Abstract
Bovine coronavirus (BCoV) is found worldwide and causes respiratory infections and diarrhoea in calves and adult cattle. In order to investigate the molecular epidemiology of BCoV, 27 reverse transcription polymerase chain reaction (RT-PCR) positive samples from 25 cattle herds in different parts of Sweden were analysed. A 1038-nucleotide fragment was PCR amplified and directly sequenced.
The analysed BCoV strains showed a high sequence identity, regardless of whether they were obtained from outbreaks of respiratory disease or diarrhoea or from calves or adult cattle. Circulation of an identical BCoV strain during a 4-month period was demonstrated in calves in one dairy herd. In a regional epizootic of winter dysentery in Northern Sweden, highly similar BCoV strains were detected. In the Southern and Central regions, several genotypes of BCoV circulated contemporaneously, indicating that in these regions, which had a higher density of cattle than the Northern regions, more extensive transmission of the virus was occurring. Identical BCoV sequences supported the epidemiological data that inter-herd contact through purchased calves was important. Swedish BCoV strains unexpectedly showed a high homology with recently detected Italian strains. This study shows that molecular analysis of the spike (S) glycoprotein gene of BCoV can be a useful tool to support or rule out suspected transmission routes.
Keywords: Molecular epidemiology, Phylogeny, Bovine coronavirus, Transmission, Respiratory
Introduction
Bovine coronavirus (BCoV) belongs to the family Coronaviridae, order Nidovirales and has a single-stranded, positive-sense RNA genome 31 kb in length, including 13 open reading frames (Clark, 1993, Saif, 2004). Variations in host range and tissue tropism of coronaviruses are attributed to the spike (S) glycoprotein (Gallagher and Buchmeier, 2001). This protein is cleaved by an intracellular protease into two functional domains (Gallagher and Buchmeier, 2001). The S1 subunit is peripheral and responsible for virus binding to host-cell receptors (Kubo et al., 1994), induction of neutralizing antibodies (Yoo and Deregt, 2001), and haemagglutination activity (Schultze et al., 1991). The S1 sequence is variable, and mutations in this region have been associated with altered antigenicity and virus pathogenicity (Fazakerley et al., 1992, Ballesteros et al., 1997). The sequence of the S2 subunit is more conserved and this subunit is responsible for cell membrane fusion activity (Yoo et al., 1991). The S1 gene has been exploited as a target to study the molecular epidemiology of BCoV infection (Hasoksuz et al., 2002a, Jeong et al., 2005, Liu et al., 2006).
BCoV infection is found worldwide and causes calf diarrhoea (CD), winter dysentery (WD) in adult cattle, and respiratory infections in cattle of all ages (Clark, 1993, Paton et al., 1998, Saif, 2004). Infection has a high morbidity but a low mortality (Alenius et al., 1991, Saif, 2004). Outbreaks typically occur in autumn and winter, probably in association with housing, but severe outbreaks in adult cattle have been reported in summer (Park et al., 2006, Decaro et al., 2008a). Economic losses can be heavy due to a marked reduction in milk yield (Tråvén et al., 2001, Saif, 2010). Calf diarrhoea and respiratory disease also cause significant losses (Svensson et al., 2003, Torsein et al., 2011), but the role of BCoV in these diseases has not been fully evaluated.
Several serological studies have shown that BCoV infections are highly prevalent in Swedish cattle herds, causing disease in both calves and adults (Tråvén et al., 1999, Hägglund et al., 2006, Bidokhti et al., 2009). In a previous study we showed that all cattle sampled during a herd outbreak of BCoV were carrying identical BCoV sequences (Liu et al., 2006). The objective of the present study was to investigate the molecular epidemiology of BCoV by analysing the polymorphic region of the S gene from herds with outbreaks of diarrhoea or respiratory disease and to determine if this method could be used to support or rule out suspected transmission routes between herds.
Materials and methods
Samples
Faecal and nasal swab samples were collected from housed cattle in 23 dairy herds and two feedlot herds (Table 1 ) in different parts of Sweden over a 4 year period. Samples were collected October 2005 to May 2006, October 2006 to May 2007, October 2007 to May 2008, and October 2008 to July 2009. Sampled cattle in all herds were showing clinical signs of BCoV infection and the presenting symptoms are summarised in Table 1.
Table 1.
Location of herds positive for BCoV, sampling time, animals sampled, symptoms and type of sample.
| Herd | Type | Year/month | Age (symptoms b) | Sample type | County |
|---|---|---|---|---|---|
| AC1 | Dairy | 2006/11 | Adult (WD) | Faecal | Västerbotten |
| AC2 | Dairy | 2007/11 | Adult (WD) | Faecal | Västerbotten |
| C1 | Dairy | 1992/02 | Adult (WD) | Faecal | Uppland |
| C2 | Dairy | 2007/01 | Adult (WD) | Faecal | Uppland |
| C3 | Dairy | 2007/11 | Adult (WD) | Faecal | Uppland |
| C4 | Dairy | 2007/12 | Adult (WD) | Faecal | Uppland |
| C5 | Dairy | 2008/01 | Adult (WD) | Faecal | Uppland |
| C6 | Feedlot | 2009/03 | Calf (RD) | Nasal | Uppland |
| I1 | Dairy | 2007/02 | Adult (WD) | Faecal | Gotland |
| I2 | Dairy | 2007/02 | Adult (WD) | Faecal | Gotland |
| I3 | Dairy | 2007/03 | Adult (WD) | Faecal | Gotland |
| I4 | Dairy | 2008/10 | Adult (WD) | Faecal | Gotland |
| M1 | Dairy | 2006/02 | Calf (CD) | Faecal | Skåne |
| M2a | Dairy | 2006/05 | Calf (CD) | Faecal | Skåne |
| N1 | Dairy | 2005/11 | Calf (CD) | Faecal | Halland |
| N2a | Dairy | 2006/03 | Calf (CD) | Faecal | Halland |
| N3 | Dairy | 2006/05 | Calf (CD) | Faecal | Halland |
| N4 | Dairy | 2009/03 | Adult (WD) | Faecal | Halland |
| P1 | Dairy | 2009/02 | Adult (RD) | Nasal | Västergötland |
| U1 | Feedlot | 2009/07 | Calf (RD) | Nasal | Västmanland |
| Y1 | Dairy | 2008/01 | Adult (WD) | Faecal | Ångermanland |
| Z1 | Dairy | 2007/11 | Adult (WD) | Faecal | Jämtland |
| Z2 | Dairy | 2007/11 | Adult (WD) | Faecal | Jämtland |
| Z3 | Dairy | 2007/11 | Adult (WD) | Faecal | Jämtland |
| Z4 | Dairy | 2007/11 | Adult (WD) | Faecal | Jämtland |
| Z5 | Dairy | 2007/11 | Adult (WD) | Faecal | Jämtland |
| Z6 | Dairy | 2007/12 | Adult (WD) | Faecal | Jämtland |
Herds M1and N1 visited on separate occasions, designated M2 and N2.
WD, winter dysentery; CD, calf diarrhoea; RD, respiratory disease.
The old Swedish BCoV strain, C1-9202, which was detected in 1992 from a WD outbreak in Uppland and previously used to experimentally reproduce disease (Tråvén et al., 2001, Niskanen et al., 2002), was also sequenced in this study for phylogenetic comparison. Data on BCoV vaccination status was not available for the herds sampled in this study, but such vaccine is rarely used in Swedish cattle herds.
RNA extraction, synthesis of cDNA and PCR
RNA extraction with TRIzol LS reagent (Invitrogen) and cDNA synthesis with random priming was performed as described by Liu et al. (2006). Polymerase chain reaction (PCR) was performed as described in that paper with minor modifications. Briefly, 5 μL of cDNA sample was added to a tube containing 45 μL of PCR mixture, comprising 5 μL of 10 × PCR buffer, 1 μL of 10 mM dNTP mix, 5 μL of 1 mg/mL bovine serum albumin, 1.5 μL of each primer (10 μM), 5 μL of 25 mM MgCl2, 1 U of Taq DNA polymerase (AmpliTaq; Perkin–Elmer) and 24 μL of water. Two primers, S1AF and S1CR (Hasoksuz et al., 2002b) were used to amplify a 1550-nucleotide (nt) fragment using Biometra T3000 Thermocycler (LABREPCO). The thermocycling profile included 35 cycles of denaturation at 94 °C for 45 s, annealing at 50 °C for 60 s, and extension at 72 °C for 200 s, and a final extension at 72 °C for 7 min.
Nested and semi-nested PCR (N- and SN-PCR) assays were developed to increase the sensitivity of the PCR method for clinical detection by amplifying 5 μL of the PCR products under the same reaction conditions described above, but with a 120 s extension time, to amplify the target fragments using published primers (Hasoksuz et al., 2002b); S1AF/S1AR primer pair for the initial 654-nt fragment (fragment A) as SN-PCR, S1BF/S1BR primer pair for the second 490-nt fragment (fragment B) as N-PCR.
DNA sequencing and phylogenetic analysis
The PCR products were purified and subsequently sequenced using the same primers as for PCR and an ABI PRISM BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) as previously described (Liu et al., 2006). Sequencing was performed at least twice in each direction in an ABI 3100 genetic analyser (Applied Biosystems). The sequencing reads for each sample were assembled to a final 1038-nt fragment of the S1 subunit spanning nucleotides 23641–24679 (amino acid residues 1–346 of the S glycoprotein) of BCoV strain Mebus (GenBank accession number U00735). Multiple sequence alignment was performed with software BioEdit version 7.0.9.0 (Hall, 1999) with an engine based on the ClustalW algorithm.
Maximum likelihood analysis was performed with software PHYML (Guindon and Gascuel, 2003), as previously described (Liu et al., 2009). The General Time Revisable (GTR) with substitution rate heterogeneity and proportion of invariable sites ‘GTR + I + G’ was selected for inferring BCoV phylogeny, and 1000 non-parametric bootstrapping analyses were performed to evaluate the robustness of the tree (Guindon and Gascuel, 2003). The dataset included the reference strains of BCoV: Mebus (GenBank accession number U00735); Quebec (AF220295); Germany/V270 (EF193075); alleged respiratory bovine coronavirus strains: LSU2 (AF058943) and OK-3 (AF058944); alleged enteric bovine coronavirus strains: F15 (D00731) and LY138 (AF058942). The analysis also included recently published sequences of three Italian strains 179/07-11 (EU019216), 438/06-TN (EU814647), and 339/06 (EF445634); two Korean strains: KWD19 (DQ389660) and KCD10 (DQ389641), and one Japanese strain: Kakegawa (AB354579). One Swedish strain Rc1N-99 (DQ121649) and two Danish strains Ac1F-03 (DQ121619) and Hc1F-01 (DQ121628) from the previous study (Liu et al., 2006) were included in the analysis, where 414 nt were treated as missing data. The analysis also included Human coronaviruses: Hu-OC43 (Z32769) and Hu-4408 (L07748); Giraffe coronavirus (EF424624) and Alpaca coronavirus (DQ915164).
Results
Genetic variation of the S gene
Comparative analysis of the 1038 nt sequence showed that all the 26 Swedish strains obtained in 2005–2009 shared a high identity both at the nt level (>97.7%) and at the deduced amino acid (aa) level (>96.7%). Compared with the reference strain Mebus, 29–36 nt substitutions were identified, resulting in 16–20 aa changes that were clustered around residues 11–40, 88–115, 143–188, and 241–257.
More genetic variation was observed within Southern strains (23 nt changes) than within Northern and Central strains (9 nt changes). Sequences from five herds (Z1–Z5) in the Northern region (Jämtland) sampled during the third year were identical. These sequences were also identical to the sequence U1-0907 obtained from a herd in the Central region (Västmanland) that had an outbreak of respiratory disease in July 2009. The two sequences N1-0511 and N2-0603 obtained from the same herd with a 4-month interval showed 100% identity, while 6 nt changes were found between sequences M1-0602 and M2-0605 obtained with a 3-month interval from another herd.
The BCoV strains obtained from respiratory and enteric disease did not show any consistent differences in the sequenced S1 region. Strain C1-9202 shared a 97.0–98.2% identity at the nt level, 96.2–97.4% at the deduced aa level, with the Swedish strains detected in 2005–2009. GenBank accession numbers for the BCoV sequences obtained in this study are JN795141–JN795167.
Phylogenetic analysis
The Swedish BCoV strains obtained for this study fell into four clusters in the consensus tree based on maximum likelihood analysis of the 1038-nt fragment of the S gene (Fig. 1 ). Cluster I included 20 strains from all over Sweden: three from Halland and one from Västergötland in the South; 2 from Gotland, an island in the east; 5 from Uppland and 1 from Västmanland in the Central region; 6 from Jämtland and one each from Västerbotten and Ångermanland in the North. Cluster II contained 4 Swedish strains: AC1-0611 from the Northern region Västerbotten and M1-0602, M2-0605 and N4-0903 from the Southern regions Skåne and Halland, and a Danish strain AcF-03 (Liu et al., 2006). This cluster also included the Italian strain Bubalus/Italy/179/07, a BCoV strain detected in buffalo calves with severe enteritis (Decaro et al., 2008b). Cluster III contained the two Swedish strains I2-0702 and I4-0801 from Gotland and the strains Italy/438/06 and Italy/339/06 that were isolated from an outbreak of enteric and respiratory disease in dairy cattle (Decaro et al., 2008a). Cluster IV contained C1-9202, the oldest Swedish strain from Uppland, together with the Swedish strain Rc1N-99 and the Danish strain HcF-01 that were collected in 1999 and 2001, respectively. The grouping of these three sequences was supported by a bootstrapping value of 62%.
Fig. 1.
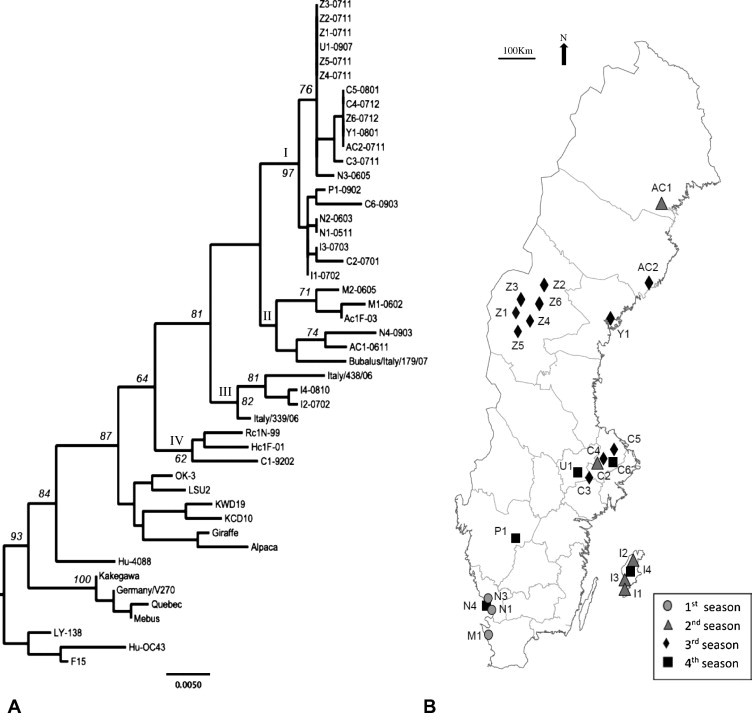
A majority-rule consensus tree (A) of maximum likelihood analysis of the Swedish BCoV strains sampled across the country (B) during four in-stable seasons. Four clusters are labelled I, II, III, and IV, respectively. Bootstrapping values (italic) are placed next to the major clades. Bar indicates changes per site. The tree is presented as rooted with LY-138, F15 and Hu-OC43. Herd C1 is not included on the map.
Discussion
Twenty-seven samples from 25 herds located in different parts of Sweden were positive for BCoV and comparative sequence analysis showed a high identity between the strains. There were no particular aa residues of the initial 346 aa of the S1 subunit that could differentiate strains obtained from cattle with respiratory disease from those obtained from calves or adult cattle with diarrhoea. The results suggest that the same strains of BCoV cause natural outbreaks of respiratory disease, CD and WD, which is in agreement with previous studies (Tråvén et al., 2001, Niskanen et al., 2002, Saif, 2010).
The occurrence, in this study, of a BCoV outbreak in summer confirms previous reports from South Korea and Italy (Park et al., 2006, Decaro et al., 2008a) that BCoV circulates between cattle herds all the year round. Furthermore, the identical 1038-nt sequences of strain U1-0907, obtained from an outbreak of respiratory disease in calves, and strains Z1-0711–Z5-0711, obtained from WD outbreaks in Jämtland, indicate that one BCoV strain may circulate in the cattle population for more than a year. Herd U1 is a feedlot which regularly purchases calves from Jämtland. Analysis of the whole S gene (4092 nt) of strains Z2-0711 and U1-0907 confirmed an identity of 99.6%, suggesting that sequence analysis of the S gene can be used to support or rule out suspected transmission routes of BCoV between regions.
The finding of identical 1038-nt sequences of two strains obtained 4 months apart from young calves in a large dairy herd (N1, >200 cows) indicates, for the first time, that a BCoV strain may circulate for an extended period of time within large herds. On the other hand, strains from another large herd (M1) obtained 3 months apart had several mutations, suggesting introduction of a new strain into that herd.
All strains obtained from Northern and Central Sweden during the third year were highly similar (nt identity >99.7%). These strains were obtained from a regional epizootic of WD, i.e. a series of outbreaks close in time and space. These regions were shown to have a low herd immunity to BCoV; 70% of the herds had BCoV seronegative primiparous cows in the Northern regions compared to 10% in the Southern parts in 2007 (Ohlson et al., 2010; A. Ohlson et al., unpublished data). This may be related to a lower density of herds in Northern regions, less transportation of animals, and/or a difference in biosecurity between the regions (Ohlson et al., 2010). The low herd immunity suggests that BCoV is not continuously circulating in the Northern parts of Sweden, and that virus causing regional epizootics is introduced from other regions. Such a transmission is also suggested by the finding of strain N3-0605 in Southern Sweden prior to and highly identical (nt identity >99.8%) to the strain causing the Jämtland outbreaks of WD (Z1–Z5).
The molecular pattern of clusters II, III and IV suggests that the BCoV strains circulating in Swedish herds are closely related to strains present in other parts of Europe. While it is easy to understand the clustering of strains from the Central and Southern parts of Sweden with strains from Denmark, which borders with Sweden, grouping of Swedish strains with Italian strains was surprising. As there were no records of animals purchased from Italy, it is unlikely that the virus was introduced to Sweden by transportation of animals directly from Italy. There have, however, been animals exported from Sweden to Central and Southern Europe in recent years. Samples sequenced from other European countries are needed in order to trace the origin and/or spread of Swedish strains and molecular analysis of future BCoV outbreaks from different parts of the world will increase the understanding of BCoV epidemiology, e.g. direct and indirect transmission routes between herds, regions or even countries.
Conclusions
In this study we showed that molecular analysis of the S gene of BCoV was a useful tool for studying the epidemiology of this virus. The analysed strains showed a high sequence identity, regardless of whether they were obtained from outbreaks of respiratory disease or diarrhoea or from calves or adults. Circulation of a BCoV strain during an extended period of time was demonstrated in calves in a large dairy herd. In Northern Sweden, a regional epizootic of highly similar BCoV strains was detected, while in the Southern and Central regions more extensive transmission of the virus was indicated. Finally, identical BCoV sequences supported epidemiological data on inter-herd contact through purchased calves.
Conflict of interest statement
None of the authors of this paper has a financial or personal relationship with other people or organisations that could inappropriately influence or bias the content of the paper.
Acknowledgments
This study was supported by Grants from The Swedish Farmers’ Foundation for Agricultural Research (V0630010, V0830402). We thank Sven-Åke Bergkvist for valuable technical assistance.
References
- Alenius S., Niskanen R., Juntti N., Larsson B. Bovine coronavirus as the causative agent of winter dysentery – Serological evidence. Acta Veterinaria Scandinavica. 1991;32:163–170. doi: 10.1186/BF03546976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros M.L., Sanchez C.M., Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidokhti M.R., Tråvén M., Fall N., Emanuelson U., Alenius S. Reduced likelihood of bovine coronavirus and bovine respiratory syncytial virus infection on organic compared to conventional dairy farms. The Veterinary Journal. 2009;182:436–440. doi: 10.1016/j.tvjl.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.A. Bovine coronavirus. British Veterinary Journal. 1993;149:51–70. doi: 10.1016/S0007-1935(05)80210-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Desario C., Campolo M., Elia G., Martella V., Greco G., Cirone F., Colaianni M.L., Cordioli P., Buonavoglia C. Severe outbreak of bovine coronavirus infection in dairy cattle during the warmer season. Veterinary Microbiology. 2008;126:30–39. doi: 10.1016/j.vetmic.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Martella V., Elia G., Campolo M., Mari V., Desario C., Lucente M.S., Lorusso A., Greco G., Corrente M., Tempesta M., Buonavoglia C. Biological and genetic analysis of a bovine-like coronavirus isolated from water buffalo (Bubalus bubalis) calves. Virology. 2008;370:213–222. doi: 10.1016/j.virol.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley J.K., Parker S.E., Bloom F., Buchmeier M.J. The V5A13.1 envelope glycoprotein deletion mutant of mouse hepatitis virus type-4 is neuroattenuated by its reduced rate of spread in the central nervous system. Virology. 1992;187:178–188. doi: 10.1016/0042-6822(92)90306-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hägglund S., Svensson C., Emanuelson U., Valarcher J.F., Alenius S. Dynamics of virus infections involved in the bovine respiratory disease complex in Swedish dairy herds. The Veterinary Journal. 2006;172:320–328. doi: 10.1016/j.tvjl.2005.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hasoksuz M., Hoet A.E., Loerch S.C., Wittum T.E., Nielsen P.R., Saif L.J. Detection of respiratory and enteric shedding of bovine coronaviruses in cattle in an Ohio feedlot. Journal of Veterinary Diagnostic Investigation. 2002;14:308–313. doi: 10.1177/104063870201400406. [DOI] [PubMed] [Google Scholar]
- Hasoksuz M., Sreevatsan S., Cho K.O., Hoet A.E., Saif L.J. Molecular analysis of the S1 subunit of the spike glycoprotein of respiratory and enteric bovine coronavirus isolates. Virus Research. 2002;84:101–109. doi: 10.1016/S0168-1702(02)00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J.H., Kim G.Y., Yoon S.S., Park S.J., Kim Y.J., Sung C.M., Jang O.J., Shin S.S., Koh H.B., Lee B.J., Lee C.Y., Kang M.I., Kim H.J., Park N.Y., Cho K.O. Detection and isolation of winter dysentery bovine coronavirus circulated in Korea during 2002–2004. Journal of Veterinary Medical Sciences. 2005;67:187–189. doi: 10.1292/jvms.67.187. [DOI] [PubMed] [Google Scholar]
- Kubo H., Yamada Y.K., Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. Journal of Virology. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Hägglund S., Hakhverdyan M., Alenius S., Larsen L.E., Belák S. Molecular epidemiology of bovine coronavirus on the basis of comparative analyses of the S gene. Journal of Clinical Microbiology. 2006;44:957–960. doi: 10.1128/JCM.44.3.957-960.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Xia H., Wahlberg N., Belák S., Baule C. Phylogeny, classification and evolutionary insights into pestiviruses. Virology. 2009;385:351–357. doi: 10.1016/j.virol.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Niskanen R., Lindberg A., Tråvén M. Failure to spread bovine virus diarrhoea virus infection from primarily infected calves despite concurrent infection with bovine coronavirus. The Veterinary Journal. 2002;163:251–259. doi: 10.1053/tvjl.2001.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson A., Heuer C., Lockhart C., Tråvén M., Emanuelson U., Alenius S. Risk factors for seropositivity to bovine coronavirus and bovine respiratory syncytial virus in dairy herds. Veterinary Record. 2010;167:201–206. doi: 10.1136/vr.c4119. [DOI] [PubMed] [Google Scholar]
- Park S.J., Jeong C., Yoon S.S., Choy H.E., Saif L.J., Park S.H., Kim Y.J., Jeong J.H., Park S.I., Kim H.H., Lee B.J., Cho H.S., Kim S.K., Kang M.I., Cho K.O. Detection and characterization of bovine coronaviruses in fecal specimens of adult cattle with diarrhea during the warmer seasons. Journal of Clinical Microbiology. 2006;44:3178–3188. doi: 10.1128/JCM.02667-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton D.J., Christiansen K.H., Alenius S., Cranwell M.P., Pritchard G.C., Drew T.W. Prevalence of antibodies to bovine virus diarrhea virus and other viruses in bulk tank milk in England and Wales. Veterinary Record. 1998;142:385–391. doi: 10.1136/vr.142.15.385. [DOI] [PubMed] [Google Scholar]
- Saif L.J. Bovine coronavirus infection. In: Coetzer J.A.W., Tustin R.C., editors. Infectious Diseases of Livestock. Second Ed. Oxford University Press; Oxford, UK: 2004. pp. 795–802. [Google Scholar]
- Saif L.J. Bovine respiratory coronavirus. The Veterinary Clinics of North America, Food Animal Practice. 2010;26:349–364. doi: 10.1016/j.cvfa.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B., Gross H.J., Brossmer R., Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. Journal of Virology. 1991;65:6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson C., Lundborg K., Emanuelson U., Olsson S.O. Morbidity in Swedish dairy calves from birth to 90 days of age and individual calf-level risk factors for infectious diseases. Preventive Veterinary Medicine. 2003;58:179–197. doi: 10.1016/s0167-5877(03)00046-1. [DOI] [PubMed] [Google Scholar]
- Torsein M., Lindberg A., Hallén Sandgren C., Persson Waller K., Törnquist M., Svensson C. Risk factors for calf mortality in large Swedish dairy herds. Preventive Veterinary Medicine. 2011;99:136–147. doi: 10.1016/j.prevetmed.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tråvén M., Björnerot L., Larsson B. Nationwide survey of antibodies to bovine coronavirus in bulk milk from Swedish dairy herds. Veterinary Record. 1999;144:527–529. doi: 10.1136/vr.144.19.527. [DOI] [PubMed] [Google Scholar]
- Tråvén M., Näslund K., Linde N., Linde B., Silván A., Fossum C., Hedlund K.O., Larsson B. Experimental reproduction of winter dysentery in lactating cows using BCV – Comparison with BCV infection in milk-fed calves. Veterinary Microbiology. 2001;81:127–151. doi: 10.1016/S0378-1135(01)00337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D., Parker M.D., Babiuk L.A. The S2 subunit of the spike glycoprotein of bovine coronavirus mediates membrane fusion in insect cells. Virology. 1991;180:395–399. doi: 10.1016/0042-6822(91)90045-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D., Deregt D. A single amino acid change within antigenic domain II of the spike protein of bovine coronavirus confers resistance to virus neutralization. Clinical and Diagnostic Laboratory Immunology. 2001;8:297–302. doi: 10.1128/CDLI.8.2.297-302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


