Abstract
Macro domains are ancient, highly evolutionarily conserved domains that are widely distributed throughout all kingdoms of life. The ‘macro fold’ is roughly 25 kDa in size and is composed of a mixed α–β fold with similarity to the P loop-containing nucleotide triphosphate hydrolases. They function as binding modules for metabolites of NAD+, including poly(ADP-ribose) (PAR), which is synthesized by PAR polymerases (PARPs). Although there is a high degree of sequence similarity within this family, particularly for residues that might be involved in catalysis or substrates binding, it is likely that the sequence variation that does exist among macro domains is responsible for the specificity of function of individual proteins. Recent findings have indicated that macro domain proteins are functionally promiscuous and are implicated in the regulation of diverse biological functions, such as DNA repair, chromatin remodeling and transcriptional regulation. Significant advances in the field of macro domain have occurred in the past few years, including biological insights and the discovery of novel signaling pathways. To provide a framework for understanding these recent findings, this review will provide a comprehensive overview of the known and proposed biochemical, cellular and physiological roles of the macro domain family. Recent data that indicate a critical role of macro domain regulation for the proper progression of cellular differentiation programs will be discussed. In addition, the effect of dysregulated expression of macro domain proteins will be considered in the processes of tumorigenesis and bacterial pathogenesis. Finally, a series of observations will be highlighted that should be addressed in future efforts to develop macro domains as effective therapeutic targets.
Abbreviations: ADPR, ADP-ribose; APLF, aprataxin PNK-like factor; bAREs, bacterial-produce ADP-ribosylating exotoxins; mARTs, mono-ADP-ribosyltransferases; AR, androgen receptor; BER, base excision repair; BCL2, B cell lymphoma 2; CoaSt6, Collaborator of Stat6; CHFR, checkpoint protein with FHA and RING domain; CBP, CREB-binding protein; CHIP, chromatin immunoprecipitation; DSBs, DNA double-stranded breaks; 3D, three-dimensional; DT, diphtheria toxin; ERα, estrogen receptor α; ETA, exotoxin A; EMT, epithelial–mesenchymal transition; ITC, isothermal titration calorimetry; IAPs, inhibitors of apoptosis; KS, Kabuki syndrome; MSCI, meiotic sex chromosome inactivation; NF-κB, nuclear factor-kappaB; OAADPR, O-acetyl-ADP-ribose; PAR, poly(ADP-ribose); PARPs, PAR polymerases; PARG, PAR glycohydrolase; PARylation, poly(ADP-ribosyl)ation; PARBMs, PAR binding motifs; PTMs, posttranslational modifications; PBZ, PAR binding zinc finger; RNAP II, RNA polymerase II; STAT6, signal transducer and activator of transcription-6; SFV, Semliki Forest Virus; SSBR, single-strand break repair; STS, staurosporine; TNF, tumor necrosis factor; TRF1, telomeric repeat binding factor-1
Keywords: Macro domain family, Structural feature, Posttranslational modifications, Biological function, Disease association
1. Introduction
Members of the macro domain family are conserved throughout evolution, with homologues identified in viruses (coronaviruses, alphaviruses), archaea (Archaeoglobus fulgidus), bacteria (Escherichia coli), invertebrates (Drosophila melanogaster), amphibians (Xenopus laevis), mammals (humans, mice), and plants (Arabidopsis thaliana, Oryza sativa; Fig. 1 ). Macro domain proteins all contain at least one copy of an approximately 130–190 amino acid conserved domain, the ‘macro domain’ [1], [2], [3] which allows them to bind various forms of ADP-ribose (ADPR) [4], [5], [6], [7], such as PAR. Little is known about the function or regulation of this domain, but its evolutionary conservation implies that it has a fundamental role in diverse organisms. Intriguingly, previous studies have indicated that the poly(ADP-ribosyl)ation (PARylation) of proteins plays a fundamental role in the cell and has the potential to orchestrate various chromatin-based biological tasks [8], [9], [10].
Fig. 1.
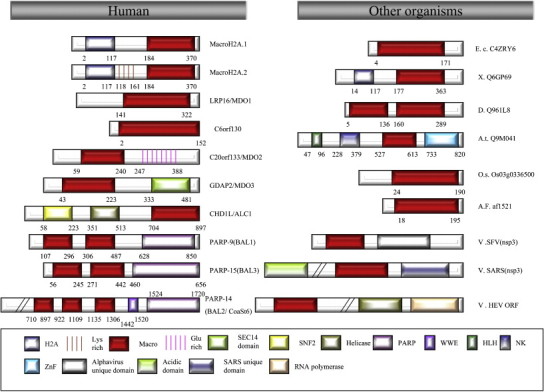
The macro domain family. The structures of the macro domain proteins are depicted, showing the conserved macro domains, as well as other domains found in selected members of the family, such as the histone H2A domain; Lys-rich domain; Glu-rich domain; SEC14 domain, a lipid-binding domain found in SEC14p and other proteins; SNF2 domain, the SNF2 helicase-like domain; PARP domain, poly(ADP-ribose)polymerase domain; WWE domain, a protein–protein interaction domain containing conserved Trp and Glu residues; HLH domain, helix–loop–helix DNA binding domain; ZnF domain, ubiquitin-binding zinc finger domain. Please note that several members of the macroPARPs (PARP-9/BAL1; PARP-14/BAL2/CoaSt6; PARP-15/BAL3) display splicing variants, but for simplicity, only one variant is illustrated. Human macro domain proteins are presented on the left and homologues from other organisms are shown on the right, including macro domain proteins from Escherichia coli (E.c), Drosophila melanogaster (D), Xenopus laevis (X), viruses (V), Archaeoglobus fulgidus (A.F), Arabidopsis thaliana (A.t) and Oryza sativa (O.s). Numbers refer to amino acid positions in the proteins.
Humans contain at least 10 genes that encode 11 members of the macro domain family, which includes macroH2A (and its various isoforms including macroH2A1/macroH2A2), MACROD1 (LRP16), MACROD2 (C20orf133), C6orf130, MACROD3 (GDAP2), ALC1 (CHD1L, CHDL), and macroPARPs (PARP-9; PARP-14; PARP-15; Table 1 ). All of these proteins contain a macro domain near either their N-terminus or C-terminus, except macroPARPs in which 2–3 putative macro domains are linked. In addition to the conserved macro domain, macro domain proteins also contain a variety of additional domains, which allow them to interact with specific target proteins or target them to specific nucleic acid regions (Fig. 1). For example, macroPARPs also contain a PARP catalytic domain, and are the only described proteins with both a PARP-like domain and macro domain. Although the function of the PARP-like domain is not fully understood, its conservation in the macroPARP homologues of Caenorhabditis elegans, Schizosaccharomyces pombe, and A. thaliana implies that it plays an important role in some aspect of either the function of this protein or its regulation. In this regard, the PARP-like domain can be used either to control protein modification or as a protein-interaction domain that mediates binding to other proteins, including transcription cofactor. Indeed, a recent report suggested that the PARP-like domain within PARP-14, which is also known as Collaborator of Stat6 (CoaSt6), might contribute to transcriptional regulation via its ability to catalyze the PARylation of p100, a co-activator recruited by signal transducer and activator of transcription-6 (STAT6) [11], [12]. Most other members of the macro domain family also contain additional domains that mediate protein–protein (the WWE domain) [13] or protein–lipid (the SEC14 domain) [14], [15] interactions, as well as chromatin remodeling (the SNF2 domain) [16], [17]. Interestingly, the presence of the macro domain in the histone protein macroH2A and in proteins that contain DNA and RNA binding motifs would suggest an essential role in nucleic acid recognition.
Table 1.
Human macro domain family protein.
| Member | Other names | Locus linka | Chromosome | Reported functions | Ref. |
|---|---|---|---|---|---|
| MacroH2A1 | MacroH2A1.1 MacroH2A1.2 |
#9555 | 5q31.3-q32 | Transcriptional regulation, genome silencing, X-chromosome inactivation | Buschbeck et al. [42] |
| Costanzi et al. [43] | |||||
| Nusinow et al. [45] | |||||
| Hoyer-Fender et al. [46] | |||||
| Sporn et al. [60] | |||||
| Angelov et al. [66] | |||||
| MacroH2A2 | H2AFY2 | #55506 | 10q22 | Transcriptional regulation, developmental roles | Costanzi et al. [62] |
| Changolkar et al. [70] | |||||
| MACROD1 | LRP16 | #28992 | 11q11 | Transcriptional co-activator | Han et al. [63] |
| Han et al. [64] | |||||
| Yang et al. [65] | |||||
| MACROD2 | C20orf133 | #140733 | 20q12.1 | Developmental disorders and Kabuki syndrome | Maas et al. [41] |
| MACROD3 | GDAP2, FLJ20142, dJ776p7.1 | #54834 | 1p12 | Ganglioside-induced differentiation-associated protein | Neuvonen et al. [7] |
| ALC1 | CHD1L, CHDL, FLJ22530 | #9557 | 1q12 | Oncogene, ATP-dependent chromatin remodeler, epithelial–mesenchymal transition (EMT) | Gottschalk et al. [23] |
| Ahel et al. [24] | |||||
| Chen et al. [50] | |||||
| Chen et al. [73] | |||||
| Chen et al. [83] | |||||
| PARP-9 | BAL, BAL1, DKFZp666B0810 | #83666 | 3q21 | Modulates transcription and promotes B-cell lymphoma migration | Aguiar et al. [19] |
| Hakmé et al. [39] | |||||
| Aguiar et al. [79] | |||||
| PARP-14 | BAL2, CoaSt6, KIAA1268, pART8 | #54625 | 3q21.1 | Transcriptional cofactor and development | Goenka et al. [11] |
| Goenka et al. [12] | |||||
| Hakmé et al. [39] | |||||
| PARP-15 | BAL3, FLJ40196, FLJ40597, MGC126750, MGC126752, pART7 | #165631 | 3q21.1 | Transcriptional co-repressor | Aguiar et al. [19] |
| C6orf130 | MGC19570, dJ34B21.3 | #221443 | 6q21.1 | Serologic marker | Marina et al. [82] |
Gene locus as indicated at http://www.ncbi.nlm.nih.gov/locuslink.
Although speculative, the topology of macro domain proteins, which consists of diverse domains flanked by N- and C-terminal tails together with the conserved potential ligand-binding macro domain, indicates important and varying roles for these proteins in the regulation of diverse cellular functions. The macro domain proteins might be viewed as molecular bridges that bring together target proteins, via interactions with the variable domains, and metabolites of NAD+, including PAR, via binding to the conserved macro domain. Here, we review our current understanding of the high level of structural similarity among macro domains, and then focus on recent advances in understanding of the biological mechanisms that underlie the different functions of macro domain proteins. Finally, we explore how dysregulation of these proteins leads to human diseases, including cancer, and discuss efforts to develop drugs that target the macro domain to treat these conditions.
2. Structure of the macro domain: implications for ADPR affinity
Three-dimensional (3D) structures of the ADPR binding fragments of macro domains have been solved recently, which has permitted comparisons to be made with previously published members of the macro domain family and has provided additional evidence of similarities in the structure of macro domain proteins [18]. The determination of the 3D structures of the macro domains of archaea Af1521 and human macroH2A1.1 showed that these proteins have structural homology within the binding site for ADPR (Fig. 2A). The structure of the macro domain includes approximately 130–190 amino acid residues that fold into a globular mixed α-helix and β-sheet structure that contains a deep groove, a potential ligand-binding pocket ([5], [6]; as in Fig. 2A and B). Although there is a relatively high degree of sequence similarity (approximately 30–40%) between any two macro domains [6], [19], the substantial sequence variation between domains is probably responsible for the selectivity of different macro domains for specific binding partners. Recently, isothermal titration calorimetry (ITC) experiments have indicated that many proteins that contain macro domains can bind various forms of ADPR, such as mono-ADPR, PAR, poly (A), and the SIRT1 metabolite O-acetyl-ADP-ribose (OAADPR) [5], [6], [7], [18]. For example, the gene macroH2A1 contains two mutually exclusive exons, and alternative splicing generates two isoforms: macroH2A1.1 and macroH2A1.2. Moreover, Gly223 and Gly224 in macroH2A1.1 are replaced by larger residues (Lys226 and Asp227) in macroH2A1.2. Although the structural differences between the two isoforms of macroH2A are small, they do differ in their affinity for various forms of ADPR, the small structural changes completely abolish interaction with both OAADPR and ADPR [20]. The macro domain of Semliki Forest Virus (SFV) binds PAR well, but ADPR only poorly (for review [7]). However, surprisingly, GDAP2 binds both PAR and ADPR inefficiently, confirming the hypothesis that sequence alterations in the ligand-binding pocket of this protein which was compared to other macro domain proteins, might be related to different substrate specificities (nucleic acid recognition versus PAR binding) [7]. Whereas MACROD1 – rather than ADPR-1′′P hydrolytic enzymes acting on PAR – not only is specific ADPR binding module, but also is PAR binding module [7]. The significance of these different interactions remains unknown and presumably must await determination of the functions of individual macro domains.
Fig. 2.
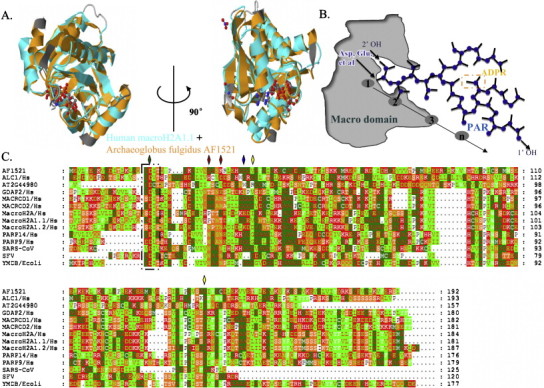
Macro domains are highly conserved structural domains that bind ADPR. (A) X-ray crystal structures of the macro domains from Archaeoglobus fulgidus (archaeal) Af1521 protein [18] (Protein Data Bank (PDB) accession code 2BFQ) and human macroH2A1.1 [22] bound to ADP-ribose (PDB accession code 3IID). The two views are rotated 90° relative to each other. (B) Schematic illustration of the proposed 2′ OH PAR capping function of macro domains, some amino-acids in conserved residues of macro domain proteins can serve as PAR acceptors, such as Asp, Glu. The square (orange) represents a mono-ADP-ribose. (C) Multiple amino acid sequence alignment of macro domain-containing proteins derived from SARS-CoV, SFV, Escherichia coli, Arabidopsis thaliana, and Archaeoglobus fulgidus with human macroH2A, MACROD1, MACROD2, GDAP2, PARP-9, PARP-14, and ALC1. The protein name is followed by the species abbreviation. Uniprot codes: AF1521 (Archaeoglobus fulgidus, O28751); AT2G44980 (Arabidopsis thaliana, Q3E6Q7); ymdB (Escherichia coli, C4ZRY6); ALC1 (human, Q86WJ1); GDAP2 (human, Q9NXN4); MACROD1 (human, Q9BQ69); MACROD2 (human, A1Z1Q3); PARP-9 (human, Q8IXQ6); PARP-14 (human, Q460N5); macroH2A1.1 and macroH2A1.2 (human, O75367); macroH2A (human, Q9P0M6). Conserved residues are colored according to their chemical properties. The black square represents the conserved motif (GDI/VT) among these different macro domain proteins. The bottle green diamond on top of the sequence indicates the amino acid that was mutated in AF1521 and ALC1, and red diamonds indicate the amino acids that were mutated in SARS-CoV. The blue diamond indicates the amino acid that was mutated in macroH2A1.1, and yellow diamonds indicate the amino acids that were mutated both in SFV and in MACROD1.
As summarized in Fig. 2C, multiple sequence alignment of macro domain proteins has indicated that there is a high level of sequence homology among viral, bacterial, archaea, and eukaryotic proteins. Most of the conserved residues are located within the ligand-binding pocket, which suggests that they are functionally important, and this region of the protein is a good candidate for the active site. Indeed, mutagenesis studies have demonstrated that some conserved residues play an important role in the ability of the domain to bind ADPR. For example, the mutations Gly182Tyr and Gly270Tyr in MACROD1 inactivate ADPR binding and the hydrolysis of ADPR-1′′P, and the corresponding mutations in the SFV macro domain protein (Gly32Tyr and Gly112Tyr) also totally abolish ADPR-1′′P hydrolysis, but none of the mutations affect the binding of PAR [7]. Similar effects may be observed for other macro domain proteins. Mutations of amino acids 10 and 24 from Asn to Ala in the ADPR-binding region of SARS-Cov macro domain did not induce a significant decrease in PAR-binding either [21]. In contrast, recent studies have determined the crystal structure of the macroH2A1.1 macro domain–ADPR complex and model PAR into the binding pocket, which allows them to identify residues (for example, Gly224) whose mutation abolishes binding of ADPR and PAR [20], [22]. An Asp20 to Ala mutation in AF1521, a macro domain protein from A. fulgidus, was found to reduce substantially the affinity of this protein for ADPR [5]. It is tempting to speculate that the Asp residue of the GDI(V)T motifs seen in recently published macro domain structures binds ADPR in an analogous manner. Indeed, two recent independent studies have supported the possibility that this Asp residue is essential for the binding of PAR by some macro domains; for instance, the macro domain of amplified in liver cancer 1 (ALC1) is necessary and sufficient for PAR binding, and PAR binding is reduced greatly in the ALC1 Asp723Ala mutant [23], [24].
3. Macro domain proteins and PARylation
Posttranslational modifications (PTMs) play a key role in regulating diverse biological functions. One of the oldest and least understood PTMs is the PARylation of proteins, including histones. PARylation is mediated by PAR polymerases (PARPs), PARP-1 and PARP-2, which use NAD+ as a substrate. PARPs catalyze the covalent attachment of ADPR units to Glu or Asp residues on target proteins to generate long, linear and branched PAR chains, which are synthesized and degraded rapidly. This reaction is reversible and dynamic process, as indicated by the short-half life of the polymer. PAR glycohydrolase (PARG) and ADP-ribosyl protein lyase catabolize PAR; the former cleaves the ribose–ribose bonds of both the linear and branched portions of PAR, whereas the latter removes the protein-proximal ADPR monomer [25]. Nuclear PARP-1 itself acts as the main PAR acceptor via auto-modification, and its activity is induced by stress-response pathways, such as responses to DNA lesions and metabolic stress ([26], [27], [28]; Fig. 3 ). Recent genetic and biochemical data indicate that PARylation has important roles in many physiological and pathophysiological processes [8], [29], [30], [31]. However, despite the important functions of PARylation, it remains poorly understood how these PTMs are recognized by other proteins.
Fig. 3.
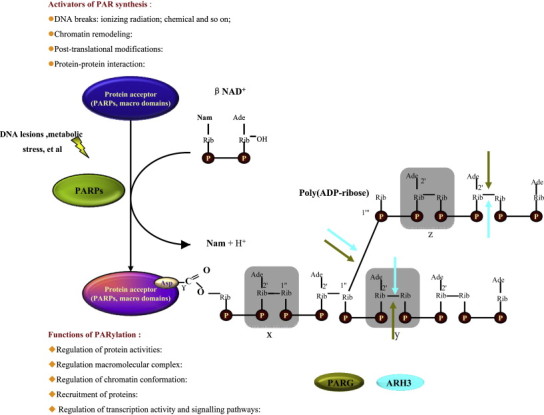
Metabolism of poly(ADP-ribose). PARPs hydrolyze NAD+, releasing nicotinamide (Nam) and one proton (H+), and catalyze the successive transfer of the ADP-ribose moiety to nuclear protein acceptors. The reaction is initiated by the formation of an ester bond between the amino-acid acceptor (Glu, Asp or COOH-Lys) and the first ADP-ribose. Polymer elongation involves the catalysis of a 2′–1′′ glycosidic bond. PAR is heterogeneous in size and complexity, as indicated by the shade labels (x, y, z labels) that represent values from 0 to more than 200. PARG and ARH3 can both hydrolyze PAR at the indicated positions. Activators and functions of PAR synthesis are indicated. Ade, adenine; PAR, poly(ADP-ribose); PARG, poly(ADP-ribose) glycohydrolase; ARH3, ADP-ribosyl hydrolase-3; PARP, poly(ADP-ribose) polymerase; P, phosphate; Rib, ribose.
Studies over recent decades have begun to identify and characterize the proteins that bind to PAR. Studies have demonstrated that most macro domain proteins could serve as a receptor of PAR in living cells [22], [23], [24]. These findings provide new insights into the role of the PAR-binding macro domain in diverse biological functions and show that PARylated macro domain proteins have the potential to orchestrate various chromatin-based biological tasks, including DNA repair and chromatin remodeling (for review [22], [23], [24]).
How widespread is the interaction of macro domains with PAR? So far only 10 human proteins containing macro domains have been reported (according to the NCBI database). Moreover, it has been shown that only a few of them bind PAR, the low number strongly suggests that other domains that bind PAR may exist. Indeed, in addition to macro domains, another two such motifs have been described and derived potential consensus sequences for proteins with this capacity. One is found in several important DNA damage checkpoint proteins such as p53, MSH6, histones, DNA–PKcs, Ku70, XRCC1 and telomerase, and is characterized by a 20 amino acid motif that contains two conserved regions: (i) a cluster rich in basic residues and (ii) a pattern of hydrophobic amino acids interspersed with basic residues [32], [33], [34]. The second characterized motif is the PAR-binding zinc finger (PBZ), which is also associated with DNA repair and checkpoint control. Recent study has demonstrated interaction of PAR with this motif in two representative human proteins, APLF (aprataxin PNK-like factor) and CHFR (checkpoint protein with FHA and RING domains) [35]. Analysis of the primary sequence of CHFR revealed a conserved putative C2H2 zinc-finger motif at its carboxy terminus. The putative C2H2 zinc-finger that is referred to as PBZ, is separated by a 6–8 amino acid spacer and has the consensus [K/R]xxCx[F/Y]-GxxCxbbxxxxHxxx[F/Y]xH (for review [35]). Study has established the functional importance of the PBZ motif, demonstrating that specific PBZ-targeted mutations abrogate their PAR-binding capacity and functions in the antephase checkpoint [35]. Collectively, the identification of specific PAR-binding sites in several proteins of the cellular signal network suggests that these proteins may be interaction partners of the PARP protein family. By targeting specific domains in these proteins, PAR could regulate protein–protein or protein–DNA interactions, protein localization, or protein degradation. PAR could also play a chaperone function in the DNA damage signal network by facilitating the temporary formation of multiprotein complexes.
It is possible that PARylated proteins act as an essential scaffold for the efficient recruitment of components of the DNA damage responses, and this is supported by a recent study that suggests that PARylated PARP-1 serves as a molecular bridge in the rapid assembly of a novel signaling complex following DNA damage in the nucleus [36]. Does this mean that the PARylation of proteins that contain one of these three PAR-binding motifs (PARBMs) provides specific interaction platforms for the recruitment of repair proteins involved in the pathways of single-strand break repair (SSBR) and base excision repair (BER)? It has not been explained exactly how DNA damage-inducing agents cause the PARP-1-mediated PARylation of PARBMs that serve as a scaffold for the recruitment of DNA damage response proteins. Whatever the mechanism, it is clear that the PARylation of proteins has a key role in diverse cellular functions, including DNA damage response and transcriptional regulation. Both inactivation of the catalytic activity of PARP-1 and the use of macro domains that cannot bind PAR abrogate macro domain-mediated chromatin rearrangement and DDR completely [22], [23], [24]. Collectively, the specific targeting of proteins to these sites of PAR accumulation depends on the recognition of PAR by defined PARBMs.
Recent evidence strongly suggests that not all of PARP family members are able to function as polymerases but instead are mono-ADP-ribosyltransferases (mARTs) [37]. It is tempting to speculate that intracellular mono-ADP-ribosylation has been widely used as a mechanism to regulate many different aspect of cell physiology. Might these three motifs also recognize mono-ADP-ribosylated substrates? Crystallographic and calorimetric studies have demonstrated that the macro domain binds to the terminal ADPR of PAR, and the recent work strongly shows that this binding is efficiently competed by an excess of free ADPR. To date, there is no clear-cut evidence that eukaryotic macro domains bind to mono-ADP-ribosylated proteins. At the least, the E988K mutant of PARP-1, which lacks intrinsic PARP activity but is capable of auto-mono-ADP-ribosylation, fails to recruit macroH2A1.1 [22]. However, a recent report suggests that Af1521 can potentially interact with mono-ADP-ribosylated proteins, which can then be identified by mass spectrometry [38]. Currently, it is unknown if indeed mono-ADP-ribosylation is a broadly used PTM and whether macro domains or other PAR-binding elements interact with a particular protein sequence motif that carries ADPR. So far no evidence supports this presumption. Thus it seems likely that separate domains recognize mono-ADP-ribosylation versus PARylation and the above findings also indicate a potential mechanism by which cells use modification-dependent interactions to orchestrate the assembly of regulatory pathways.
4. Widespread biological functions of macro domain proteins
4.1. The developmental roles of macro domain proteins
Macro domain proteins are expressed ubiquitously in adult tissues, but the physiological and cellular functions of these proteins remain elusive. Of the mammalian macro domain proteins, only the potential developmental roles of macroH2A and the macroPARPs have been investigated. The role of macroH2A in development is characterized better than that of other macro domain proteins, possibly because macroH2A was the first of these proteins to be described and is the most intensively studied.
The differential distribution of several macroPARPs at different stages of development hints at a possible physiological role in development. The first important observation was that the expression levels of different macroPARPs vary significantly during mouse embryogenesis and in adult tissues [39]. PARP-9 is developmentally regulated, prominently expressed in the thymus, in specific regions of the central nervous system and of the gut. This regionalized expression pattern during mouse organogenesis suggests that PARP-9 could have a function in lymphogenesis, neurogenesis, and development of the intestine. In the adult mouse, the highest levels of PARP-9 transcripts were found in the medulla of the thymus, suggesting a role for PARP-9 in thymocytes maturation. PARP-14 also likely plays a role during thymic development and function, because this organ is the major site of PARP-14 expression, although at low levels [39]. However, PARP-14 knockout mice presented no overt developmental abnormalities and displayed normal Mendelian genetics [40]. Interestingly, human PARP-9 and mouse PARP-14 were reported to act in the transcriptional regulation of gene expression activated by IFNγ and IL-4, respectively [11], [12]. These two cytokines can antagonize each other's function in thymocytes maturation and macrophage activation during the immune response, raising the hypothesis of a possible antagonistic function for PARP-9 and PARP-14 in the immune response. PARP-9 was also expressed at higher levels in the enterocytes of the intestine, suggesting specific functions that could be related to homeostasis, nutrient digestion, and absorption, or to the barrier and defense function against toxic compounds or pathogenic microoranisms.
In addition, previous studies have indicated that C20orf133 is a causative gene for Kabuki syndrome (KS), which is a rare congenital malformation. Interestingly, the pattern of expression of C20orf133 during mouse embryonic development supports its importance for the development of various tissues and organs [41]. Furthermore, changes in the expression of the subtypes of macroH2A during development suggest that macroH2A plays an important role in the developmental regulation of chromatin structure. Further studies have indicated that macroH2A, as an epigenetic regulator of development and cell fate decisions, is involved in the concerted regulation of gene expression programs during cellular differentiation and vertebrate development [42]. As macroH2A1.1 is expressed in metabolic tissues, including pancreatic beta islets, and macroH2A1-deficient in mice revealed metabolic disorders (e.g. dysregulation of fat and sugar metabolism), rather than obvious DNA damage phenotypes [43]. The spatial expression of subtypes of macroH2A is not uniform, which suggests that macroH2A subtypes have specific functions in a subset of developmental processes and particular tissues.
Another role of macroH2A in cellular differentiation and development relates to the inactivation of one X chromosome in the somatic cells of females and to male meiotic sex chromosome inactivation (MSCI) [44], [45], [46]. MSCI is an evolutionarily driven process in which both the X and the Y chromosome become heterochromatic and transcriptionally inactive in males during prophase I at pachytene [47], but unlike X-inactivation in somatic cells, MSCI does not require Xist RNA (a transcript specific to X-chromosome inactivation) [48]. In addition, the location of macroH2A in somatic cells changes in a manner that depends on the cell cycle [49]. These studies on macroH2A have shown that dramatic changes could occur in chromatin during cellular differentiation and development.
Emerging evidence has indicated that the different subtypes of macroH2A might compensate for each other functionally, for instance, knockout mice that lack macroH2A1 develop normally [43], but macroH2A-deficient zebrafish, which express only one form of macroH2A, macroH2A2, show developmental defects [42]. Therefore, comparing to the two completely different results, there might be additional regulatory pathways that compensate for the loss of macroH2A function. It remains to be seen how a double knockout of both forms of macroH2A would affect mammalian development; however, one might speculate that the two forms of macroH2A would compensate for each other. Taken together, these results demonstrate that macro domain function is required for proper development, and during developmental processes, different macro domains might compensate for each other functionally.
4.2. The macro domain as an anti-apoptotic factor
Many studies in cell-based and animal models have shed important light on the potential involvement of members of the macro domain family in some apoptotic signaling pathways. The overexpression of macro domain proteins in various cell lines has been shown to protect against multiple apoptotic signals, such as staurosporine (STS), camptothecin, phleomycin, and ionizing radiation [24], [50]; Furthermore, knockdown of the expression of macro domain proteins in various cell lines results in increased apoptosis [24], [50]. The antiapoptotic activity of overexpressed macro domain proteins requires an intact macro domain, because deletion of this domain abrogates the ability of these proteins to antagonize apoptosis [50].
4.2.1. Regulation of apoptosis via PAR-dependent pathways
Recently, several studies have demonstrated that macro domain proteins can also inhibit apoptosis in a PAR-dependent manner [24], [40]. Growing evidence has demonstrated a role for macro domain protein in the regulation of cell apoptosis that occurs in response to biological, chemical or physical stimuli, via at least two non-exclusive mechanisms in PAR-dependent manners: through the modulation of chromatin structure, or through direct interaction with transcription factors and/or cofactors. On the one hand, after DNA damage, macro domain can inhibit apoptosis by mediating a PAR-dependent chromatin-remodeling activity and facilitate DNA repair reactions within a chromatin context [24]. On the other hand, the mechanism by which PARP-14 can mediate inhibition of cell apoptosis is by interacting directly with transcription cofactors (such as p100) [40]. Thereby, PARP-14 mediates interleukin (IL)-4 regulation of the expression of genes determining cell survival. Intriguingly, the intrinsic PARP activity of PARP-14 was shown to be required for this regulation event, these results indicate that PAR polymerization mediates a survival signal in cells. More recent evidence for the essential role of PAR in the efficient management of apoptotic pathways has been provided by the IL-4-induced protection of B cells against apoptosis is impaired significantly by the absence of automodification PARP-14 or the inactivation of its intrinsic PARP catalytic activity (for review [40]). Collectively, recent studies have led to a greater interest in the biology of PAR, but the emphasis so far has been largely on the identification of PAR-binding proteins [5], [7], [31], [32], [34], [35]. Many studies go beyond PAR binding to assign specific functional outcomes for the binding events in the nucleus, revealing the essential role of these interactions between macro domain proteins and PAR in the inhibition of cell apoptosis process [23], [24].
4.2.2. Regulation of apoptosis via Nur77-associated pathway
Another protein that can explain the important role of intact macro domain within ALC1 in cell apoptosis is Nur77, which is also known as NGFI-B or TR3, is a unique transcription factor belonging to orphan nuclear receptor superfamily [51], [52], [53]. However, the role of Nur77 in apoptosis is still not completely understood, because Nur77 has been reported both to enhance and to suppress apoptosis [53]. Nur77 mediated apoptosis involves both its transcriptional activities to up-regulate the gene expressions responsible for promoting apoptosis [54] and its translocation from the nucleus to the cytosol to convert Bcl2 function from antiapoptosis to proapoptosis [52]. Interestingly, Nur77 functions in both positive and negative regulation of apoptosis depending on the cellular context and different external signals. This notion has been confirmed by previous studies showing that, Nur77 survives cells in tumor necrosis factor (TNF) induced cell apoptosis [55] and prevents A20B cells from ceramide-induced cell death [56]. It is most likely that Nur77 exerts its antiapoptotic effects by functioning in the nucleus [55], [57]. However, the mechanism by which the nucleus to the mitochondria translocation of Nur77 is manipulated is still unclear. Notably, a study from Chen et al. demonstrated that the intact C-terminal macro domain of ALC1 (aa600–897) is responsible for the protein's antiapoptotic activity [50]. The proapoptotic activity of Nur77 is abrogated in cells that are overexpressing ALC1, which interacts with Nur77 and inhibits its translocation from the nucleus to the mitochondria [50]. This novel role for macro domains in regulating apoptotic pathway extends the physiological functions of these structures beyond the control of transcription factor activity.
Previous study has established that macro domain protein has an essential role for mediating inhibition of cell death through caspases rather than the caspase-independent pathway [40], [50]. Macro domain participates in apoptotic signals by several means: by regulating the transactivation of transcription factors and by inhibiting the nucleus to the mitochondria translocation of apoptosis-associated proteins, and also by protecting against DNA damage. Notably, the macro domain mediates protein–protein interactions and is also essential for the binding of PAR. Besides these mechanistic insights, the most convincing piece of evidence for antiapoptotic function of macro domain proteins is the fact that cells derived from PARP-14-knockout mice have profound defects in executing cell survival by different stimuli [40]. Collectively, the anti-apoptotic activity of macro domain proteins might depend on the structural and biochemical features of this domain that allow interaction with other transcription factors involved in the regulation of apoptosis.
4.3. The DNA damage response involves macro domain proteins
A role for macro domains in mediating DNA-damage responses is strongly implied by a series of observations: after DNA damage, macro domains can sense PARP-1 activation in vivo by PAR-dependent manners [22]; they co-localize with sites of DNA repair or sites of DNA single/double strand lesions [22], [23], [24]; and many proteins linked to DNA repair and checkpoint dynamically interact with macro domain through PARylated PARP-1[22].
One of the first pieces of evidence that suggested a role of macro domain proteins in the DNA damage response was the cytological observation that, following DNA damage, macro domain protein localizes at damage induced foci (also known as ionizing radiation induced foci) [22], [23], [24], which co-localize with foci where the DNA repair proteins accumulates. A comprehensive summary of the proteins that co-localize with macro domains before and after DNA damage was recently published by several laboratories [22], [23], [24] and portrays an extremely complex set of interactions. Many of these proteins linked to DNA repair, such as DNA–PKcs, Ku70–Ku80, XRCC1, APLF and PARP-1, co-localize with macro domain after DNA damage. These interactions are dependent on PARP-1 enzymatic activity, which suggests that macro domain localizes at DNA damage induced foci through PARylated PARP-1. The DNA damage induced foci, marked by the histone variant H2AX phosphorylated on Ser139 (known as γH2AX), represent sites of DNA breaks [58]. γH2AX is essential for the accumulation of numerous DNA damage repair factors at sites of DNA breaks, suggesting that γH2AX is one of initial recruiting factors for various checkpoint and DNA repair proteins to DNA breaks. Specifically, in cells expressing macroH2A1.1, γH2AX increased at the laser cut relative to the surrounding chromatin [22]. Thus, the transient compaction of macroH2A1.1 chromatin upon PARP-1 activation can dynamically modulate DNA damage responses.
Despite having conserved macro domain, macro domain containing protein does not bind directly to γH2AX. The localization of macro domain proteins to damage induced foci occurs in PARP-1-dependent manner, but is independent of another PARP activity: PARP-2 [22]. So how does macro domain localize to damage induced foci? Mass spectrometry analysis and affinity purification approaches identified the PARP-1 protein as a macro domain binding protein [22]. Following DNA damage, PARP-1 was activated, providing a convenient readout for transient PAR accumulation within a spatially defined region (the ‘foci’) in vivo. Interestingly, macro domain proteins were rapidly recruited to PARP-1 activation sites and also identified as a component of PARP-1, Ku70–Ku80 and DNA–PKcs complex. Detailed analyses indicate that PARP-1 bridges the interaction between macro domain protein and Ku70–Ku80–DNA–PKcs and mediates the localization of macro domain protein to sites of DNA damage (Fig. 4 ). The finding that PARP-1 and its enzymatic activity are required for proper macro domain proteins localization following DNA damage suggested the existence of a PAR-dependent signaling pathway that controls the retention of the Ku70–Ku80, DNA–PKcs, PARP-1 and macro domain complex at DNA double-stranded breaks (DSBs) (for review [22]). This was supported by the observation that the recruitment of macro domain proteins to the sites of DNA damage is abrogated completely by using PARP inhibitors or PAR-binding deficient macro domain.
Fig. 4.
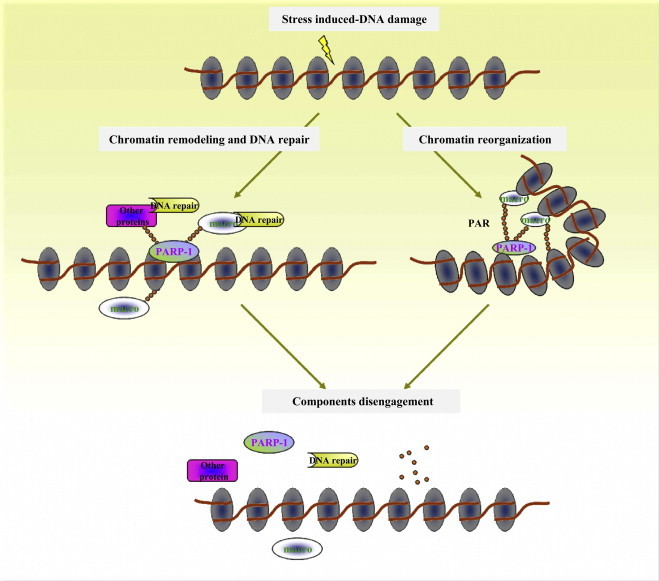
Macro domain proteins and the DNA damage response. DNA damage results in the recruitment and activation of PARP-1. Subsequently, PARP-1 catalyzes the synthesis of PAR on itself and other proteins. These polymers serve as molecular bridges for proteins that contain PAR-binding domains (such as macro domain proteins, PBZ proteins, and other proteins), and thereby contribute to chromatin remodeling and DNA repair, when the DNA has been repaired, these components are disengaged from the nucleosomes.
The functional consequences of this complex set of interactions have not been completely elucidated. It is clear that macro domain proteins mediate checkpoint responses and the inhibition of apoptosis after DNA damage, as discussed above. However, do they also have a role in DNA repair? A couple of observations suggest that this is likely: the co-localization of many DNA repair factors with macro domain proteins occurs mostly at early time points after DNA damage [22], [23], [24], and activation of PARP-1 results in the co-localization of macroH2A1.1, XRCC1, APLF and γH2AX, which indicates a PARP-1 dependent accumulation of DNA repair machinery in response to DNA damage [22]. These observations imply that macro domain protein is specifically targeted to sites of DNA damage through interaction with PAR and functions to regulate compaction of chromatin during DNA repair. What might be the functional consequences of this chromatin compaction? Recent studies have shown that it inhibits the recruitment of Ku70, a protein involved in DNA repair, and increases the phosphorylation of H2AX, both of which suggest a possible role for macro domain in regulating DNA-damage responses [22], [24]. Thus, the transient compaction of chromatin induced by macro domain upon PARP-1 activation can dynamically modulate DNA damage responses. Moreover, using RNA mediated interference induced knockdown of PARP-1 or treatment with PARP inhibitors, the efficient recruitment of macro domain at the foci is inhibited or blocked (for review [22]). Thus, it seems possible that macro domain, perhaps by facilitating access of the DNA repair machinery to chromatin, might modulate proper DNA damage responses.
In conclusion, macro domain proteins might regulate DNA damage responses in different ways: by mediating the rearrangement of chromatin and transiently affect the DNA damage response by PAR-dependent manners; by actively regulating DNA repair; and/or by integrating DNA repair with checkpoint responses. All of these scenarios are possible and not mutually exclusive, and further work is needed to understand the role of macro domain proteins in DNA damage responses.
4.4. The macro domain and the modulation of chromatin structure
At sites of DNA breakage, the chromatin structure is opened up by the removal of histones as a result of their non-covalent association with PARylated PARP-1 and their PARylation by PARP-1 [8], [9]. One important function of histone modification is the ordered recruitment of chromatin remodeling activities that recognize modified histones via specific domains (methylation is recognized by chromo-domains and PHD domains, acetylation is recognized by bromo-domains, PARylation is recognized by macro domains and PBZ domains) (for review [22], [35], [59]).
As mentioned above, in response to DNA damage, PARylated macro domains are recruited rapidly to PARP-1 activation sites (for review [22]). Does this mean that the macro domain might serve as a modulator of chromatin structure? Indeed, most evidence suggests that most macro domain proteins contribute to the assembly of chromatin by one of two different typical patterns. The first mode is exemplified by ALC1, which is a member of the SNF2 superfamily of ATPases and which contributes to the regulation of chromatin via an ATP-dependent chromatin remodeling pathway [23], [24]. Interestingly, recent study strongly showed that the ATPase and nucleosome-remodeling activities of ALC1 are dependent on NAD+-dependent PAR synthesis by PARP-1 and the macro domain of ALC1 and also suggested a coupling of ATPase and PAR binding activities [23]. Surprisingly, ATPase activity depends on an intact macro domain, exemplified by ALC1 (D723A), which does not bind PAR, lacks ATPase activity in either the presence or absence of PARP-1 and NAD+. However, free PAR or ADPR are unable to activate ATPase and nucleosome-remodeling activities of ALC1, which strongly suggests that ALC1 ATPase activity depends on auto-modification of PARP-1 and/or on PARylation of ALC1 itself (for review [23]). Unlike other chromatin remodeling and modifying enzymes and complexes, ALC1 lacks targeting domains, such as bromo- or chromo-domains, however, recent findings provided strongly evidence that nucleosomes are the relevant substrate for ALC1 and raised the possibility that ALC1 could be targeted to chromatin by PARylation via its macro domain [23], [24]. In the second mode, the PARylation of macro domain proteins might contribute to the epigenetic modification of histones [22]. Physiological PARP activation, such as PARP-1 and PARP-2, may result in transient, macroH2A1.1-dependent chromatin changes, which might be relevant for the proper tuning of local chromatin architecture (for review [22]). This effect requires an intact macroH2A1.1 macro domain (that is, PAR binding) and catalytically active PARP-1. This result indicates that macro domain in macroH2A1.1 can be recruited to sites of PAR synethesis in the nucleus and that the recruitment is dependent on PAR binding. Interestingly, in both typical patterns, the PARylation of macro domains plays a foundational role in chromatin remodeling, because the mutation and deletion of the macro domain in macroH2A1.1 totally abrogates the ability of these proteins to modulate chromatin structure. Notably, the macroH2A1.2 variant of macroH2A, which is deficient for PAR binding, cannot sense PARP-1 activation or mediate chromatin remodeling [22]. The different isoforms of macroH2A show distinct expression patterns [60], and the dichotomy between macroH2A1.1 and macroH2A1.2 function correlates with their expression. Whereas macroH2A1.2 is expressed widely, macroH2A1.1 is detected in post-mitotic and senescent cells [61], [62], which suggests that cell-type specific expression of macro domain proteins could contribute to chromatin plasticity. Taken together, these findings show that PAR-binding macro domains mediate the rearrangement of chromatin and lead to chromatin relaxation, which has a transient effect on the DNA damage response; they provide a key insight into the molecular consequences of the macro domain, and emphasize the importance of chromatin reorganization in genome stability.
5. The transcriptional roles of macro domain proteins
Although the biochemical function of macro domain proteins remains largely unknown, consistent evidence is accumulating for a role for most macro domain proteins in transcriptional regulation (Table 1). As mentioned previously, the macro domain, which is an evolutionarily conserved domain, is found in proteins that are involved in diverse biological functions, including the regulation of transcription. Remarkably, the macro domain can activate transcription by functioning as a co-activator of specific transcription factors [11], [12], [63], [64], [65]. Conversely, the macro domain can also bind DNA directly; when tethered to the promoter area macro domains display a cryptic transcriptional repression activity that depends on the presence of an intact domain [19], [66]. This suggests that the conformation of the macro domain and/or its interactions with other proteins determine its effect upon transcription.
In agreement with this idea, certain macro domain proteins have been found to act as both transcriptional co-activators and co-repressors (Fig. 5 ). CoaSt6/PARP-14 can act as a co-activator in the Stat6 possibly through their interaction with the transcriptional co-activator p100 with PARylation modification catalyzed by its intrinsic PARP activity [11], [12], [40]. Similar effects may be observed for other macro domain proteins, MACROD1 contributes to elevated nuclear-factor (NF)-κB activity by acting as its essential co-activator (our unpublished observations), and it also interacts directly with nuclear receptors. For example, MACROD1 acts as a potential co-activator to amplify the transactivation activity of nuclear receptors, such as estrogen receptor α (ERα) and androgen receptor (AR), through its conserved domain under conditions of receptor stimulation [63], [64], [65]. These findings are supported by the analysis of PARP-14−/− mice [40]. Inactivation of PARP-14 in these mice blocks the IL-4-induced protection of B cells against apoptosis after irradiation or growth factor withdrawal, and also impairs IL-4-dependent transcriptional activation. Furthermore, the induction of several B-cell survival factors (e.g. Pim-1 [67]; Mcl-1 [68]) by IL-4 also depends on PARP-14 [40]. Unlike bona fide co-activators such as CREB-binding protein (CBP) and p300, macro domain proteins do not possess intrinsic histone acetylase activity. However, they can regulate transcriptional activity and interfere with p300-dependent histone acetylation [69].
Fig. 5.
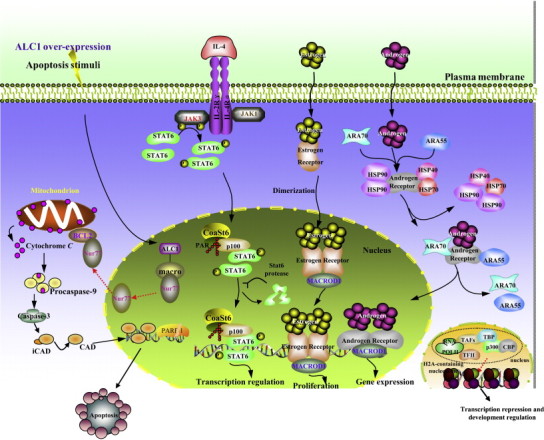
Differential cellular roles of macro domain proteins. Some examples of the participation of macro domain proteins in cellular pathways linked to cell proliferation and cell death. Collectively, macro domain family proteins are involved in the inactivation of chromosomes and transcriptional repression. See main text for details, the red arrows represent repression and black arrows represent activation.
As mentioned above, macro domain proteins can also act as co-repressors of transcription (Table 1); For example, the BAL family proteins repress transactivation when tethered to a promoter [19]. Furthermore, the macro domain of macroH2A has been implicated in the direct silencing of transcription by interfering with the binding of NF-κB to its cognate sequence [66]. Interestingly, the H2A-like domain of macroH2A does not affect p300-dependent RNA polymerase II (RNAP II) transcription [69], but does interfere with SWI/SNF nucleosome mobilization [66]. MacroH2A exhibits some redundancy in function with respect to nucleosome remodeling because each individual domain of macroH2A (either non-histone region or H2A-like) when fused to H2A can impair nucleosome remodeling [69]. It is tempting to speculate that in vivo macroH2A could contribute to the repression of transcription by affecting at least two different pathways: histone acetylation and chromatin remodeling. In addition, macroH2A1 is required for the transcriptional silencing of endogenous murine leukaemia viruses found in the mouse genome [70]. Although most of the current literature has focused on the role of macroH2A1 in the repression of gene expression, recent evidence suggests that transcriptional repression might not be the only function of this histone variant. For example, phosphorylated macroH2A1 is excluded from the transcriptionally inert inactive X chromosome [71]. In addition, one group has documented an unexpected role for macroH2A1 in enhancing the transcription of a subset of autosomal genes [72]. These findings indicate that the macro domain might have functional versatility in the regulation of transcription. The ability of macro domain proteins to interact with co-activators such as p100 suggests that the macro domain could co-activate transcription through its ability to stabilize co-activator–transcription factor complexes. By contrast, the mechanisms by which some macro domains act as co-repressors remain unclear, although it is tempting to speculate that macro domain proteins could also participate in and stabilize co-repressor–transcription factor complexes.
Current understanding of some of the molecular mechanisms that underlie transcriptional regulation suggests that many of the biological functions of the macro domain might depend on its ability to bind PAR. For instance, PARP-14 can regulate the activity of Stat6 in a ligand-dependent manner by PARylating and interacting with p100, a cofactor for Stat6, and in PARP-14−/− mice, IL-4-induced protection of B cells against apoptosis, which depends on Stat6, is impaired profoundly. However, it is unclear, whether or not PARylation plays a fundamental role in other types of transcriptional regulation.
The data gathered so far support a model in which the macro domain exerts its regulatory activity on transcription in the nucleus, where it regulates the proper assembly of transcriptional complexes. Therefore, it is possible that macro domain proteins are recruited transiently to transcription factors and cofactors, or to their proximity, either to take part in transcription or to sever as modifiers (for example by PARylation) (Fig. 5). In the case of nuclear receptors, this process could depend on their ligand-binding by macro domain proteins, as suggested by the fact that the interaction between MACROD1 and ERα or AR is dependent on both intact macro domain and receptor stimulation [63], [64], [65]. However, chromatin that contains macroH2A1 and has been assembled in vitro is more repressive to transcription than canonical chromatin; it specifically blocks transcriptional initiation, and not elongation [69]. The models proposed here also suggest that some macro domain proteins regulate the transcriptional activity of particular transcription factors and their target genes through special ways respectively.
6. Links between macro domain proteins and diseases
6.1. Macro domains in cancer and degenerative diseases
It is now well established that various members of the macro domain family are overexpressed in a range of human tumors [73], [74], [75], [76], [77], [78], [79], [80]. Generally, MACROD1 appears to be the family member most widely overexpressed in human cancers, with high levels of expression observed in endometrial carcinoma, gastric carcinoma, colorectal carcinoma, and breast carcinoma [74], [75], [76], [77], [78]. ALC1 is most widely overexpressed in hepatocellular carcinoma (HCC) [80], [81], [83]. Recent studies have now begun to delve into the more substantial problems, including: whether the overexpression of macro domain proteins affects the differentiation state, growth rate or metastatic potential of a tumor cell; what the immediate downstream consequences of macro domain proteins overexpression are; and what the prospects are for inhibiting macro domains or its downstream targets in the tumor cell.
Overexpression of macro domain protein has been correlated with the histological grade of a cancer cell in some tumor types. In HCC, ALC1 is expressed at higher levels in higher tumor grades [81] and, in gastric carcinoma, high MACROD1 expression has been correlated with poorly differentiated histological grade [76]. High MACROD1 expression is associated with poor or moderate histological grade in invasive ductal breast carcinoma and a poor prognostic outcome [77]. MACROD1 overexpression is also reported to correlate with poor prognostic outcome and to associate with poor or moderate histological grade in colorectal cancer [78]. However, further studies which allow us to better define the MACROD1 functional importance in various cancers and to determine whether MACROD1 serves as a new molecular marker to assess the prognosis of carcinomas will be needed.
Important molecular marker correlations are also now beginning to be drawn. Clinical research has indicated that the expression level of some of macro domain proteins in carcinoma is significantly higher than that in matched normal tissues and is correlated significantly with shortened survival in patients with cancer [76], [77], [78], [79], [81]. In addition, macro domain proteins could become useful biomarkers to predict the risk of recurrence of some tumors. For instance, studies have indicated that the human histone variant macroH2A can predict lung cancer recurrence and therefore could serve as a useful prognostic biomarker [60]. Moreover, another macro domain protein, C6orf130, which is a B-cell antigen, represents a promising biomarker of effective anti-CLL immunity [82].
How can the effect of macro domain proteins on the state of a tumor cell be established more definitively? Overexpression of MACROD1 in endometrial cancer cell lines has been shown to increase the invasiveness of these cells in tissue culture [75]. In contrast, knockdown of MACROD1 in prostate cancer cell lines has been shown to decrease the growth of these cells in vitro [65]. Macro domain protein has an important role in enabling cancer cells to adapt their metabolism to cope with the demands of enhanced migration and metastasis, PARP-9, for instance, was found to be overexpressed in aggressive diffuse large B-cell lymphomas (DLB-CL), and its ectopic over-expression promotes the migration of lymphocytes in vitro, which indicates that PARP-9 might promote the dissemination of malignant B cells in high-risk DLB-CL [79]. ALC1 is likely to have key roles in the initiation and progression of HCC, an assertion that is supported by overexpression data for a range of HCC cell lines. This involvement has been illustrated by the susceptibility of transgenic mice that are ubiquitously expressing ALC1 to various types of tumor [73]. In primary HCC, overexpression of ALC1 was significantly associated with tumor microsatellite formation, advanced tumor stage, overall survival time [81]. To explore its oncogenic mechanisms, in vitro and in vivo functional studies in mice showed that ALC1 contributed to tumor cell migration, invasion, and metastasis by increasing cell motility and inducing filopodia formation and epithelial–mesenchymal transition (EMT) [83]. Whether macro domain is required for the maintenance of transformed state or tumor cell aggressiveness can also be tested by crossing tumor prone strains (in which macro domains are expressed in the primary tumor) to animals with reduced macro domain dosages. It is these types of animal model systems that will ultimately allow us to determine the precise role of macro domain overexpression in the generation, establishment or progression of various tumor types.
In addition to its effects on the tumor cell, macro domain protein has been shown to be crucial for degenerative diseases. Recent studies on macro domain proteins have suggested a role for these proteins in chromatin biology [22], [23], which in turn suggests that the genes for these proteins might be involved in congenital malformation syndromes. At present, several congenital malformation syndromes have been shown to be caused by haploinsufficiency of a gene involved in chromatin remodeling [84], [85]. One such syndrome is KS and mutations in the macro domain gene C20orf133 have been identified in patients with KS. However, both the identification of different chromosomal rearrangements in patients with KS features and the absence of C20orf133 mutations in a large number of patients with KS suggest that KS is genetically heterogeneous [41].
What are the downstream consequences of macro domain loss that lead to the observed malignant phenotypes defects in human cancers? Because macro domain proteins control the transcription of other genes, it will be important to determine both the immediate early transcriptional effects of macro domain loss and the secondary transcriptional effects to understand the phenotype fully. Recently, an indirect effect of macro domain loss on ARHGEF9 expression in ALC1-silenced HCC cell line has been reported, and the analysis extended to include a role for ARHGEF9 in mediating the ALC1 loss phenotype in HCC [83]. Moreover, strongly evidence was reported to support that the transcriptional regulator ALC1 upregulates ARHGEF9 transcription, which subsequently increases Cdc42 activity, causing filopodia formation, EMT, and finally HCC invasion and metastasis [83]. However, it is no direct evidence whether macro domain in ALC1 plays a major role in the regulation of primary tumor malignant phenotype. According to previous study, macro domain in ALC1 has an essential role for inhibition of cell death in HCC cell line [50]. It is likely that other effectors of macro domain loss are also mediating the effects on tumor cells and a more in depth analysis is now required.
6.2. PARP inhibitors in cancer therapy
Other than surgery, the most common cancer treatments are radiotherapy and chemotherapies that function by generating DNA damage [86], [87]. DNA repair represents a common mechanism for resistance to cancer therapy, thus the resistance of cancer cells to radiation and chemotherapy might reflect specific properties of the DDR of these cells ([88]; Fig. 6A). PARP-1 has been implicated in DNA repair and the maintenance of genomic integrity. This ‘guardian angel’ function of PARP is evidenced by a series of molecular mechanisms which are involved in the regulation of the DNA BER pathway and the high frequency of sister chromatid exchange in PARP-1−/− mice after exposure to IR or alkylating agents [89]. Therefore, it has been speculated that inhibition of the DDR might enhance the effectiveness of radiotherapy and chemotherapy and, indeed, more and more attention has been paid to the clinical potential of small molecule inhibitors in cancer therapy. To date, studies have indicated that inhibitors of PARPs might be effective as therapeutic agents for the treatment of multi-tissue tumors. As mentioned previously, PARP plays a role in the response of cells to stress-induced DNA single-strand breaks (SSBs) and forms part of the BER pathway [90]. In both cultured human cancer cells and xenograft mouse models, PARP inhibitors have been shown to enhance the cytotoxicity of the DNA-methylating agent temozolomide, ionizing radiation, and the topoisomerase-I inhibitors irinotecan and topotecan [10], [91], [92]. The combination of doxorubicin and PARP inhibitors especially sensitizes p53-deficient breast cancer cells to apoptosis [93].
Fig. 6.
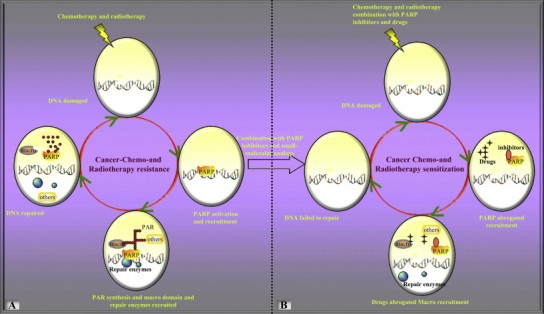
Presumed targeting macro domain in cancer therapy. Schematic illustration of possibly machinery involved in resistance to cancer radiotherapy and chemotherapy and the potential application of small molecular analogues of ADP-ribose in combination with PARP inhibitors in cancer therapy.
In this context, another recently recognized potency of PARP inhibitors could be in some case of enhancing the ability to kill tumor-cells deficient in homologous recombination. Recently, two studies from Bryant et al. and Farmer et al. have demonstrated that PARP inhibitors strongly enhance apoptosis in cancer cells that are deficient in either of the tumor suppressors BRCA1 and BRCA2, which are encoded by the most commonly mutated genes in familial breast cancer and are involved in homologous recombination [94], [95]. A final potential application of PARP inhibitors in tumor therapy might involve enhancement of the anti-tumor effects of radiotherapy [96]. In vivo, a preclinical study on the efficiency of PARP inhibitors to enhance radiotherapy has been reported recently [92].
A number of PARP inhibitors have entered the clinic trials in both intravenous and oral formulations (Table 2 ). To date, these PARP inhibitors have entered phase II trials; further phase II trials are currently underway that will help elucidate further the role and potential for this new targeted therapy. However, from phase II to phase III trials, it is a very long and difficult process. The initial findings from ongoing clinical studies of PARP inhibitors have confirmed the preclinical data. However, it is our opinion that in order for the full potential of PARP inhibitors to realize two key questions must be addressed by these studies. The first is how to identify those tumors that will benefit most from these new drugs. BRCA mutation is not restricted to triple-negative breast cancers and can occur in other subtypes. In addition, BRCA mutation has been observed in other tumor types, such as head and neck squamous-cell carcinomas, uterine cervical carcinomas and non-small-cell lung cancers. A major challenge in the coming years will be to identify which tumors the BRCA mutation precisely corresponds to. The second question is two-fold and involves determining how exactly PARP inhibitors exert their beneficial effects in tumor cells and whether different PARP inhibitors are equivalent in terms of suppression of PARP activity in cells and inhibition of polymer synthesis in patients.
Table 2.
PARP inhibitors in clinical trialsa.
| Drug | Company | Clinical trials | Phase | Ref. |
|---|---|---|---|---|
| AG014699 | Pfizer | BRCA1- or BRCA2-mutant tumors | Phase II | Plummer et al. [97] |
| Daniel et al. [98] | ||||
| Thomas et al. [99] | ||||
| ABT-888 | Abbott | Glioblastoma multiforme (with temozomide) | Phase II | Albert et al. [100] |
| Clarke et al. [101] | ||||
| Donawho et al. [102] | ||||
| Solid tumors and leukaemia (various combinations) | Phase I | Horton et al. [103] | ||
| Liu et al. [104] | ||||
| BRCA1- or BRCA2-mutant tumors | Phase I | Liu et al. [105] | ||
| Penning et al. [106] | ||||
| BSI-201 | Sanofi-Aventis | Ovarian cancer, glioblastoma multiforme, and uterine cancer (various combinations) | Phase II | O'Shaughnessy et al. [107] |
| Triple-negative breast cancer (with gemcitabine and carboplatin) | Phase III | Ossovskaya et al. [108] | ||
| BRCA2-mutant pancreatic cancer (various combinations) | Phase 1b | Mendeleyev et al. [109] | ||
| Other solid tumors | Phase I/II | |||
| AZD2281 | Astra Zeneca | BRCA1- or BRCA2-mutant tumors (with carboplatin) | Phase II | Dungey et al. [110] |
| Platin sensitive ovarian cancer | Phase II | Dungey et al. [111] | ||
| Triple-negative breast cancer (single agent or with carboplatin) | Phase II | Evers et al. [112] | ||
| Other solid tumors | Phase I/II | Hay et al. [113] | ||
| Rottenberg et al. [114] | ||||
| Menear et al. [115] | ||||
| CEP-8983/CEP-9722 (prodrug) | Cephalon | Solid tumors (with temozolomide) | Phase I | Miknyoczki et al. [116] |
| MK-4827 | Merck | Solid tumors and ovarian cancer | Phase I | Jones et al. [117] |
aBased on information obtained from http://www.clinicaltrials.gov. PARP, poly(ADP-ribose) polymerase.
In summary, research has established that PARP inhibitors are active anticancer agents in BRCA mutant tumors. Although these results are exciting, there is still much work to be done to translate them into clinical practice. It will be important to determine whether preclinical models have accurately predicted the activity of PARP inhibitors in settings beyond BRCA1- and BRCA2-deficient tumors. Although there is still much to be learnt about PARPs and PARP inhibitors, the recent tantalizing results suggest that further basic and translational studies are likely to be informative and rewarding.
6.3. Macro domain in infectious diseases
Pathogens have developed sophisticated mechanisms to either block or subvert normal host immune (clearance) processes, thereby enhancing pathogenesis and affecting disease outcome. Pathogens produce multiple virulence factors whose actions manifest in clinically recognized symptom profiles of infection. Their diverse functions and interplay with bacterial and host mechanisms confound attempts to precisely define the contribution of each virulence factors to the bacterium's pathogenesis [118]. Despite the complexity of bacterial pathogenesis, several bacterially-produced ADP-ribosylating exotoxins (bAREs) have been shown to contribute to the onset and progression of clinically relevant infections [119]. Studies have characterized that some of these bAREs ADP-ribosylate eukaryotic proteins that are important components of host cellular physiology. For example, diphtheria toxin (DT) from Corynebacterium diphtheria and exotoxin A (ETA) from Pseudomonas aeruginosa, directly inhibit translation elongation factor 2 (eEF2) [120], [121], [122], thereby blocking its downstream interactions with the ribosome and inhibiting protein synthesis in the host cell [123], [124]. In addition, cholera toxin and pertussis toxin (PT) are able to ADP-ribosylate the α-subunits of the heterotrimeric G proteins, which in turn perturbs normal signal transduction [119], [125]. Still other toxins can disrupt the eukaryotic cytoskeleton by ADP-ribosylating either the monomeric GTP-binding proteins of Rho family or actin [119].
As mentioned previously, macro domains are found in organisms ranging from viruses and bacteria to yeast and humans. Moreover, biochemical analysis has revealed that macro domains can bind with ADP-ribose metabolites [7], but the precise functional role of the bacterial macro domains remains elusive. It is possible that macro domains may interact with ADP-ribosylated proteins, since many bacterial mARTs have been identified [119]. Whether the bacterial macro domain effectively contributes to pathogenesis, however, has not yet been clearly defined. Interestingly, a recent study demonstrated that the macro domain was able to recognize protein targets within a host cell that had been ADP-ribosylated by bacterial exotoxins and by endogenous mARTs [38]. It is tempting to speculate that bARE activity may be able to modulate the biological activity of bacterial macro domains via mono-ADP-ribosylation. Mono-ADP-ribosylation could act as a signal termination mechanism for βγ; when an activated G protein-coupled receptor induces dissociation of the α- and βγ-subunits of the G protein, it also initiates a signal termination process by inducing mono-ADP-ribosylation of the active βγ dimer. The modified dimer is unable to interact with the effector and, in time, will be de-ADP-ribosylated, thereby allowing it to reassociate with the α-subunit [126], [127]. Hence, mono-ADP-ribosylation of macro domain by bacterial exotoxins might act as a ‘signaling’ function that mediates microorganism activities and facilitates its effects on host cells. Until now, the precise regulatory mechanisms of macro domains in infectious diseases remain largely uncharacterized. Further studies of the ADP-ribosylation machinery will not only increase our understanding of the functional role of macro domains, in such processes as signaling, immune response and membrane trafficking, but will also help to identify new targets for drug development.
6.4. The macro domain as a new potential therapeutic target in diseases
The macro domain family is conserved almost universally across all three domains of life: bacteria, archaea, and eukaryotes. The wide distribution of this protein family suggests that it is involved in an important and ubiquitous cellular process. The macro domain is also found as a single copy or as multiple copies in combination with a number of otherwise unrelated domains, which shows that gene multiplication through evolution has been accompanied by structural and functional diversification. The remarkable conservation of different macro domains indicates that the basic functions of this protein family have been conserved during several hundred million years of evolution. It is tempting to speculate that this conserved domain has helped to maintain the stability of chromatin in most organisms during their adaptation to the surrounding environment in evolution and development. It is doubtful that the role of macro domain proteins in cancer would have attracted so much attention if these proteins themselves, or their upstream or downstream effectors, were not thought to be attractive targets for the design of anti-cancer drugs. It has been consistently shown that the expression of macro domain proteins is higher in cancer cells than in normal cells, which suggests that these proteins might be a useful tissue biomarker for the diagnosis of cancer and their levels in serum might be a useful marker for prognosis [60], [82]. Presumably, the unique DDR machinery that is regulated by macro domain proteins provides a common mechanism for resistance to cancer therapy. Therefore, it has been speculated that therapy that targets macro domain proteins might enhance the effectiveness of radiotherapy and DNA-damaging chemotherapies.
The macro domain is the first globular protein module known to bind ADPR, metabolites of NAD+, and its derivatives. Interestingly, a study by Durkacz et al. has demonstrated that one function of homopolymer chains of ADPR is to participate in the cellular recovery from DNA damage. Thereby, the rejoining of DNA strand breaks caused by dimethyl sulphate and cytotoxicity is prevented by specific inhibitors of PARP and is also prevented by nutritionally depleting the cells of NAD+ [128]. However, many mutagenesis studies have indicated that the binding of ADPR to macro domains depends on a limited number of amino acid residues, which might represent what are known as ‘hot spots’ in terms of drug design. Generally, good drug targets correspond to surfaces with hot spots that can be covered by a drug-sized molecule. Therefore, it is tempting to speculate that small molecular analogues of ADPR that bind within the ligand pocket of macro domains might be of therapeutic value in a number of areas of medical interest (Fig. 6B). The problem with targeting ADPR binding sites is that, because ADPR chains commonly serve as a protein-interaction scaffold, such drugs would affect numerous ADPR-binding domain interactions and signaling pathways, which would lead to side effects. However, it is expected that by targeting specific effector proteins that contain ADPR-binding domains, instead of a broad spectrum of ADPR binding proteins, it will be possible to manipulate specific cellular processes. A number of potential target proteins are particularly interesting. Firstly, recent evidence has shown that the macro domain has an important role in PARP-1-mediated DNA damage recognition and repair [22], [23], [24], therefore, molecules targeting macro domains might enhance the effectiveness of radiotherapy and chemotherapy and restrict other human disease. Notably, a paper from Chen et al. strongly suggest that silencing macro domain protein expression in HCC by the corresponding shRNA has a great therapeutic potential in HCC treatment, especially to increase the chemosensitivity combined with chemotherapy [81]. Secondly, a large number of viruses and microbial parasites contain macro domain proteins, and some of these proteins are necessary for host cell infection and replication. The nsp3 macro domain has an essential role for sindbis virus (SINV) replication and age-dependent susceptibility to encephalomyelitis [129]. Unexpectedly, mutations in SINV macro domain profoundly impaired SINV replication and viral RNA synthesis particularly in neurons. Thirdly, macroH2A1.1 has been found to be enriched in post-mitotic and senescent cells, which suggests a role for this protein in chromatin biology [61], [62]. It remains to be seen whether the level of macroH2A can be correlated with the proliferation state of a cell and thus, potentially play a role in tumor biology. Finally, macro domains might show an association with the sirtuin family of enzymes because of their ability to bind the ADPR-related derivatives that are produced by sirtuins. Recently, it was shown that sirtuins play important roles in the aging process and in diseases such as cardiovascular disorders [130], [131], [132], [133], [134]. In response to DNA damage and oxidative stress, SIRT1 directly interacts with and deacetylates p53, which promotes cell survival by specifically repressing p53-dependent apoptotic response and the possible effect in cancer therapy [133], [134]. Therefore, the manipulation of sirtuin activities is appealing as a novel therapeutic strategy for the treatment of currently human diseases, such as cancers.
Encouragingly, over the past few years, progress in the field of structure-based drug design has indicated that it is pharmacologically possible to disrupt protein–protein interactions with small molecules; this has been exemplified by the development of small peptidomimetic inhibitors that target proteins that control apoptotic pathways in cancer cells such as inhibitors of apoptosis (IAPs) and B cell lymphoma 2 (BCL2) (for review [135]). These approaches require new strategies for the chemical synthesis of “peptidomimetic-like” compounds. It is possible that the molecular targeting of macro domain proteins will contribute to the future restriction of human diseases, including cancer, and the pharmacological development and usage of such modern therapeutics are promising.
7. Conclusions and future perspectives
On the basis of what we have discussed here, it is apparent that macro domains are unique evolutionarily conserved domains that regulate functions as diverse as the inhibition of apoptosis and the regulation of development, and that this is achieved by different biochemical means, including transcriptional regulation and PTMs of proteins, as well as modification or maintenance of chromatin domains in PAR-dependent manners.
Two questions immediately come to mind. First, how can macro domains carry out so many functions? And second, what is the evolutionary advantage of concentrating such a plethora of diverse functions into macro domains?
In response to the first question, perhaps we are not confronted with alone macro domain, but rather with diverse macro domain containing proteins—there is in fact evidence to suggest that not all macro domain proteins are created equal. We have described above how macro domain proteins may have different functions. In humans, at least ten genes encoding macro domain proteins are found, each protein contains from one to three macro domain. Often just a few macro domain proteins per cell are found to associate with specific proteins partners, other transcriptional factors or chromatin regions. Finally, different macro domains can bind various metabolites of NAD+, including PAR. Throughout this review, the notion that not only structural but also functional heterogeneity could exist among macro domains was raised. In the future, therefore, it will be of great importance to understand how different macro domain proteins might regulate different functions and if this is achieved is an integrated fashion.
With respect to the second question, we propose that the apparently diverse functions of macro domain proteins are in fact coherent, in that they allow macro domain to oppose and restrict tumor cell apoptosis and DNA damage at multiple levels. Therefore, it can be speculated that macro domains have evolved to carry out and perhaps coordinate tumorigenesis activities. Alternatively, macro domain may have initially evolved to regulate a more fundamental biology function (for example, DNA repair) and only later diversified into several tumor controlling activities. Understanding when macro domains have emerged during evolution might shed light on the ancestral scope and fundamental function of this intriguing ancient domain.
Another important and still unexplored area of macro domain research is whether macro domain proteins can transfer PAR to their interaction partners. Some macro domains can also hydrolyze phosphate groups from nucleotides or ADPR derivatives. The specific roles of the binding and enzymatic activities of macro domains, however, have remained elusive. This hypothesis appears to be reasonable in that PAR has been found to be transferred from PAR -binding proteins to partner proteins. For example, tankyrase, which is a member of the PARP superfamily, has been found to transfer PAR to its interacting protein telomeric repeat binding factor-1 (TRF1); ADP-ribosylation of TRF1 diminishes its ability to bind to telomeric DNA [136].
A tremendous amount of work has been done over the last decade to decipher the physiological and pathophysiological roles of macro domain proteins on molecular level. The research about functions of macro domain proteins, initially an esoteric field involving only a small community of researchers, is currently a hot topic. Many groups with a wide range of expertise have become involved in the biological functions of macro domain proteins research. However, despite the progress made in recent years in biochemistry, molecular biology, physiology, and pathophysiology of ADP-ribosylation of proteins, no unified picture of the physiology and pathophysiology roles of distinct PARylation reactions has yet emerged. Despite notable progress of new genetic tools and the availability of new techniques such as mass spectrometry, in vitro chromatin reconstitution systems, or chromatin immunopreciptation (CHIP) technologies in vivo, many basic questions about proteins ADP-ribosylation reactions remain unanswered, including the following. Can proteins really be covalently modified by PARylation, or are the PAR polymers just non-covalently associated with proteins in vivo? By what mechanisms are chromatin structures modulated through PARylation of PAR-binding domains? What is the functional relevance of PARylation in transcription, DNA repair and chromatin rearrangement? Can PAR have an influence on the histone code? How is the histone code modulated by mono-ADP-ribosylation of histones? Can mono-ADP-ribose serve as a histone modification marker for DNA repair and chromatin remodeling? Might mono-ADP-ribose or OAADPR function as a competitive inhibitor of the binding of PAR to macro domains in vivo?
One major future challenge is to understand in more detail how the PARylation of macro domain proteins is controlled. An enormous barrier is that the PARylation of proteins cannot be detected easily in cells by common laboratory methods, and thus might represent a vast area within the proteome that has been largely overlooked. Although technically difficult, the question of whether proteins are covalently or simply noncovalently modified by PARylation has to be addressed urgently by biochemical approaches combined with mass spectrometry techniques. The answer will undoubtedly change the field, and if PARylation could be confirmed in vitro and in vivo, it will certainly provide opportunities for exciting new research. Such knowledge will not only enhance our appreciation of the functions of macro domains but will undoubtedly present exciting opportunities to improve the understanding and management of human health and disease. It remains to be seen whether these observations will reveal new avenues for drug discovery, such as the use of analogues of ADPR, but they will surely teach us much about an aspect of protein regulation that remains only sparsely investigated to date.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Acknowledgements
We thank our colleagues for stimulating discussions and anonymous reviewers for their helpful comments. We apologize to colleagues whose work we could not cite due to space restrictions. This work is supported in part by grants from National Natural Science Foundation of China (grants 81071617 and 30870507) and Ministry of Science and Technology of China (2010CB912802).
Contributor Information
Weidong Han, Email: hanwdrsw69@yahoo.com.
Xiaobing Fu, Email: fuxiaobing@vip.sina.com.
References
- 1.Pehrson J.R., Fried V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 2.Pehrson J.R., Fuji R.N. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 1998;26:2837–2842. doi: 10.1093/nar/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ladurner A.G. Inactivating chromosomes: a macro domain that minimizes transcription. Mol. Cell. 2003;12:1–3. doi: 10.1016/s1097-2765(03)00284-3. [DOI] [PubMed] [Google Scholar]
- 4.Martzen M.R., McCraith S.M., Spinelli S.L., Torres F.M., Fields S., Grayhack E.J., Phizicky E.M. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 5.Karras G.I., Kustatscher G., Buhecha H.R., Allen M.D., Pugieux C., Sait F., Bycroft M., Ladurner A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., Ahola T., Canard B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neuvonen M., Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 2008;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirier G.G., de Murcia G., Jongstra-Bilen J., Niedergang C., Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Realini C.A., Althaus F.R. Histone shuttling by poly(ADP-ribosylation) J. Biol. Chem. 1992;267:18858–18865. [PubMed] [Google Scholar]
- 10.Yung T.M., Sato S., Satoh M.S. Poly(ADP-ribosyl)ation as a DNA damage-induced posttranslational modification regulating poly(ADP-ribose) polymerase-1–topoisomerase I interaction. J. Biol. Chem. 2004;279:39686–39696. doi: 10.1074/jbc.M402729200. [DOI] [PubMed] [Google Scholar]
- 11.Goenka S., Cho S.H., Boothby M. Collaborator of Stat6 (CoaSt6)-associated poly(ADP-ribose) polymerase activity modulates Stat6 dependent gene transcription. J. Biol. Chem. 2007;282:18732–18739. doi: 10.1074/jbc.M611283200. [DOI] [PubMed] [Google Scholar]
- 12.Goenka S., Boothby M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4210–4215. doi: 10.1073/pnas.0506981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravind L. The WWE domain: a common interaction module in protein ubiquitination and ADP-ribosylation. Trends Biochem. Sci. 2001;26:273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- 14.Schaaf G., Ortlund E.A., Tyeryar K.R., Mousley C.J., Ile K.E., Garrett T.A., Ren J., Woolls M.J., Raetz C.R., Redinbo M.R., Bankaitis V.A. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Angelo I., Welti S., Bonneau F., Scheffzek K. A novel bipartite phospholipids-binding module in the neurofibromatosis type 1 protein. EMBO Rep. 2006;7:174–179. doi: 10.1038/sj.embor.7400602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen J.A., Sweder K.S., Hanawalt P.C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flaus A., Martin D.M., Barton G.J., Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen M.D., Buckle A.M., Cordell S.C., Löwe J., Bycroft M. The crystal structure of AF1521a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 2003;330:503–511. doi: 10.1016/s0022-2836(03)00473-x. [DOI] [PubMed] [Google Scholar]
- 19.Aguiar R.C., Takeyama K., He C., Kreinbrink K., Shipp M.A. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly (ADP-ribose) polymerase activity. J. Biol. Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 20.Kustatscher G., Hothorn M., Pugieux C., Scheffzek K., Ladurner A.G. Splicing regulates NAD+ metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 21.Malet H., Coutard B., Jamal S., Dutartre H., Papageorgiou N., Neuvonen M., Ahola T., Forrester N., Gould E.A., Lafitte D., Ferron F., Lescar J., Gorbalenya A.E., de Lamballerie X., Canard B. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 2009;83:6534–6545. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timinszky G., Till S., Hassa P.O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E.H., Scheffzek K., Hottiger M.O., Ladurner A.G. A macrodomain-containing histone rearranges chromatin upon sensing PARP-1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 23.Gottschalk A.J., Timinszky G., Kong S.E., Jin J., Cai Y., Swanson S.K., Washburn M.P., Florens L., Ladurner A.G., Conaway J.W., Conaway R.C. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahel D., Horejsí Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S.C., Jackson S.P., Owen-Hughes T., Boulton S.J. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidovic L., Vodenicharov M., Affar E.B., Poirier G.G. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp. Cell Res. 2001;268:7–13. doi: 10.1006/excr.2001.5263. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson M.K., Jacobson E.L. Discovering new ADP-ribose polymer cycles: protecting the genome and more. Trends Biochem. Sci. 1999;24:415–417. doi: 10.1016/s0968-0004(99)01481-4. [DOI] [PubMed] [Google Scholar]
- 27.Hassa P.O., Haenni S.S., Elser M., Hottiger M.O. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber V., Dantzer F., Ame J.C., de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 29.Andrabi S.A., Kim N.S., Yu S.W., Wang H., Koh D.W., Sasaki M., Klaus J.A., Otsuka T., Zhang Z., Koehler R.C., Hurn P.D., Poirier G.G., Dawson V.L., Dawson T.M. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quénet D., El Ramy R., Schreiber V., Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int. J. Biochem. Cell Biol. 2009;41:60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 31.Yu W., Ginjala V., Pant V., Chernukhin I., Whitehead J., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Oshimura M., Feinberg A.P., Lobanenkov V., Klenova E., Ohlsson R. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 32.Pleschke J.M., Kleczkowska H.E., Strohm M., Althaus F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 33.Gagné J.P., Hunter J.M., Labrecque B., Chabot B., Poirier G.G. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose) binding proteins. Biochem. J. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanai M., Hanashiro K., Kim S.H., Hanai S., Boulares A.H., Miwa M., Fukasawa K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol. 2007;9:1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 35.Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 36.Stilmann M., Hinz M., Arslan S.C., Zimmer A., Schreiber V., Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IκB kinase activation. Mol. Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 37.Kleine H., Poreba E., Lesniewicz K., Hassa P.O., Hottiger M.O., Litchfield D.W., Shilton B.H., Lüscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Dani N., Stilla A., Marchegiani A., Tamburro A., Till S., Ladurner A.G., Corda D., Di Girolamo M. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hakmé A., Huber A., Dollé P., Schreiber V. The macroPARP genes Parp-9 and Parp-14 are developmentally and differentially regulated in mouse tissues. Dev. Dyn. 2008;237:209–215. doi: 10.1002/dvdy.21399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho S.H., Goenka S., Henttinen T., Gudapati P., Reinikainen A., Eischen C.M., Lahesmaa R., Boothby M. PARP-14, a member of the B aggressive lymphoma family, transduces survival signals in primary B cells. Blood. 2009;113:2416–2425. doi: 10.1182/blood-2008-03-144121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maas N.M, Van de Putte T., Melotte C., Francis A., Schrander-Stumpel C.T., Sanlaville D., Genevieve D., Lyonnet S., Dimitrov B., Devriendt K., Fryns J.P., Vermeesch J.R. The C20orf133 gene is disrupted in a patient with Kabuki syndrome. J. Med. Genet. 2007;44:562–569. doi: 10.1136/jmg.2007.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buschbeck M., Uribesalgo I., Wibowo I., Rué P., Martin D., Gutierrez A., Morey L., Guigó R., López-Schier H., Di Croce L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 2009;16:1074–1079. doi: 10.1038/nsmb.1665. [DOI] [PubMed] [Google Scholar]
- 43.Changolkar L.N., Costanzi C., Leu N.A., Chen D., McLaughlin K.J., Pehrson J.R. Developmental changes in histone macroH2A1-mediated gene regulation. Mol. Cell. Biol. 2007;27:2758–2764. doi: 10.1128/MCB.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costanzi C., Pehrson J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 45.Nusinow D.A., Hernández-Muñoz I., Fazzio T.G., Shah G.M., Kraus W.L., Panning B. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, macroH2A, and contributes to silencing of the inactive X chromosome. J. Biol. Chem. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
- 46.Hoyer-Fender S., Costanzi C., Pehrson J.R. Histone macroH2A1.2 is concentrated in the XY-body by the early pachytene stage of spermatogenesis. Exp. Cell Res. 2000;258:254–260. doi: 10.1006/excr.2000.4951. [DOI] [PubMed] [Google Scholar]
- 47.Khil P.P., Smirnova N.A., Romanienko P.J., Camerini-Otero R.D. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- 48.Turner J.M., Mahadevaiah S.K., Elliott D.J., Garchon H.J., Pehrson J.R., Jaenisch R., Burgoyne P.S. Meiotic sex chromosome inactivation in male mice with targeted disruptions of Xist. J. Cell Sci. 2002;115:4097–4105. doi: 10.1242/jcs.00111. [DOI] [PubMed] [Google Scholar]
- 49.Chadwick B.P., Willard H.F. Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome. J. Cell Biol. 2002;157:1113–1123. doi: 10.1083/jcb.200112074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L., Hu L., Chan T.H., Tsao G.S., Xie D., Huo K.K., Fu L., Ma S., Zheng B.J., Guan X.Y. Chromodomain helicase/adenosine triphosphatase DNA binding protein 1-like (CHD1L) gene suppresses the nucleus-to-mitochondria translocation of Nur77 to sustain hepatocellular carcinoma cell survival. Hepatology. 2009;50:122–129. doi: 10.1002/hep.22933. [DOI] [PubMed] [Google Scholar]
- 51.Mangelsdorf D.J., Evans R.M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 52.Lin B., Kolluri S.K., Lin F., Liu W., Han Y.H., Cao X., Dawson M.I., Reed J.C., Zhang X.K. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–540. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 53.Moll U.M., Marchenko N., Zhang X.K. p53 and Nur77/TR3—transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 54.Rajpal A., Cho Y.A., Yelent B., Koza-Taylor P.H., Li D., Chen E., Whang M., Kang C., Turi T.G., Winoto A. Transcriptional activation of known and novel apoptotic pathways by Nur77 orphan steroid receptor. EMBO J. 2003;22:6526–6536. doi: 10.1093/emboj/cdg620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki S., Suzuki N., Mirtsos C., Horacek T., Lye E., Noh S.K., Ho A., Bouchard D., Mak T.W., Yeh W.C. Nur77 as a survival factor in tumor necrosis factor signaling. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8276–8280. doi: 10.1073/pnas.0932598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brás A., Albar J.P., Leonardo E., de Buitrago G.G., Martínez-A C. Ceramide-induced cell death is independent of the Fas/Fas ligand pathway and is prevented by Nur77 overexpression in A20 B cells. Cell Death Differ. 2000;7:262–271. doi: 10.1038/sj.cdd.4400653. [DOI] [PubMed] [Google Scholar]
- 57.de Léséleuc L., Denis F. Inhibition of apoptosis by Nur77 through NF-κB activity modulation. Cell Death Differ. 2006;13:293–300. doi: 10.1038/sj.cdd.4401737. [DOI] [PubMed] [Google Scholar]
- 58.Srivastava N., Gochhait S., de Boer P., Bamezai R.N. Role of H2AX in DNA damage response and human cancers. Mutat. Res. 2009;68:180–188. doi: 10.1016/j.mrrev.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 60.Sporn J.C., Kustatscher G., Hothorn T., Collado M., Serrano M., Muley T., Schnabel P., Ladurner A.G. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene. 2009;28:3423–3428. doi: 10.1038/onc.2009.26. [DOI] [PubMed] [Google Scholar]
- 61.Pehrson J.R., Costanzi C., Dharia C. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J. Cell. Biochem. 1997;65:107–113. doi: 10.1002/(sici)1097-4644(199704)65:1<107::aid-jcb11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 62.Costanzi C., Pehrson J.R. MACROH2A2, a new member of the MACROH2A core histone family. J. Biol. Chem. 2001;276:21776–21784. doi: 10.1074/jbc.M010919200. [DOI] [PubMed] [Google Scholar]
- 63.Han W.D., Zhao Y.L., Meng Y.G., Zang L., Wu Z.Q., Li Q., Si Y.L., Huang K., Ba J.M., Morinaga H., Nomura M., Mu Y.M. Estrogenically regulated ERα target gene LRP16 interacts with ERα and enhances the receptor's transcriptional activity. Endocr. Relat. Cancer. 2007;14:741–753. doi: 10.1677/ERC-06-0082. [DOI] [PubMed] [Google Scholar]
- 64.Han W.D., Mu Y.M., Lu X.C., Xu Z.M., Li X.J., Yu L., Song H.J., Li M., Lu J.M., Zhao Y.L., Pan C.Y. Up-regulation of LRP16 mRNA by 17β-estradiol through activation of estrogen receptor α (ERα) but not ERβ, and promotion of human breast cancer MCF-7 cell proliferation: a preliminary report. Endocr. Relat. Cancer. 2003;10:217–224. doi: 10.1677/erc.0.0100217. [DOI] [PubMed] [Google Scholar]
- 65.Yang J., Zhao Y.L., Wu Z.Q., Si Y.L., Meng Y.G., Fu X.B., Mu Y.M., Han W.D. The single-macro domain protein LRP16 is an essential cofactor of androgen receptor. Endocr. Relat. Cancer. 2009;16:139–153. doi: 10.1677/ERC-08-0150. [DOI] [PubMed] [Google Scholar]
- 66.Angelov D., Molla A., Perche P.Y., Hans F., Côté J., Khochbin S., Bouvet P., Dimitrov S. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell. 2003;11:1033–1041. doi: 10.1016/s1097-2765(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 67.van Lohuizen M., Verbeek S., Krimpenfort P., Domen J., Saris C., Radaszkiewicz T., Berns A. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56:673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 68.Opferman J.T., Letai A., Beard C., Sorcinelli M.D., Ong C.C., Korsmeyer S.J. Development and maintenance of B and T lymphocytes requires anti-apoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 69.Doyen C.M., An W., Angelov D., Bondarenko V., Mietton F., Studitsky V.M., Hamiche A., Roeder R.G., Bouvet P., Dimitrov S. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol. Cell. Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Changolkar L.N., Singh G., Pehrson J.R. MacroH2A1-dependent silencing of endogenous murine leukemia viruses. Mol. Cell. Biol. 2008;28:2059–2065. doi: 10.1128/MCB.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bernstein E., Muratore-Schroeder T.L., Diaz R.L., Chow J.C., Changolkar L.N., Shabanowitz J., Heard E., Pehrson J.R., Hunt D.F., Allis C.D. A phosphorylated subpopulation of the histone variant macroH2A1 is excluded from the inactive X chromosome and enriched during mitosis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1533–1538. doi: 10.1073/pnas.0711632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gamble M.J., Frizzell K.M., Yang C., Krishnakumar R., Kraus W.L. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 2009;24:21–32. doi: 10.1101/gad.1876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen M., Huang J.D., Hu L., Zheng B.J., Chen L., Tsang S.L., Guan X.Y. Transgenic CHD1L expression in mouse induces spontaneous tumors. PLoS One. 2009;4:e6727. doi: 10.1371/journal.pone.0006727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liao D.X., Han W.D., Zhao Y.L., Pu Y.D., Mu Y.M., Luo C.H., Li X.H. The expression and clinical significance of LRP16 gene in human breast cancer. Ai Zheng. 2006;25:866–870. [PubMed] [Google Scholar]
- 75.Meng Y.G., Han W.D., Zhao Y.L., Huang K., Si Y.L., Wu Z.Q., Mu Y.M. Induction of LRP16 gene by estrogen promotes the invasive growth of Ishikawa human endometrial cancer cells through down-regulation of E-cadherin. Cell Res. 2007;17:869–880. doi: 10.1038/cr.2007.79. [DOI] [PubMed] [Google Scholar]
- 76.Li Y.Z., Zhao P., Han W.D. Clinicopathological significance of LRP16 protein in 336 gastric carcinoma patients. World J. Gastroenterol. 2009;15:4833–4837. doi: 10.3748/wjg.15.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao P., Lu Y., Han W.D. Clinicopathological significance and prognostic value of leukemia related protein 16 (LRP16) expression in invasive ductal breast carcinoma. Cancer Sci. 2010;101:2262–2268. doi: 10.1111/j.1349-7006.2010.01658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xi H.Q., Zhao P., Han W.D. Clinicopathological significance and prognostic value of LRP16 expression in colorectal carcinoma. World J. Gastroenterol. 2010;16:1644–1648. doi: 10.3748/wjg.v16.i13.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aguiar R.C., Yakushijin Y., Kharbanda S., Salgia R., Fletcher J.A., Shipp M.A. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood. 2000;96:4328–4334. [PubMed] [Google Scholar]
- 80.Ma N.F., Hu L., Fung J.M., Xie D., Zheng B.J., Chen L., Tang D.J., Fu L., Wu Z., Chen M., Fang Y., Guan X.Y. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology. 2008;47:503–510. doi: 10.1002/hep.22072. [DOI] [PubMed] [Google Scholar]
- 81.Chen L, Yuan Y.F., Li Y., Chan T.H., Zheng B.J., Huang J., Guan X.Y. Clinical significance of CHD1L in hepatocellular carcinoma and therapeutic potentials of virus-mediated CHD1L depletion. Gut. 2010:224071. doi: 10.1136/gut.2010.224071. [DOI] [PubMed] [Google Scholar]
- 82.Marina O., Hainz U., Biernacki M.A., Zhang W., Cai A., Duke-Cohan J.S., Liu F., Brusic V., Neuberg D., Kutok J.L., Alyea E.P., Canning C.M., Soiffer R.J., Ritz J., Wu C.J. Serologic markers of effective tumor immunity against chronic lymphocytic leukemia include nonmutated B-cell antigens. Cancer Res. 2010;70:1344–1355. doi: 10.1158/0008-5472.CAN-09-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen L., Chan T.H., Yuan Y.F., Hu L., Huang J., Ma S., Wang J., Dong S.S., Tang K.H., Xie D., Li Y., Guan X.Y. CHD1L promotes hepatocellular carcinoma progression and metastasis in mice and is associated with these processes in human patients. J. Clin. Invest. 2010;120:1178–1191. doi: 10.1172/JCI40665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tartaglia M., Mehler E.L., Goldberg R., Zampino G., Brunner H.G., Kremer H., van der Burgt I., Crosby A.H., Ion A., Jeffery S., Kalidas K., Patton M.A., Kucherlapati R.S., Gelb B.D. Mutations in PTPN11,encoding the protein tyrosine phosphatase SHP-2, cause noonan syndrome. Nat. Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 85.Petrij F., Giles R.H., Dauwerse H.G., Saris J.J., Hennekam R.C., Masuno M., Tommerup N., van Ommen G.J., Goodman R.H., Peters D.J. Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 86.Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 87.Espinosa E., Zamora P., Feliu J., González Barón M. Classification of anticancer drugs—a new system based on therapeutic targets. Cancer Treat. Rev. 2003;29:515–523. doi: 10.1016/s0305-7372(03)00116-6. [DOI] [PubMed] [Google Scholar]
- 88.Khanna K.K., Jackson S.P. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 89.de Murcia J.M., Niedergang C., Trucco C., Ricoul M., Dutrillaux B., Mark M., Oliver F.J., Masson M., Dierich A., LeMeur M., Walztinger C., Chambon P., de Murcia G. Requirement of poly(ADP-ribose)polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 91.Tentori L., Portarena I., Vernole P., De Fabritiis P., Madaio R., Balduzzi A., Roy R., Bonmassar E., Graziani G. Effects of single or split exposure of leukemic cells to temozolomide, combined with poly(ADP-ribose) polymerase inhibitors on cell growth, chromosomal aberrations and base excision repair components. Cancer Chemother. Pharmacol. 2001;47:361–369. doi: 10.1007/s002800000248. [DOI] [PubMed] [Google Scholar]
- 92.Calabrese C.R., Almassy R., Barton S., Batey M.A., Calvert A.H., Canan-Koch S., Durkacz B.W., Hostomsky Z., Kumpf R.A., Kyle S., Li J., Maegley K., Newell D.R., Notarianni E., Stratford I.J., Skalitzky D., Thomas H.D., Wang L.Z., Webber S.E., Williams K.J., Curtin N.J. Anticancer chemosensitization and radiosensitization by the novel poly (ADP-ribose) polymerase-1 inhibitor AG14361. J. Natl. Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 93.Muñoz-Gámez J.A., Martín-Oliva D., Aguilar-Quesada R., Cañuelo A., Nuñez M.I., Valenzuela M.T., Ruiz de Almodóvar J.M., De Murcia G., Oliver F.J. PARP inhibition sensitizes p53-deficient breast cancer cells to doxorubicin-induced apoptosis. Biochem. J. 2005;386:119–125. doi: 10.1042/BJ20040776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumors with inhibitors of poly (ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 95.Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., Martin N.M., Jackson S.P., Smith G.C., Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 96.Chalmers A.J. Poly(ADP-ribose) polymerase-1 and ionizing radiation: sensor, signaller and therapeutic target. Clin. Oncol. (R. Coll. Radiol.) 2004;16:29–39. doi: 10.1016/s0936-6555(03)00223-1. [DOI] [PubMed] [Google Scholar]
- 97.Plummer R., Jones C., Middleton M., Wilson R., Evans J., Olsen A., Curtin N., Boddy A., McHugh P., Newell D., Harris A., Johnson P., Steinfeldt H., Dewji R., Wang D., Robson L., Calvert H. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin. Cancer Res. 2008;14:7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Daniel R.A., Rozanska A.L., Thomas H.D., Mulligan E.A., Drew Y., Castelbuono D.J., Hostomsky Z., Plummer E.R., Boddy A.V., Tweddle D.A., Curtin N.J., Clifford S.C. Inhibition of poly(ADP-ribose) polymerase-1 enhances temozolomide and topotecan activity against childhood neuroblastoma. Clin. Cancer Res. 2009;15:1241–1249. doi: 10.1158/1078-0432.CCR-08-1095. [DOI] [PubMed] [Google Scholar]
- 99.Thomas H.D., Calabrese C.R., Batey M.A., Canan S., Hostomsky Z., Kyle S., Maegley K.A., Newell D.R., Skalitzky D., Wang L.Z., Webber S.E., Curtin N.J. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol. Cancer Ther. 2007;6:945–956. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- 100.Albert J.M., Cao C., Kim K.W., Willey C.D., Geng L., Xiao D., Wang H., Sandler A., Johnson D.H., Colevas A.D., Low J., Rothenberg M.L., Lu B. Inhibition of poly(ADP-ribose) polymerase enhances cell death and improves tumor growth delay in irradiated lung cancer models. Clin. Cancer Res. 2007;13:3033–3042. doi: 10.1158/1078-0432.CCR-06-2872. [DOI] [PubMed] [Google Scholar]
- 101.Clarke M.J., Mulligan E.A., Grogan P.T., Mladek A.C., Carlson B.L., Schroeder M.A., Curtin N.J., Lou Z., Decker P.A., Wu W., Plummer E.R., Sarkaria J.N. Effective sensitization of temozolomide by ABT-888 is lost with development of temozolomide resistance in glioblastoma xenograft lines. Mol. Cancer Ther. 2009;8:407–414. doi: 10.1158/1535-7163.MCT-08-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Donawho C.K., Luo Y., Luo Y., Penning T.D., Bauch J.L., Bouska J.J., Bontcheva-Diaz V.D., Cox B.F., DeWeese T.L., Dillehay L.E., Ferguson D.C., Ghoreishi-Haack N.S., Grimm D.R., Guan R., Han E.K., Holley-Shanks R.R., Hristov B., Idler K.B., Jarvis K., Johnson E.F., Kleinberg L.R., Klinghofer V., Lasko L.M., Liu X., Marsh K.C., McGonigal T.P., Meulbroek J.A., Olson A.M., Palma J.P., Rodriguez L.E., Shi Y., Stavropoulos J.A., Tsurutani A.C., Zhu G.D., Rosenberg S.H., Giranda V.L., Frost D.J. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models, Clin. Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 103.Horton T.M., Jenkins G., Pati D., Zhang L., Dolan M.E., Ribes-Zamora A., Bertuch A.A., Blaney S.M., Delaney S.L., Hegde M., Berg S.L. Poly(ADP-ribose) polymerase inhibitor ABT-888 potentiates the cytotoxic activity of temozolomide in leukemia cells: influence of mismatch repair status and O6-methylguanine-DNA methyltransferase activity. Mol. Cancer Ther. 2009;8:2232–2242. doi: 10.1158/1535-7163.MCT-09-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu S.K., Coackley C., Krause M., Jalali F., Chan N., Bristow R.G. A novel poly(ADP-ribose) polymerase inhibitor, ABT-888, radiosensitizes malignant human cell lines under hypoxia. Radiother. Oncol. 2008;88:258–268. doi: 10.1016/j.radonc.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 105.Liu X., Shi Y., Guan R., Donawho C., Luo Y., Palma J., Zhu G.D., Johnson E.F., Rodriguez L.E., Ghoreishi-Haack N., Jarvis K., Hradil V.P., Colon-Lopez M., Cox B.F., Klinghofer V., Penning T., Rosenberg S.H., Frost D., Giranda V.L., Luo Y. Potentiation of temozolomide cytotoxicity by poly(ADP)ribose polymerase inhibitor ABT-888 requires a conversion of single-stranded DNA damages to double-stranded DNA breaks. Mol. Cancer Res. 2008;6:1621–1629. doi: 10.1158/1541-7786.MCR-08-0240. [DOI] [PubMed] [Google Scholar]
- 106.Penning T.D., Zhu G.D., Gandhi V.B., Gong J., Liu X., Shi Y., Klinghofer V., Johnson E.F., Donawho C.K., Frost D.J., Bontcheva-Diaz V., Bouska J.J., Osterling D.J., Olson A.M., Marsh K.C., Luo Y., Giranda V.L. Discovery of the poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide(ABT-888) for the treatment of cancer. J. Med. Chem. 2009;52:514–523. doi: 10.1021/jm801171j. [DOI] [PubMed] [Google Scholar]
- 107.O'Shaughnessy J., Osborne C., Pippen J., Yoffe M., Patt D., Monaghan G., Monaghan G., Rocha C., Ossovskaya V., Sherman B., Bradley C. Efficacy of BSI-201, a poly(ADP-ribose) polymerase-1(PARP1) inhibitor, in combination with gemcit–abine/carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): results of a randomized Phase II trial. J. Clin. Oncol. 2009;27:3. [Google Scholar]
- 108.Ossovskaya V., Li L., Broude E.V., Lim C., Roninson I.B., Bradley C., Sherman B. BSI-201 enhances the activity of multiple classes of cytotoxic agents and irradiation in triple negative breast cancer. Abstract 5552. Proc. Annu. Meet. Am. Assoc. Cancer Res. 2009 [Google Scholar]
- 109.Mendeleyev J., Kirsten E., Hakam A., Buki K.G., Kun E. Potential chemotherapeutic activity of 4-iodo-3-nitrobenzamide. Metabolic reduction to the 3-nitroso derivative and induction of cell death in tumor cells in culture. Biochem. Pharmacol. 1995;50:705–714. doi: 10.1016/0006-2952(95)00189-7. [DOI] [PubMed] [Google Scholar]
- 110.Dungey F.A., Caldecott K.W., Chalmers A.J. Enhanced radiosensitization of human glioma cells by combining inhibition of poly(ADP-ribose) polymerase with inhibition of heat shock protein 90. Mol. Cancer Ther. 2009;8:2243–2254. doi: 10.1158/1535-7163.MCT-09-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dungey F.A., Löser D.A., Chalmers A.J. Replication-dependent radiosensitization of human glioma cells by inhibition of poly(ADP-ribose) polymerase: mechanisms and therapeutic potential. Int. J. Radiat. Oncol. Biol. Phys. 2008;72:1188–1197. doi: 10.1016/j.ijrobp.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 112.Evers B., Drost R., Schut E., de Bruin M., van der Burg E., Derksen P.W., Holstege H., Liu X., van Drunen E., Beverloo H.B., Smith G.C., Martin N.M., Lau A., O’Connor M.J., Jonkers J. Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin. Cancer Res. 2008;14:3916–3925. doi: 10.1158/1078-0432.CCR-07-4953. [DOI] [PubMed] [Google Scholar]
- 113.Hay T., Matthews J.R., Pietzka L., Lau A., Cranston A., Nygren A.O., Douglas-Jones A., Smith G.C., Martin N.M., O’Connor M., Clarke A.R. Poly(ADP-ribose) polymerase-1 inhibitor treatment regresses autochthonous Brca2/p53 mutant mammary tumors in vivo and delays tumor relapse in combination with carboplatin. Cancer Res. 2009;69:3850–3855. doi: 10.1158/0008-5472.CAN-08-2388. [DOI] [PubMed] [Google Scholar]
- 114.Rottenberg S., Jaspers J.E., Kersbergen A., van der Burg E., Nygren A.O., Zander S.A., Derksen P.W., de Bruin M., Zevenhoven J., Lau A., Boulter R., Cranston A., O’Connor M.J., Martin N.M., Borst P., Jonkers J. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Menear K.A., Adcock C., Boulter R., Cockcroft X.L., Copsey L., Cranston A., Dillon K.J., Drzewiecki J., Garman S., Gomez S., Javaid H., Kerrigan F., Knights C., Lau A., Jr., Loh V.M., Matthews I.T., Moore S., O’Connor M.J., Smith G.C., Martin N.M. 4-[3-(4-Cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J. Med. Chem. 2008;51:6581–6591. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- 116.Miknyoczki S., Chang H., Grobelny J., Pritchard S., Worrell C., McGann N., Ator M., Husten J., Deibold J., Hudkins R., Zulli A., Parchment R., Ruggeri B. The selective poly(ADP-ribose) polymerase-1(2) inhibitor, CEP-8983, increases the sensitivity of chemoresistant tumor cells to temozolomide and irinotecan but does not potentiate myelotoxicity. Mol. Cancer Ther. 2007;6:2290–2302. doi: 10.1158/1535-7163.MCT-07-0062. [DOI] [PubMed] [Google Scholar]
- 117.Jones P., Altamura S., Boueres J., Ferrigno F., Fonsi M., Giomini C., Lamartina S., Monteagudo E., Ontoria J.M., Orsale M.V., Palumbi M.C., Pesci S., Roscilli G., Scarpelli R., Schultz-Fademrecht C., Toniatti C., Rowley M. Discovery of 2-{4-[(3S)-piperidin-3-yl] phenyl}-2H-indazole-7-carboxamide (MK-4827): a novel oral poly(ADP-ribose) polymerase (PARP) inhibitor efficacious in BRCA-1 and -2 mutant tumors. J. Med. Chem. 2009;52:7170–7185. doi: 10.1021/jm901188v. [DOI] [PubMed] [Google Scholar]
- 118.Popoff M.R. Bacterial exotoxins. Contrib. Microbiol. 2005;12:28–54. doi: 10.1159/000081688. [DOI] [PubMed] [Google Scholar]
- 119.Krueger K.M., Barbieri J.T. The family of bacterial ADP-ribosylating exotoxins. Clin. Microbiol. Rev. 1995;8:34–47. doi: 10.1128/cmr.8.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yates S.P., Jorgensen R., Andersen G.R., Merrill A.R. Stealth and mimicry by deadly bacterial toxins. Trends Biochem. Sci. 2006;31:123–133. doi: 10.1016/j.tibs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 121.Collier R.J., Cole H.A. Diphtheria toxin subunit active in vitro. Science. 1969;164:1179–1181. doi: 10.1126/science.164.3884.1179. [DOI] [PubMed] [Google Scholar]
- 122.Jørgensen R., Ortiz P.A., Carr-Schmid A., Nissen P., Kinzy T.G., Andersen G.R. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat. Struct. Biol. 2003;10:379–385. doi: 10.1038/nsb923. [DOI] [PubMed] [Google Scholar]
- 123.Collier R.J. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon. 2001;39:1793–1803. doi: 10.1016/s0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- 124.Jørgensen R., Merrill A.R., Yates S.P., Marquez V.E., Schwan A.L., Boesen T., Andersen G.R. Exotoxin A–eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature. 2005;436:979–984. doi: 10.1038/nature03871. [DOI] [PubMed] [Google Scholar]
- 125.Cassel D., Pfeuffer T. Mechanism of cholera toxin action: covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc. Natl. Acad. Sci. U. S. A. 1978;75:2669–2673. doi: 10.1073/pnas.75.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lupi R., Corda D., Di Girolamo M. Endogenous ADP-ribosylation of the G protein β subunit prevents the inhibition of type 1 adenylyl cyclase. J. Biol. Chem. 2000;275:9418–9424. doi: 10.1074/jbc.275.13.9418. [DOI] [PubMed] [Google Scholar]
- 127.Lupi R., Dani N., Dietrich A., Marchegiani A., Turacchio S., Berrie C.P., Moss J., Gierschik P., Corda D., Di Girolamo M. Endogenous mono-ADP-ribosylation of the free G βγ prevents stimulation of phosphoinositide 3 kinase-γ and phospholipase C-β2 and is activated by G-protein-coupled-receptors. Biochem. J. 2002;367:1–7. doi: 10.1042/BJ20020660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Durkacz B.W., Omidiji O., Gray D.A., Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 129.Park E., Griffin D.E. The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology. 2009;388:305–314. doi: 10.1016/j.virol.2009.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rogina B., Helfand S.L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tissenbaum H.A., Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 132.Schwer B., Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 133.Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 134.Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A. hSIR2SIRT1 functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 135.Hoeller D., Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 136.Smith S., Giriat I., Schmitt A., de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]


