production of a large number of infectious particles in the culture fluid within 3 days post-infection. Upon subsequent culturing of the remaining adherent cells, the cells gradually proliferated and recovered normal morphology similar to that of the parental cells, and continued to produce large numbers of infectious viral particles during the observation period of 5 months. Among a total of 87 cell clones obtained from the persistently infected Vero E6, only four cell clones (named #13, #18, #21, and #34) were positive for viral RNA. Clones #13, #18, and #34 shifted to viral RNA-negative during subsequent cultures, while #21 continuously produced infectious particles at a high rate. The SARS-CoV receptor, angiotensin-converting enzyme 2, was almost completely down regulated from the cell surface of persistently infected cells. Western blot analysis as well as electron microscopy indicated that the ratios of spike to nucleocapsid protein in clone #21 as well as its parental persistently infected cells were lower than that in the cells in the acute phase of infection. These Vero E6 cells persistently infected with SARS-CoV may be useful for clarifying the mechanism of the persistent infection and also for elucidating the possible pathophysiologic significance of such long-term maintenance of this virus.
Keywords: ACE2, Cell clone, Coronavirus, Down-regulation, Nucleocapsid protein, Persistent infection, SARS, Spike protein, Vero
Abbreviations: ACE2, angiotensin-converting enzyme 2; CoV, coronavirus; dpi, day post-infection; E, envelope; ELISA, enzyme-linked immunosorbent assay; FITC, fluorescein isothiocyanate; GAPDH, glyceraldehydes-3-phosphate dehydrogenase gene; HRP, horseradish peroxidase; IF, immunofluorescence; M, membrane; MAb, monoclonal antibody; MHV, murine hepatitis virus; MOI, multiplicity of infection; N, nucleocapsid; PAb, polyclonal antibody; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; RT, reverse transcriptase; S, spike; SARS, severe acute respiratory syndrome; TCID, tissue culture infectious dose; TTBS, Tris-buffered saline containing Tween 20
1. Introduction
Severe acute respiratory syndrome (SARS)-associated coronavirus (CoV), a newly recognized member of the family Coronaviridae, causes a deadly acute infectious respiratory disorder [1], [2], [3], [4]. CoVs are enveloped, positive-strand RNA viruses. CoVs have ~30 kb genomes that encode polymerase and structural proteins such as spike (S), envelope (E), membrane (M), and nucleocapsid (N), as well as several nonstructural proteins. The SARS-CoV is distinct on the basis of its genome sequence from other CoVs known to infect humans or animals [5], [6].
Recently, angiotensin-converting enzyme 2 (ACE2) was identified as the host cell receptor for infection of SARS-CoV [7]. The S protein of this virus was shown to be sufficient for the binding to this receptor molecule [8], [9]. ACE2 is expressed in many tissues, including epithelia of the lung and small intestine [10], [11]. Indeed, pathological characterization of several autopsied tissues from SARS-CoV-infected individuals also revealed viral signals only in the restricted tissues such as lung and small intestine [11], [12]. Interestingly, the SARS-CoV-infected ileal and colonic mucosa appeared normal at the microscopic level and no viral inclusion bodies could be detected in the intestinal biopsies obtained from a SARS patient [13] or in fatal cases of SARS [14]. Recent examination of seven ACE2-positive cell lines of human intestinal origin for SARS-CoV infection revealed that only one cell line (LoVo) was sensitive to infection without CPE [15]. This culture seems to accord with the histopathologic findings in intestinal samples from patients [13], [14]. In sharp contrast to the LoVo cell line, the Vero E6 cell line undergoes a lytic infection with characteristic refractile rounding CPE [16], [17] that seem to correspond to the features of multinucleated pneumocytes resulting from local replication of SARS-CoV in the lungs of patients [11], [12].
It may be worthwhile to attempt the establishment of persistent infection of SARS-CoV even in the highly sensitive Vero E6 cells, because CoVs other than SARS-CoV cause a variety of diseases and can also be used to establish persistent infections even in highly sensitive host cells. Murine CoVs, especially mouse hepatitis virus (MHV), induce hepatitis, enteritis, and encephalitis in mice. The pathogenic effects vary with the virus strain, route of infection, and the strain, age, and immune status of the host mice [18], [19], [20]. Persistent infection of MHV can be established in immunosuppressed or neonatal animals [21], [22], [23], [24]. Also, MHV causes both lytic and persistent infections in a variety of murine cultured cell lines and in these persistent infections, the cells produce virus variants with defective or aberrant phenotypes [25], [26], [27], [28], [29], [30], [31]. On the other hand, human CoVs such as 229E and OC43 are also responsible not only for common colds [32], [33] but also pneumonia, meningitis, diarrhea [34], [35], as well as central nervous disorders [36], [37]. Persistent infections of these human CoVs can be established in human neuronal cell lines [38], [39].
SARS-CoV can replicate in Vero E6 cells and produce a large amount of the progeny particles. After the production of the progeny particles, the host cells die via an apoptotic mechanism [40], [41]. This host cell killing in the acute phase seems to be the typical course for this virus. In this study, we examined the possible establishment of persistent infection of SARS-CoV in Vero E6 cells. The results showed that Vero E6 could survive the acute infection with SARS-CoV, and the survivor cells allowed long-term persistence of this virus, although serial continuous passages resulted in a gradual decrease of the viral RNA-positive cell population. Upon cell cloning of the persistently infected cell population, only 1 out of 87 cell clones obtained was a long-term producer of infectious virus particles at a high rate, with aberrant expression of S protein.
2. Materials and methods
2.1. Cells and virus
The Vero E6 cells were maintained in MEM (GIBCO BRL) supplemented with 10% fetal bovine serum (ICN Flow), 100 U/ml penicillin and 100 μg/ml streptomycin (GIBCO BRL) (complete medium) passage every 3 days. The CaCo-2 cells were maintained in DMEM (GIBCO BRL) supplemented with 10% fetal bovine serum (ICN Flow), 100 U/ml penicillin, 100 μg/ml streptomycin (GIBCO BRL), and MEM non-essential amino acids solution (GIBCO BRL) (complete medium).
For virus propagation, SARS-CoV (Frankfurt-1 strain) [42] was grown in Vero E6 cells in T-75 flasks in 5% CO2 in air at 37 °C. The culture supernatants were obtained 4 days post-infection (dpi) and filtered through a cell strainer to remove cell debris, and then aliquot stored at –80 °C. The virus titers were determined as tissue culture infectious dose (TCID)50/ml. Ninety-six-well microplates with monolayers of Vero E6 cells that had been seeded at 4 × 105 cells per ml (0.1 ml per well) and cultured for 1 day were infected with 100 μl of serial 10-fold dilutions of the culture media of infected cells (4 wells per each dilution). After incubation for 3 days, the wells were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) overnight, then stained with crystal violet. Based on the presence or absence of residual cells, virus infectivity by TCID50 was calculated by Karber's method [43].
For infection with SARS-CoV, cells were infected at a multiplicity of infection (MOI) of 1. After adsorption for 1 h, the cells were cultured in complete medium. Persistently infected cells were obtained by passaging every 3 days.
2.2. Cell cloning
The persistently SARS-CoV-infected Vero E6 cells were subjected to limiting dilution in 96-well microplates (1 cell per well in MEM supplemented with 20% fetal bovine serum). After incubation for 2 weeks with no medium change, the wells containing a single colony per well were carefully identified by microscopic observation and the cells in each well were transferred to 24-well microplates and gradually scaled up to propagation in a large scale culture.
2.3. Reverse transcriptase (RT)-polymerase chain reaction (PCR) for SARS-CoV RNA
TRIzol (Invitrogen) was used to extract total RNA according to the manufacturer's instructions. To detect genomic RNA of SARS-CoV, RNA was reverse-transcribed with SuperScript III RT (Invitrogen) using the primer 5′-TCTTGATGGATCTGGGTAAGGC-3′ (complementary to nucleotides 15857–15878 at open reading frame 1ab). In the first PCR, the cDNAs were amplified with ExTaq (Takara, Kyoto, Japan) using primers 5′-GAATCCTGACATCTTACGCG-3′ at nucleotides 13871–13890 and 5′-TGTTAGGCATGGCTCTGTCA-3′ at nucleotides 15255–15236 (94 °C for 1 min; 35 cycles of 94 °C for 30 s, 61 °C for 30 s, 72 °C for 90 s; and finally 72 °C for 10 min). Second-round PCR was performed using nested primers (5′-TGCGTGATGCAGGCATTGTA-3′ at nucleotides 13957–13976 and 5′-CATAGCTGGATCAGCAGCAT-3′ at nucleotides 14510–14491) with ExTaq (94 °C for 1 min; 35 cycles of 94 °C for 30 s, 61 °C for 30 s, 72 °C for 45 s; and finally 72 °C for 5 min). The primers were prepared according to the genome sequence of SARS-CoV in the previous reports [5], [6].
For the mRNA levels for SARS-CoV S and N genes, semi-quantitative RT-PCR was performed. For the S gene, cDNAs prepared with oligo(dT) primer (Invitrogen) were amplified using primers 5′-GGAAAAGCCAACCAACCTCGATCTC-3′ (nucleotides 23–47, corresponding to the common leader sequence) and 5′-ACTACATCTATAGGTTGATAGCCCT-3′ (nucleotides 22084–22108). For the N gene, the same cDNA was amplified using the same leader primer and the primer 5′-AGGAAGTTGTAGCACGGTGGCAGC-3′ (nucleotides 28585–28608). As a control for the input RNA, levels of a housekeeping gene, glyceraldehydes-3-phosphate dehydrogenase gene (GAPDH) were also assayed using forward primer 5′-ACCACAGTCCATGCCATCAC-3′ and reverse primer 5′-TCCACCACCCTGTTGCTGTA-3′. The intensity of each band was quantified with NIH image software.
2.4. Determination of full-length sequences at SARS-CoV S and N genes
For direct sequencing of SARS-CoV S and N genes, the cDNA synthesized as described above was used for amplification of the region containing the full-length S gene using primers 5′-CTGCTGTAATGTCTCTTAAGGAG-3′ at nucleotides 21358–21380 and 5′-CTGGCTGTGCAGTAATTGATCC-3′ at nucleotides 25298–25319, as well as for amplification of the region containing the full-length N gene using primers 5′-CATGAAGGTCACCAAACTGC-3′ at nucleotides 28053–28072 and 5′-CATCTGCCTTGTGTGGTCAT-3′ at nucleotides 29398–29417. These PCR products after purification were subjected to direct sequencing by ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). The nucleotide sequences were aligned and then translated to amino acid sequences with GENETYX-MAC, version 9.0 (Software Development, Tokyo, Japan).
2.5. Antibodies specific to SARS-CoV
The crude fraction of SARS-CoV particles was prepared by centrifugation of the conditioned medium of infected Vero E6 cells 3 dpi through 20% (w/v) sucrose in PBS in a Beckman SW28 rotor at 25,000 rpm for 1 h. The emulsion of the resulting precipitate with complete Freund's adjuvant (WAKO, Tokyo, Japan) was used for intraperitoneal immunization of BALB/c mice for monoclonal antibody (MAb) preparation (2G3 and 3A2) and intramuscular immunization of guinea-pigs for polyclonal antibody (PAb) preparation. The hybridomas producing MAb were prepared as described previously [44] using the murine myeloma PAI cell line. In addition, following anti-SARS-CoV PAbs were purchased: rabbit PAb to the N-terminus region of SARS-CoV S protein (Cat. No. AP6009b; ABGENT, San Diego, USA) and rabbit PAb to the C-terminus region of SARS-CoV N protein (Cat. No. AP6005b; ABGENT).
2.6. Antibodies specific to ACE2
For detection of cell surface ACE2, goat ACE2 PAb (anti-human ACE2 ectodomain antibody; Research and Diagnostic Systems, Inc.) was used.
2.7. Indirect immunofluorescence (IF)
Vero E6 cells (uninfected or infected with SARS-CoV) in 8-chamber glass slides with covers (Lab-Tek II Chamber Slide System; Nalge Nunc International) were washed with PBS, and then fixed with cold acetone. Alternatively, the cells were trypsinized and the resulting single cells were smeared and then fixed with cold acetone. The fixed cells were reacted with murine MAb against SARS-CoV (3A2) for 15 min at room temperature. After staining with the second antibody, fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, USA), the cells were further stained for 30 s with Hoechst 33258 at a final concentration of 1 μM in PBS. After washing, the cells were observed under an IF microscope.
2.8. Flow cytometry and MIF
For flow cytometry and MIF, the trypsinized, uninfected or infected Vero E6 cells without fixation were reacted with anti-SARS-CoV MAb (3A2) or ACE2 PAb for 15 min at room temperature, as for indirect IF test, and then finally fixed with 4% formaldehyde in PBS, and subjected to flow cytometry (FACSCalibur; Becton–Dickinson) and IF microscope.
2.9. Antigen-capture enzyme-linked immunosorbent assay (ELISA)
Antigen-capture ELISA was carried out according to the method previously described [45]. The 96-well ELISA assay plates (flat bottom; Falcon Becton–Dickinson) were coated with 50 mM carbonate buffer containing guinea-pig anti-SARS-CoV PAb for 1 h at 37 °C. The plates were blocked with 1% bovine serum albumin in PBS for 16 h at 4 °C. After washing with 0.05% Tween-20 in PBS (PBST), the wells were incubated for 1 h at 37 °C with the samples, which were the infected cell culture media supplemented with 0.1% Nonidet-P 40. The plates were washed with PBST and incubated with murine MAb to SARS-CoV S protein (2G3) for 1 h at 37 °C. After washing with PBST, the plates were incubated with horseradish peroxidase (HRP)-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories) for 1 h at 37 °C. Finally, the plates were incubated with 0.4 mg/ml o-phenylenediamine in citrate–phosphate buffer (pH 5.4) with 0.012% hydrogen peroxide for 30 min, and then the reactions were stopped and the absorbance was measured at 492 nm.
2.10. Western blotting
Uninfected or infected Vero E6 cells were washed with cold PBS, then solubilized in ice-cold lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Nonidet-P 40, 0.1% deoxycholate, 0.1% SDS, 4 mM EDTA, 10 mM NaF, 2 mM Na2VO3, 2 mM phenylmethylsulfonyl fluoride) [46]. Cell lysates in Eppendorf tube were prepared by centrifugation at 15,000 rpm for 5 min at 4 °C and the protein concentrations in the lysates were adjusted, and then the samples were mixed with the same volume of 2 × SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer. The viral proteins in the virus particle fractions obtained by centrifugation of the culture media in a Beckman SW50.1 rotor at 35,000 rpm for 30 min at 4 °C were solubilized in SDS-PAGE sample buffer. The samples were subjected to 10% SDS-PAGE, then Western-blotted onto polyvinylidene difluoride membranes (Millipore, Bedford, USA). The membranes were blocked with 5% skim milk in Tris–buffered saline containing 0.05% Tween 20 (TTBS) and then incubated for 2 h with anti-SARS-CoV (guinea-pig PAb or rabbit PAb), then reacted with HRP-conjugated donkey anti-guinea-pig or anti–rabbit secondary antibodies (Jackson ImmunoResearch Laboratories), respectively, for 1 h at 37 °C. The membranes were washed with TTBS, incubated in commercial enhanced chemiluminescence reagent (ECL, Amersham Pharmacia Biotech) and exposed to X-ray film. The intensity of each band was quantified with NIH image software.
2.11. Electron microscopy
Vero E6 cells (mock-infected or infected with SARS-CoV) were suspended in 2% glutaraldehyde in PBS and collected with a plastic scraper. The cells were centrifuged at 800 rpm for 5 min, then fixed at 4 °C for 2 h. After the cells were washed five times with PBS, they were further fixed with 1% osmium tetroxide (OsO4) in the same buffer at 4 °C for 2 h, washed, dehydrated in a graded ethanol series, and embedded in epoxy resin. Ultrathin sections were made with an ultramicrotome (Reichert Ultracut S, Leica, Wien, Austria), stained doubly with uranyl acetate and lead citrate, and observed under an electron microscope (H-7100; Hitachi) at an accelerating voltage of 75 kV. Some sections were stained with 1% ruthenium red before the conventional double staining.
3. Results
3.1. Establishment of Vero E6 cells persistently infected with SARS-CoV
Monolayers of Vero E6 cells were infected with SARS-CoV at an MOI of 1. The cells and medium were harvested daily to determine cell numbers as well as virus production rates for 6 days. As shown in Fig. 1 A, the cell number was suddenly decreased on day 3. However, the cell number again increased gradually thereafter, and the cells were observed to contain vacuoles. Light microscopic observation revealed that the CPE characterized by cell rounding first appeared on day 2, and the next day most of the cells were rounded and detached from the plastic surface (Fig. 1B). When we replaced the cell culture medium by fresh complete medium on the third day, some remaining survivor cells were detected, and these vacuolated cells gradually proliferated. On day 6 post-infection, the number of round cells was decreased, and almost confluent monolayers of the survivors were obtained.
Fig. 1.
Kinetics of the appearance of CPE in Vero E6 cells after SARS-CoV infection. Monolayers of Vero E6 cells in 12-well microplates were infected with an MOI of 1 for 1 h at 37 °C. The cells were then washed with PBS and cultured in complete medium for 6 dpi at 37 °C. The culture medium was replaced with fresh complete medium on day 3 post-infection. The adherent viable cell numbers were counted daily (A). Then, the cell morphologies were observed daily under a light microscope (B).
The cell smears prepared daily from the infected cell samples described above were stained with murine MAb (3A2), which recognizes SARS-CoV S protein (data not shown). As shown in Fig. 2 , most of the Vero E6 cells were viral antigen-positive on the 1st and 2nd days. The cells that did not undergo rounding on day 3 were also viral antigen-positive, although only a fraction of the cells were positive: 70.8%, 67.4%, 45.0%, 39.0%, 39.4%, and 39.5% on days 1–6, respectively. By co-staining with Hoechst 33258, it was found that most of such surviving cells with expression of viral antigens contained no apparent apoptotic bodies inside them (Fig. 2). Next, the culture supernatants harvested daily from the infected Vero E6 cells were subjected to infectivity titration and also to antigen-capture ELISA using anti-SARS-CoV guinea-pig PAb and murine (2G3) MAb, as shown by TCID50 per ml and absorption at 492 nm, respectively (Fig. 3 ). The viral production rate was the highest (107.5 TCID50 per ml) on days 2–3 (Fig. 3A). After the cells were replaced in fresh medium on day 3, the surviving cells also continued to produce infectious virus particles at a high rate, near 107 TCID50 per ml (Fig. 3A). A similar particle production pattern was also observed by antigen-capture ELISA (Fig. 3B). The guinea-pig PAb recognizes SARS-CoV proteins, including S and N proteins (as described below), whereas murine 2G3 MAb recognizes the SARS-CoV S protein (data not shown). Thus, these surviving, viral antigen-positive Vero E6 cells were shown to be persistently infected with SARS-CoV, so this cell population was named Vero/SARS-CoV. The Vero/SARS-CoV cells came to show a similar morphology and cell proliferation rate as the parental Vero E6 cells upon subsequent continuous passaging of the cells. However, the percentage of the cells positive for SARS-CoV antigens gradually decreased to ~10% by 5 months after infection.
Fig. 2.
Kinetics of the appearance of SARS-CoV antigens in Vero E6 cells after the infection. The same infected Vero E6 cells used for the experiments shown in Fig. 1 were trypsinized, smeared, fixed with cold acetone, then strained with anti-SARS-CoV S protein murine MAb (3A2; top panels) as well as Hoechst 33258 (middle panels). The lower panels show the merged images.
Fig. 3.
Kinetics of SARS-CoV production by Vero E6 cells after infection. The culture media harvested daily from the same infected Vero E6 cultures used for the experiments shown in Fig. 1 were subjected to virus infectivity titration (A) and antigen-capture ELISA using anti-SARS-CoV guinea-pig PAb and murine MAb (2G3) (B). The results are shown by TCID50 per ml and adsorption at 492 nm, respectively. As a negative control for ELISA, complete medium was used.
3.2. Preparation of infectious virus-producing cell clones
The Vero/SARS-CoV cells were used 1 month after infection for the preparation of cell clones by limiting dilution in 96-well microplates. A total of 87 cell clones were obtained. As shown in Fig. 4 B, nested RT-PCR of the cell clones for SARS-CoV revealed only four (#13, #18, #21, and #34) cell clones to be positive for SARS-CoV RNA, while the other 83 cell clones were completely negative for SARS-CoV RNA by nested PCR analysis, as shown for #12 as a representative. In single-round PCR analysis, #21 showed a strong positive band, similar to those of acutely and persistently infected cells, while #18 showed only a faint band, and the #13 and #34 were negative (Fig. 4A). The amounts of viral RNA in cell clones #13, #18, and #34 just after their preparation were lower than that in #21, and they became negative after several further passages (Fig. 4B). Titrations of the infectivity of their culture supernatants also confirmed these shifts: we observed only 101.5–< 100.5 TCID50 per ml in #13 and 106–7–< 100.5 TCID50 per ml in #18 and #34; in contrast, 107–8 TCID50 per ml in #21 throughout the study period.
Fig. 4.
Screening of Vero/SARS-CoV cell-derived cell clones for virus particle production by RT-PCR. Total RNA extracts from Vero E6 cells that were mock-infected or acutely SARS-CoV-infected for 2 days as in Fig. 1, persistently SARS-CoV-infected (Vero/SARS-CoV) for 1 month (1 M), 2 M or 5 M, and five representative cell clones from Vero/SARS-CoV (#13, #18, #21, #34, and #12) either just after the cell cloning (0 M) or after being cultured for a subsequent 1 M, were reverse-transcribed, and then the cDNA fractions were used for single-round PCR (A). An aliquot (1/10) of the single-round PCR product was further subjected to second-round PCR (B).
Representative cell clones obtained from Vero/SARS-CoV cells that were continuously passed for 3 months after their preparation were stained with anti-SARS-CoV S MAb (3A2) (Fig. 5 ). The control cells were Vero E6 cells mock-infected or acutely infected with SARS-CoV for 2 days, as well as Vero/SARS-CoV cells that were maintained for 5 months after infection. The expression of S antigen in #21 cells was quite strong in about 10% of the cells, like that in acutely and persistently infected Vero E6 cells. In contrast, viral antigens were not detected in #13, #18, and #34 that had been maintained for 1 month after their preparation, nor in the viral RNA-negative clone #12 at the time of its preparation.
Fig. 5.
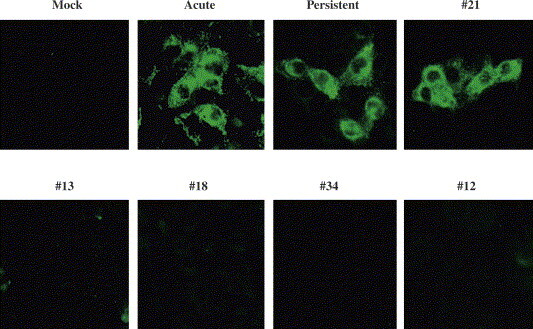
Expression of SARS-CoV antigens in the persistently infected cell clones. The Vero E6 cells that were mock-infected, acutely SARS-CoV-infected as in Fig. 1, persistently SARS-CoV-infected (Vero/SARS-CoV) for 5 months, and several representative cell clones from Vero/SARS-CoV (#21 as a virus producer; #13, #18, and #34 as cell clones that had shifted from positive to negative, as shown by RT-PCR [see Fig. 4]; and #12 as an RT-PCR-negative clone already even just after cell cloning), were similarly seeded on eight-chamber glass slides. The next day, the cells were rinsed with PBS, dried, fixed with cold acetone, and then stained with anti-SARS-CoV murine MAb (3A2).
Next, the expression of ACE2, identified as a receptor for SARS-CoV infection [7], was examined on the cell surface of the cell clones by MIF and flow cytometry (Fig. 6 ). Only 18% of the mock-infected Vero E6 cells were positive for ACE2 on their cell surface. Twelve percent of the persistently infected Vero/SARS-CoV cells that were cultured for 4 months were positive for SARS-CoV S protein. The expression of ACE2 on the persistently infected cells was almost completely down-regulated, although the expression level of ACE2 on #21 cells were heterogeneous. On the other hand, cell clones #12 and #13 which were derived from the Vero/SARS-CoV cells and negative for SARS-CoV S protein after culturing for 3 months after the primary infection, highly expressed ACE2 on their cell surface: 42% of #12 and 17% of #13.
Fig. 6.
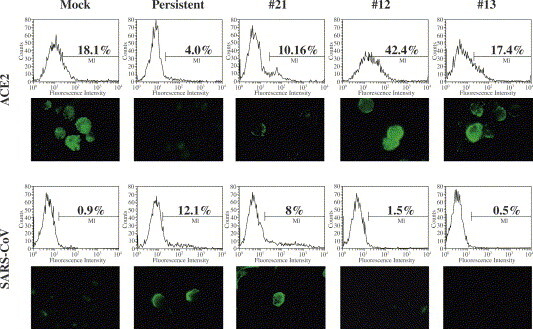
ACE2 down-regulation on the surface of Vero/SARS-CoV cells. Mock-infected Vero E6 cells and persistently infected Vero/SARS-CoV that had been cultured for 5 months, as well as cell clones #21, #12 and #13 that had also been cultured for 3 months after their preparation were stained without fixation, with anti-ACE2 PAb as well as anti-SARS-CoV S murine MAb (3A2). The stained cells were subjected to IF microscopy and flow cytometry.
Electron micrography showed the production of a large amount of viral particles from #21 cells (on 2 months after their preparation) (Fig. 7 E) as well as many viral particles inside vesicles of this cell clone (Fig. 7F), as was also seen for acutely infected Vero E6 cells (Fig. 7A, 7B). The morphology of the virions released from acutely infected Vero E6 was similar to that described previously [47]. The particle number seemed to be slightly lower in the persistently infected Vero/SARS-CoV cells that had been continuously passed for 4 months after infection (Fig. 7C, D). Interestingly, there were far fewer surface projections surrounding the particle core on Vero/SARS-CoV (Fig. 7C, D) and on #21 (Fig. 7E) than on the acutely infected Vero E6 cells (Fig. 7A).
Fig. 7.
Electron microscopic observation of Vero/SARS-CoV-derived cell clones to assess their production of viral particles. The cells used for electron microscopy were the Vero E6 cells acutely SARS-CoV-infected for 2 days (A, B), persistently SARS-CoV-infected (Vero/SARS-CoV) for 4 months (C, D), and #21 cell clone cultured for 2 months after its preparation (E, F). Bars indicate 100 nm.
In fact, direct sequencing of the PCR products including SARS-CoV S and N genes that were amplified using the RNA from #21 cells revealed that a total of five nucleotide substitutions were detected at S gene: A1325G (Y442C), C1414T (L472F), G1780T (V594F), C1921T (H641Y), and C2380T (P794S). There was no mutation at the N gene in the RNA from #21. On the other hand, there was no apparent difference in virus replication rate of the SARS-CoV from #21 cells, compared with that of the SARS-CoV from acutely infected Vero E6 cells, in both Vero E6 and CaCo-2 cells (Fig. 8 ). The Vero E6 and CaCo-2 cells were similarly infected at an MOI of 1 with SARS-CoV in the culture fluid of acutely infected Vero E6 cells and that in the culture fluid of #21 cells. The RNA extracted from the infected cells on days 0 (just after adsorption) and 10 were analyzed for viral genomic RNA levels by RT-PCR using primers at 1ab region. The results showed that the amounts of PCR products on day 10 samples were almost similar between SARS-CoV at acute and persistent phases, although those on day 0 samples were apparently higher in the SARS-CoV at acute phase than that at persistent phase in both Vero E6 and CaCo-2 cells. Thus, the adsorption rate may be different between SARS-CoV prepared at acute and persistent phases, but the subsequent replication rate of the virus from persistent phase was almost similar to that from acute phase.
Fig. 8.
Comparison of growth rates of SARS-CoVs at acute and persistent phases in Vero E6 and CaCo-2 cells. The Vero E6 and CaCo-2 cells were mock-infected (Mock) and infected at an MOI of 1 with SARS-CoV prepared from the Vero E6 cells that were acutely infected for 2 days (Acute) and from persistently SARS-CoV-infected for 5 months (Persistent). After adsorption for 1 h, the cells were washed with PBS. The total RNAs were extracted from the cells just after washing with PBS (Day 0) as a control and also from the cells that were cultured in complete medium at 37 °C for 10 dpi (Day 10). The culture medium was replaced with fresh complete medium every 2–3 days. The total RNAs were used for quantification of the SARS-CoV genomic RNA by RT-PCR using primers at the 1ab region.
3.3. Comparison of viral expression between acutely and persistently infected Vero E6 cells
We next compared the levels for expression of SARS-CoV S and N genes among Vero E6 cells acutely infected with SARS-CoV for 2 days, Vero/SARS-CoV that had been maintained for 5 months after the primary infection, and the virus producer cell clone #21 by RT-PCR (Fig. 9 A). The results by triplicate experiments showed a slightly higher ratio of the S/N ratio in acutely infected Vero E6 cells, compared with Vero/SARS-CoV and #21 cells, i.e. 0.95 ± 0.15, 0.24 ± 0.05, and 0.30 ± 0.17, respectively. Next, Western blotting was also carried out to confirm the differences in the expression of viral proteins among the same acutely infected Vero E6 cells, Vero/SARS-CoV, and #21 (Fig. 9B). The cell fractions as well as the virus particle fractions precipitated from the culture media by ultracentrifugation were characterized. Western blotting using the cell fractions and guinea-pig anti-SARS-CoV PAb revealed lower amounts of S protein in Vero/SARS-CoV and #21 compared with that in acutely infected cells. However, the amounts of N protein in Vero/SARS-CoV and #21 cells were similar to that in the acutely infected cells. Similarly, rabbit PAb to the N-terminus region of SARS-CoV S protein also detected a higher amount of the S protein in the acutely infected cells than those in #21. Rabbit PAb to the C-terminus region of SARS-CoV N protein detected the N protein in the acutely infected cells and Vero/SARS-CoV and a rather lower amount of N proteins in #21. The S/N ratios by triplicate experiments using rabbit anti-S and anti-N PAbs were 0.81 ± 8.080 in the acutely infected cells and 0.059 ± 0.014 in the Vero/SARS-CoV. In the virus particle fractions, a lower amount of S protein in #21 was detected compared with that in acute infection, but an only slightly lower amount of N protein in #21 was detected compared with that in acute infection. Thus, the S to N ratio in #21 was significantly lower than that in acutely infected cells.
Fig. 9.
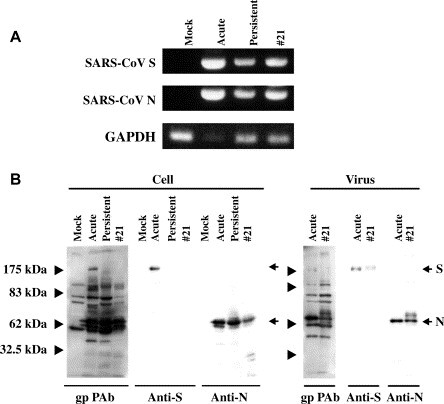
Comparison of expression levels of major SARS-CoV genes between acutely infected and persistently infected Vero E6 cells. A, Total RNAs were extracted from the Vero E6 cells that were mock-infected (Mock) or acutely SARS-CoV-infected for 2 days (Acute), and persistently SARS-CoV-infected for 5 months (Persistent), as well as cell clone #21 that had been cultured for 3 months after its preparation. The RNAs were used for amplification of the SARS-CoV mRNAs at S and N regions by semi-quantitative RT-PCR. As a control, housekeeping gene GAPDH was amplified using the same RNA preparations. B, Cell lysates were prepared from the same cells as used in figure A. The virus particle fractions were also prepared from the culture media of these cells by ultracentrifugation. The samples were electrophoresed in a 10% SDS-PAGE, then Western-blotted onto a M, and reacted with guinea-pig anti-SARS-CoV PAb, rabbit PAb to the N-terminus region of SARS-CoV S protein (Anti-S) and rabbit PAb to the C-terminus region of SARS-CoV N protein (Anti-N).
4. Discussion
In this study, persistent infection with SARS-CoV was established in Vero E6 cells, which are highly sensitive to infection with this virus [16], [17] and are generally killed via apoptosis after the infection [40], [41]. The cells that survived the infection in the acute phase proliferated at a similar rate and with similar morphology as uninfected Vero E6 cells. Nevertheless, these Vero/SARS-CoV cells maintained high production of infectious virus particles. Surprisingly, only 1 (#21) out of 87 cell clones prepared from Vero/SARS-CoV was a long-term high producer of infectious virus particles during our observation period of 5 months after the primary infection. Three cell clones were also positive for SARS-CoV RNA, but the viral RNA disappeared from these cell clones upon subsequent culturing for 1 month.
Chen and Baric [48] demonstrated with MHV that persistence selects for resistant host cells that down-regulate expression of receptor and variant viruses that grow more efficiently than wild-type in these cultures. When we examined the preparation of several Vero E6 cell clones, it was found that there was no difference in the cell surface expression levels of receptor ACE2 among the 32 cell clones obtained. However, the cell clones showed different degrees of CPE appeared by SARS-CoV infection that affected the rates to obtain the persistently SARS-CoV-infected cells (data not shown). Thus, such difference observed in MHV is not likely in SARS-CoV.
The viral expression in cell clone #21 was shown to be different with regard to the S to N ratio from that in acutely infected Vero E6 cells. This different S/N ratio was also similarly observed in uncloned, parental persistently SARS-CoV-infected Vero E6 cells, suggesting the universality of this difference in persistently infected Vero E6 cells. Similar observations have been reported for other CoVs: a number of MHV variants with deletions in the S glycoprotein have been isolated from persistently MHV-infected animals; however, the significance of generating and potentially accumulating deletion variants in the persistent viral population remains unclear, although it has been proposed that the complexity of the population of persisting viral RNAs may contribute to chronic disease [49], [50]. Interestingly, the cultured cells persistently infected with MHV were shown to be resistant to superinfection with MHV but not to infection with other unrelated viruses [51]. When the culturing of cells persistently infected with MHV was carried out for >400 passages, the yield of released virus gradually decreased and only ~10% of the cells were viral antigen-positive at passage 97 [51], which was similar to the pattern we observed for SARS-CoV in this study. In the case of MHV persistence, if fewer than 100 cells were subcultured, then the persistent infection ended and the resulting cultures no longer produced virus and became susceptible to MHV infection [51]. In contrast, we were able to obtain several virus-positive cell clones by culturing a single Vero/SARS-CoV cell in the wells of 96-well microplates. In human neuronal cell lines persistently infected with human CoV OC43 and 229E, there were no important common viral-genome mutations related to the persistent infection [38], [39]. On the other hand, using bovine CoV it was found that a translation-attenuating intraleader open reading frame was selected in this virus mRNA during persistent infection [52], [53]. In this study, we found that the persistently infected cells maintained the expression of SARS-CoV antigens for at least 5 months during maintenance without cell cloning, while most of the cell clones prepared from Vero/SARS-CoV cells were negative for virus RNA by RT-PCR. Several cell clones positive for virus RNA at early passages after the preparation also readily shifted to become negative, suggesting that a certain host cell factor(s) may be involved in the maintenance of SARS-CoV in the Vero E6 cells during the uncloned condition. In addition, such host factor, probably together with other factor(s), may be also involved in a long-term maintenance of SARS-CoV even in the cloned condition such as in #21. It is noteworthy that the SARS-CoV S to N ratio in persistently infected cells was significantly lower than that in acutely infected cells. Electron microscopic observation also supported this conclusion, as shown by the much lower density of the “knob” structures in persistently than acutely infected Vero E6 cells. In fact, we identified a total of 5 nucleotide substitutions at S gene, but no substitution at N gene in the viral RNA from #21 cell clone. However, we found no apparent difference in the replication rates in Vero E6 and CaCo-2 cells between SARS-CoV prepared from acutely and persistently infected Vero E6 cells. We will need further studies to clarify the correlation between the S/N ratio and the establishment of persistent infection with this virus.
After SARS-CoV infection, patients have been shown to develop immune responses to the virus: > 90 of the SARS patients were found to seroconvert within 20 days [54]. Another report showed that conventional serological tests such as IF, Western blotting, and ELISA using serum samples from patients with SARS that were obtained 7–35 days after the onset of fever detected specific IgG reactions in at most 89% of them [55]. However, on the basis of Western blotting using sera obtained from a total of 43 patients with SARS that were obtained 7–35 days after the onset of fever, most (79%) of the patients produced antibodies to N protein, but only 40% produced antibodies to S protein [55]. A similar estimate (83.3%) was also obtained in another study, although 25.0% had not produced antibodies to SARS-CoV by day 21, and 16.7% had not produced viral antibodies even by day 60 [56]. Thus, in a considerable number of cases it seems to be difficult to detect specific antibody responses to SARS-CoV, suggesting the possibility that such infected individuals may readily allow the virus to establish a persistent infection.
Pathological characterization of several autopsied tissues from SARS-CoV-infected individuals revealed viral signals mostly in the lung and small intestine, but not in lymphoid organs such as spleen and lymph nodes [11], [12]. Thus, lymphocytes might be resistant to SARS-CoV infection in vivo. Nevertheless, evidence of SARS-CoV infection in peripheral blood mononuclear cells (PBMC) was reported. RT-PCR is a relatively rapid and sensitive method for the diagnosis of patients with SARS symptoms using patient-derived samples such as nasopharyngeal aspirates [57], [58]. One trial to standardize the sample source for quantification of SARS-CoV used plasma and PBMC derived from patients at three SARS stages: days 1–7, 13–39, and 79–91 after fever onset [59]. The viral RNA was detectable even in PBMC, where its level was higher than that in plasma at all clinical stages. The peak of the shedding of SARS-CoV corresponded to the peak of the course of SARS-CoV infection. Surprisingly, residual SARS-CoV may persist in a patient's circulation, in most cases for a relatively long time without obvious effects on the host. The viral loads in the plasma were not significantly different in the samples obtained at days 13–39 and 79–91. Thus, some cells, including PBMC, may be a reservoir for this virus, although the pathophysiologic significance of such persistently infected cells is not clear yet. In addition, the recent finding that dendritic cells can take up SARS-CoV and transfer it to susceptible target cells seems to be important, since such dendritic cells may serve as a reservoir that might provide long-term exposure to this virus and contribute to the persistence and chronicity of infection [9]. The above observations, together with the fact that other CoVs readily establish persistent infections in vivo and in vitro, and also together with our establishment of persistent infection in the Vero E6 cell line after induction of severe CPE, suggest that complete elimination of SARS-CoV from all patients who have recovered from SARS seems to be unlikely. The pathophysiologic significance of such long-lasting persistent infection with SARS-CoV in vivo, if any, should be clarified in further studies.
Acknowledgments
We are grateful to Dr. John Ziebuhr, University of Würzburg, Germany for giving us the Frankfurt strain of SARS-CoV through Dr. Fumihiro Taguchi, National Institute of infectious Diseases, Tokyo, Japan. This work was supported in part by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the 21st Century COE Program (Combined Program on Microbiology and Immunology) from Japan Society for the Promotion of Science.
References
- 1.Dresten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Esckmann N., Esckmann N., Escrion N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Holmes K.V. SARS-associated Coronavirus. N. Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 6.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 7.Li W., Morre M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann H., Geier M., Marzi A., Krumbiegel M., Peipp M., Fey G.H., Gramberg T., Pöhlmann S. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem. Biophys. Res. Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z.-Y., Huang Y., Ganesh L., Leung K., Kong W.-P., Schwarz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-sign. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SRAS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.To K.-F., Lo A.W.I. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2) J. Pathol. 2004;203:740–743. doi: 10.1002/path.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding Y., He L., Zhang Q., Huang Z., Che X., Hou J., Wang H., Shen H., Qiu L., Li Z., Geng J., Cai J., Han H., Li X., Kang W., Weng D., Liang P., Jiang S. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung W.K., To K.-F., Chan P.K.S., Chan H.L.Y., Wu A.K.L., Lee N., Yuen K.Y., Sung J.J.Y. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.To K.-F., Tong J.H.M., Chan P.K.S., Au F.W.L., Chim S.S.C., Chan K.C.A., Cheung J.L.K., Liu E.Y.M., Tse G.M.K., Lo A.W.I., Lo D.Y.M., Ng H.-K. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J. Pathol. 2004;202:157–163. doi: 10.1002/path.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan P.K.S., To K.-F., Lo A.W.I., Cheung J.L.K., Chu I., Au F.W.L., Tong J.H.M., Tam J.S., Sung J.J.Y., Ng H.-K. Persistent infection of SARS coronavirus in colonic cells in vitro. J. Med. Virol. 2004;74:1–7. doi: 10.1002/jmv.20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng M.-L., Tan S.-H., See E.-E., Ooi E.-E., Ling A.-E. Proliferative growth of SARS coronavirus in Vero E6 cells. J. Gen. Virol. 2003;84:3291–3303. doi: 10.1099/vir.0.19505-0. [DOI] [PubMed] [Google Scholar]
- 17.Ng M.L., Tan S.H., See E.E., Ooi E.E., Ling A.E. Early events of SARS coronavirus infection in Vero cells. J. Med. Virol. 2003;71:323–331. doi: 10.1002/jmv.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compton S.R., Barthold S.W., Smith A.L. The cellular and molecular pathogenesis of coronaviruses. Lab. Anim. Sci. 1993;43:15–28. [PubMed] [Google Scholar]
- 19.Kyuwa S., Stohlman S.A. Pathogenesis of a neurotropic murine coronavirus, strain JHM in the central nervous system of mice. Semin. Virol. 1990;1:273–280. [Google Scholar]
- 20.Ohtsuka N., Taguchi F. Mouse susceptibility to mouse hepatitis virus infection is linked to viral receptor genotype. J. Virol. 1997;71:8860–8863. doi: 10.1128/jvi.71.11.8860-8863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleming J.O., Houtman J.J., Alaca H., Hinze H.C., McKenzie D., Aiken J., Bleadale J., Baker S. Persistence of viral RNA in the central nervous system of mice inoculated with MHV-4. Adv. Exp. Med. Biol. 1993;342:327–332. doi: 10.1007/978-1-4615-2996-5_50. [DOI] [PubMed] [Google Scholar]
- 22.Kyuwa S., Tagawa Y.-I., Shibata S., Doi K., Machii K., Iwakura Y. Murine coronavirus-induced subacute fatal peritonitis in C57BL/6 mice deficient in gamma interferon. J. Virol. 1998;72:9286–9290. doi: 10.1128/jvi.72.11.9286-9290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perlman S., Jacobson G., Olson A.L., Afifi A. Identification of the spinal cord as a major site of persistence during chronic infection with a murine coronavirus. Virology. 1990;175:418–426. doi: 10.1016/0042-6822(90)90426-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorenson O., Coulter-Mackie M.B., Puchalski S., Dales S. In vivo and in vitro models of demyelinating disease. IX. Progression of JHM virus infection in the central nervous system of the rat during overt and asymptomatic phases. Virology. 1984;137:347–357. doi: 10.1016/0042-6822(84)90227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gombold J.L., Hingley S.T., Weiss S.R. Fusion-defective mutants of mouse hepatitis virus A59 contain a mutation in the spike protein cleavage signal. J. Virol. 1993;67:4504–4512. doi: 10.1128/jvi.67.8.4504-4512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes K.V. In: Coronaviridae and their replication. Fields B.N., editor. Raven Press; New York: 1990. Virology 2nd ed; pp. 841–856. [Google Scholar]
- 27.Holmes K.V., Behnke J.N. In: Evolution of a Coronavirus during Persistent Infection in vitro. ter Meulen V., Siddell S., Wege H., editors. Plenum Press; New York: 1981. Biochemistry and Biology of Coronaviruses; pp. 287–300. [DOI] [PubMed] [Google Scholar]
- 28.Lavi E., Suzumura A., Hirayama M., Highkin M.K., Dambach D.M., Silberberg D.H., Weiss S.R. Coronavirus mouse hepatitis virus (MHV)-A59 causes a persistent, productive infection in primary glial cell cultures. Microb. Pathog. 1987;3:79–86. doi: 10.1016/0882-4010(87)90066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucas A., Coulter M., Anderson R., Dales S., Fintoff W. In vivo and in vitro models of demyelinating diseases. II. Persistence and host-regulated thermosensitivity in cells of neural derivation infected with mouse hepatitis and measles viruses. Virology. 1978;88:325–337. doi: 10.1016/0042-6822(78)90289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stohlman S.A., Sakaguchi A.Y., Weiner L.P. Characterization of the cold-sensitive murine hepatitis virus mutants rescued from latently infected cells by cell fusion. Virology. 1979;98:448–455. doi: 10.1016/0042-6822(79)90567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taguchi F., Ikeda T., Makino S., Yoshikura H. A murine coronavirus MHV-S isolate from persistently infected cells has a leader and two consensus sequences between the M and N genes. Virology. 1994;198:355–359. doi: 10.1006/viro.1994.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIntosh K., Becker W.B., Chanock R.M. Growth in suckling mouse brain of "IBV-like" viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. USA. 1964;58:2268–2273. doi: 10.1073/pnas.58.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myint S.H. Human Coronavirus – a brief review. Rev. Med. Virol. 1994;4:35–46. [Google Scholar]
- 34.Resta S., Luby J.P., Rosenfield C.R., Siegel J.O. Isolation and propagation of a human enteric coronavirus. Science. 1985;229:978–981. doi: 10.1126/science.2992091. [DOI] [PubMed] [Google Scholar]
- 35.Riski H., Hovi T. Coronavirus infections of man associated with diseases other than the common cold. J. Med. Virol. 1980;6:259–265. doi: 10.1002/jmv.1890060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray R.S., Brown B., Brian D., Cabirac G.F. Detection of Coronavirus RNA and antigen in multiple sclerosis brain. Ann. Neurol. 1992;31:525–533. doi: 10.1002/ana.410310511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salmi A., Ziola B., Hovi T., Reunanen M. Antibodies to coronaviruses OC43 and 229E in multiple sclerosis patients. Neurology. 1982;32:292–295. doi: 10.1212/wnl.32.3.292. [DOI] [PubMed] [Google Scholar]
- 38.Arbour N., Coté G., Lachance C., Tardieu M., Cashman N.R., Talbot P.J. Acute and persistent infection of human neural cell lines by human coronavirus OC43. J. Virol. 1999;73:3338–3350. doi: 10.1128/jvi.73.4.3338-3350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arbour N., Ekandé S., Coté G., Lachance C., Chagnon F., Tardieu M., Cashman N.R., Talbot P.J. Persistent infection of human oligodendrocytic and neuroglial cell lines by human coronavirus 229E. J. Virol. 1999;73:3326–3337. doi: 10.1128/jvi.73.4.3326-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun. 2004;319:1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan H., Xiao G., Zhang J., Hu Y., Yuan F., Cole D.K., Zhang C., Gao G.F. SARS coronavirus induces apoptosis in Vero E6 cells. J. Med. Virol. 2004;73:323–331. doi: 10.1002/jmv.20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karber J. Beitrag zur kollectiven Behandlung pharmakologische Reihenversuche. Arch. Exp. Path. Pharmak. 1931;162:480–483. [Google Scholar]
- 44.Ikuta K., Morita C., Miyake S., Ito T., Okabayashi M., Sano K., Nakai M., Hirai K., Kato S. Expression of human immunodeficiency virus type 1 (HIV-1) gag antigens on the surface of a cell line persistently infected with HIV-1 that highly expresses HIV-1 antigens. Virology. 1989;170:408–417. doi: 10.1016/0042-6822(89)90431-5. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe M., Zhang Q., Kobayashi T., Kamitani W., Tomonaga K., Ikuta K. Molecular ratio between Borna disease virus-p40 and -p24 proteins in infected cells determined by quantitative antigen capture ELISA. Microbiol. Immunol. 2000;44:765–772. doi: 10.1111/j.1348-0421.2000.tb02561.x. [DOI] [PubMed] [Google Scholar]
- 46.Koyama Y., Yoshioka Y., Hashimoto H., Matsuda T., Baba A. Endothelines increase tyrosine phosphorylation of astrocytic focal adhesion kinase and paxillin accompanied by their association with cytoskeletal components. Neuroscience. 2000;101:219–227. doi: 10.1016/s0306-4522(00)00330-4. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Q., Cui J., Huang X., Zheng H., Huang J., Fang L., Li K., Zhang J. The life cycle of SARS coronavirus in Vero E6 cells. J. Med. Virol. 2004;73:332–337. doi: 10.1002/jmv.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W., Baric R.S. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J. Virol. 1996;70:3947–3960. doi: 10.1128/jvi.70.6.3947-3960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adami C., Pooley J., Glomb J., Stecker E., Fazal F., Fleming J.O., Baker S.C. Evolution of mouse hepatitis virus (MHV) during chronic infection: quasispecies nature of the persisting MHV RNA. Virology. 1995;209:337–346. doi: 10.1006/viro.1995.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowe C.L., Baker S.C., Nathan M.J., Fleming J.O. Evolution of mouse hepatitis virus: detection and characterization of spike deletion variants during persistent infection. J. Virol. 1997;71:2959–2969. doi: 10.1128/jvi.71.4.2959-2969.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawicki S.G., Lu J.-H., Holmes K.V. Persistent infection of cultured cells with mouse hepatitis virus (MHV) results from the epigenetic expression of the MHV receptor. J. Virol. 1995;69:5535–5543. doi: 10.1128/jvi.69.9.5535-5543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hofmann M.A., Senanayake S.D., Brian D.A. A translation-attenuating intracellular open reading frame is selected on coronavirus mRNAs during persistent infection. Proc. Natl. Acad. Sci. USA. 1993;90:11733–11737. doi: 10.1073/pnas.90.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hofmann M.A., Sethna P.B., Brian D.A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 1990;64:4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., Chan K.H., Ng J.S., Zheng B.J., Ng W.L., Lai R.W., Guan Y., Yuen K.Y. HKU/UCH SARS Study Group, Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ming Leung D.T., Gang Tam F.C., Sheung Chan P.K., Ken Cheung J.L., Niu H., Lun Tam J.S., Leong Lim P. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X., Zhou B., Li M., Liang X., Wang H., Yang G., Wang H., Le X. Serology of severe acute respiratory syndrome: implications for surveillance and outcome. J. Infect. Dis. 2004;189:1158–1163. doi: 10.1086/380397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Chan-Yeung M., Lam W.K., Seto W.H., Yam L.Y., Cheung T.M., Wong P.C., Lam B., Ip M.S., Chan J., Yuen K.Y., Lai K.N. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 58.World Health Organization Multicentre Collaboration Network for Severe Acute Respiratory Syndrome Diagnosis A multicentre collaboration to investigate the cause of severe acute respiratory syndrome. Lancet. 2003;361:1730–1733. doi: 10.1016/S0140-6736(03)13376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H., Mao Y., Ju L., Zhang J., Liu Z., Zhou X., Li Q., Wang Y., Kim S., Zhang L. Detection and monitoring of SARS coronavirus in the plasma and peripheral blood lymphocytes of patients with severe acute respiratory syndrome. Clin. Chem. 2004;50:1237–1240. doi: 10.1373/clinchem.2004.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]


