Abstract
In the past two decades, humans have faced many new viral infectious agents in emerging and re-emerging infectious diseases (EIDs). Many factors contribute to the appearance of EIDs. These factors are complex but can be classified into three different categories: virus factors, human factors, and ecological factors. The factors contributing to the cause of such viral infectious diseases will be systematically reviewed in this article.
Keywords: Emerging infectious diseases, Re-emerging infectious diseases, Viral infectious diseases
1. Introduction
Emerging and re-emerging infectious diseases (EIDs) have surfaced in recent decades. An “emerging infection” is either a new infection that has never appeared before or a known infection that has a recent increase in prevalence. The immunodeficiency virus (HIV) pandemic and severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) outbreaks are prototypical examples of the emerging infectious diseases that the public did not face prior to the 1980s and 2003, respectively. A “re-emerging infection” is a familiar infection that recurs. Influenza A virus pandemics of 1918, 1957, and 1968 are prototypical examples of re-emerging infections.
Since EIDs have important public health consequences, there is a major concern regarding where viruses came from. The general forces contributing to the emergence of EIDs fall into three categories: (1) viral factors, (2) human factors, and (3) ecological factors. In fact, human factors are the key factors driving disease emergence and influencing disease patterns. This article will systematically review the causes of emergence and re-emergence of viral infectious diseases.
2. Viral factors: virus evolution and adaptation
Most of the viral agents in EID are RNA viruses ( Table 1) that have high mutation rates. This leads to their rapid evolution and environmental adaptability, so that RNA viruses can reach adaptive equilibrium within their host species very rapidly. There are three fundamental mechanisms in the genetic variation of the viral genome: (1) point mutation, (2) recombination, and (3) reassortment.
Table 1.
Some of the most important emerging and re-emerging viruses
| Family | Organization of genome | Genome segment(s) | Genus | Virus | Disease | Reservoir in nature | Vector in nature | Year isolated |
|---|---|---|---|---|---|---|---|---|
| Arenaviridae | (A)ssRNA | 2 | New World Arenavirus | Guanarito | Venezuelan HF | Rodents | – | 1989 |
| Junín | AHF | Rodents | – | 1958 | ||||
| Machupo virus | Bolivian HF | Rodents | – | 1963 | ||||
| Sabiá | Brazilian HF | Rodents | – | 1994 | ||||
| Old World Arenavirus | Lassa virus | Lassa fever | Rodents | – | 1969 | |||
| LCMV | LCM | Rodents | – | 1930s | ||||
| Bunyaviridae | (A)(−)ssRNA | 3 | Hantavirus | Hantavirus | HFRS | Rodents | – | 1977 |
| SNV | HPS | Rodents | – | 1993 | ||||
| 3 | Nairovirus | CCHF virus | CCHF | Livestock, cows | Ticks | 1981 | ||
| 3 | Phlebovirus | RVFV | RVF | Livestock | Mosquitoes | 1931 | ||
| Caliciviridae | (+)ssRNA | 1 | “Norwalk-like viruses” | NV | Gastroenteritis | Humans | ? | 1972 |
| “Sapporo-like viruses” | SV | Gastroenteritis | ? | ? | 1970s | |||
| Coronaviridae | (+)ssRNA | 1 | Coronavirus | SARS-CoV | SARS | ? | ? | 2003 |
| Filoviridae | (−)ssRNA | 1 | “Ebola-like viruses” | Ebola | Ebola HF | ? | ? | 1976 |
| “Marburg-like viruses” | Marburg | Marburg HF | ? | ? | 1967 | |||
| Flaviviridae | (+)ssRNA | 1 | Flavivirus | DENV | DF, DHF, DSS | Humans | Mosquitoes | 1950s |
| JEV | Fatal encephalitis | Birds, pigs | Mosquitoes | 1946 | ||||
| KFDV | Kyasanur Forest disease | Nonhuman primate | Ticks | 1990s | ||||
| Omsk HF virus | Omsk HF | Rodents | Ticks | 1990s | ||||
| SLE virus | SLE | Birds | Mosquitoes | 1971 | ||||
| WNV | West Nile encephalitis | Birds | Mosquitoes | 1937 | ||||
| YFV | YF | Nonhuman primate | Mosquitoes | 1944 | ||||
| Hepacivirus | HCV | Chronic hepatitis, cirrhosis | Humans | – | – | |||
| Hepadnaviridae | dsDNA | 1 | Orthohepadnavirus | HBV | Chronic hepatitis, cirrhosis | Humans | – | 1965 |
| Orthomyxoviridae | (−)ssRNA | 8 | Influenzavirus A | Human influenza A | Respiratory infection | Birds | – | 1933 |
| 8 | Influenzavirus A | Avian influenza A | Respiratory infection | Birds | – | 1997 | ||
| 8 | Influenzavirus B | Human influenza B | Respiratory infection | Birds | – | 1939 | ||
| 7 | Influenzavirus C | Human influenza C | Respiratory infection | Birds | – | 1940s | ||
| Paramyxoviridae | (−)ssRNA | 1 | Morbillivirus | hMPV | Respiratory infection | Humans | – | 2001 |
| Rubulavirus | NDV | END, VND | Birds | – | 1944 | |||
| Henipavirus | NiV | Fatal encephalitis | Bats | – | 1999 | |||
| HeV | Fatal encephalitis | Bats | – | 1994 | ||||
| Piconrnaviridae | (−)ssRNA | 1 | Aphthovirus | FMDV | FMD | Cattle, buffalo | – | 1898 |
| Hepatovirus | HAV | Hepatitis | Humans | – | 1973 | |||
| Poxviridae | dsDNA | 1 | Orthopoxvirus | Monkeypox virus | Similar to smallpox | Squirrels | ? | 1958 |
| Reoviridae | dsRNA | 10 | Orbivirus | BTV | EHD | ? | Insects | 1975 |
| 11 | Rotavirus | Rotavirus | Fever, nosocomial infections | Humans | – | 1973 | ||
| Retroviridae | (+)ssRNA | Dimer | Lentivirus | HIV | AIDS | ? | – | 1983 |
| Togaviridae | (+)ssRNA | 1 | Alphavirus | EEEV | Encephalitis | Birds | Mosquitoes | 1973 |
| VEEV | Encephalitis | Rodents | Mosquitoes | 1938 | ||||
| WEEV | Encephalitis | Birds, rabbits | Mosquitoes | 1970s |
(A)ssRNA: ambisense sense single-stranded RNA; AHF: Argentine hemorrhagic fever; AIDS: acquired immunodeficiency syndrome; BTV: blue tongue virus; CCHF: Crimean-Congo hemorrhagic fever; DENV: dengue virus; DHF: dengue hemorrhagic fever; DF: dengue fever; EEEV: Eastern equine encephatitis virus; dsDNA: double-stranded DNA; dsRNA: double-stranded RNA; DSS: dengue shock syndrome; EHD: epizzotic hemorrhagic disease; END: exotic Newcastle disease; FMD: foot-and-mouth disease; FMDV: foot-and-mouth disease virus; HAV: hepatitis A virus; HBV: hepatitis B virus; HCV: hepatitis C virus; HeV: Hendra virus; HF: hemorrhagic fever; HFRS: hemorrhagic fever with renal syndrome; HIV: human immunodeficiency virus; hMPV: human metapneumovirus; HPS: hantavirus pulmonary syndrome; JEV: Japanese encephalitis virus; KFDV: Kyasaunr Forest disease virus; LCM: lymphocytic choriomeningitis; LCMV: lymphocytic choriomeningitis virus; NDV: Newcastle disease virus; NiV: Nipah virus; NV: Norwalk virus; RVF: Rift Valley fever virus; SARS: severe and acute respiratory syndrome; SARS-CoV: SARS-associated coronavius; SFV: simian foamy virus; SIV: simian immunodeficiency virus; SLE: St. Louis encephatitis; SNV: Sin Nombre virus; ssRNA: single-stranded RNA; SV: Sapporo virus; VEEV: Venezuelan equine encephalitis virus; VND: velogenic Newcastle disease; WEEV: Western equine encephalitis virus; WNV: West Nile virus; YF: yellow fever; YFV: yellow fever virus.
2.1. Point mutation
The common adaptation strategy for virus evolution is point mutation. The high mutation rate of RNA viruses is due to the poor fidelity of the RNA-dependent RNA polymerase and RNA-dependent DNA polymerase (i.e. reverse transcriptase [RT]) in which there is an absence of the 3′ → 5′ proofreading exonuclease. Thus, the point mutation rates among RNA viruses is approximately 10−4 to 10−5, whereas in DNA viruses it is 10−8 to 10−11 [1]. In the case of human immunodeficiency virus type 1 (HIV-1), the in vivo point mutation rate is 4 × 10−5 mutations per bp per replication cycle (i.e. 2 mutations in 5 new genomes produced) [2]. The mutation rate of the SARS-CoV during the 2002–2003 epidemic was estimated to be 8.26 × 10−6 per nt per day (i.e. 3 mutations per RNA in every round of replication) [3].
Usually, viral proteins differing by as little as 1 or 2 amino acid residues confer striking differences in phenotype. For example, certain mutations in HIV-1 viral proteins or enzymes can lead to virus resistance to antiviral drugs. Mutations in the HIV-1 RT, PR, and gp41 genes lead to resistance to reverse transcription inhibitors, protease inhibitors, and entry inhibitors, respectively [4]. In the case of influenza A virus, the accumulation of a series of point mutations on the influenza virus surface glycoprotein hemagglutinin (HA) results in amino acid residue substitutions on antigenic sites of HA [5]. These changes prevent binding of host antibodies induced by the previous influenza infection in a phenomenon known as “antigenic drift”.
2.2. Recombination
The second viral mutation strategy for adaptation is recombination. Recombination allows two copies of genetic materials to exchange, producing a new “mixed” or “hybrid” genome molecule [6]. A high recombination rate occurs in coronavirus (CoV) [7], [8], Eastern equine encephalitis virus (EEEV), and Western equine encephalitis virus (WEEV) [9]. The high recombination rates of those viruses are due to (1) the large size of the genome, (2) the discontinuous transcription, and (3) the presence of transcriptionally active full and sub-genomic length RNAs, which increases the amount of template for strand switching.
SARS-CoV represents a typical case of recombination. When two CoVs infect the same cell (termed “co-infection”) ( Fig. 1A), there is the potential for two RNA molecules to cross over. In fact, phylogenetic analyses substantiate the presence of past recombination events between mammalian-like and avian-like parent viruses in the SARS-CoV evolution [10], [11], [12].
Fig. 1.
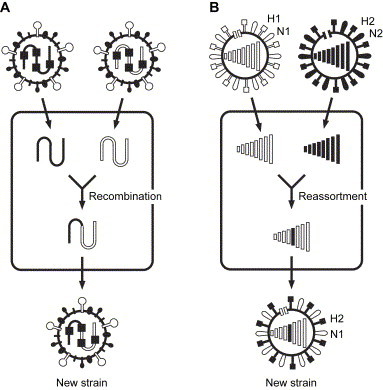
Recombination and reassortment. (A) Two CoVs co-infect in a single cell. The long viral genome may have a chance to undergo recombination. The genome of the new strain is a chimera of the parent strains. (B) Two different strains of influenza A viruses (H1N1 and H2N2, in this case) co-infect a single cell. The progeny virus combines genomic segments from parent strains to form the new strain (H2N1 in this case).
2.3. Reassortment
The third viral adaptation mechanism is gene reassortment. Gene reassortment occurs when segmented viruses, which are viruses with multiple segmented genomes (Table 1), co-infect the same cell and eventually lead to the progeny virus containing a genome set derived from the multiple parent viruses.
For example, the distinguishing feature of the influenza A virus is a genome consisting of 8 segments of ssRNA. When reassortment occurs among influenza viruses, it is called an “antigenic shift” or “genetic shift”. Antigenic shift causes dramatic changes in the influenza virus surface glycoproteins HA and/or neuraminidase (NA). As illustrated in Fig. 1B, when two different strains of influenza A viruses co-infect the same cell, progeny viruses may inherit sets of RNA segments made up of combinations of segments from either of the parent viruses. Thus, coinfection of the same cell in the host animal by different virus strains allows for widely divergent sequences to occur by recombination or reassortment.
3. Human factors: population growth and urbanization
Surprisingly, the genetic evolution of viruses does not seem to be the major cause of virus emergence. Viruses seem to be stable within their particular ecological niche. In fact, a microbe introduced into a new region may have a greater or lesser impact, depending on the susceptibility of a host population. Human factors are actually the most potent factors driving disease emergence.
3.1. Population growth
Since the pathogen of a disease needs to be introduced into the human population and then subsequently spread and maintain itself in the population, a large human population size favors the spread and perpetuation of diseases. For example, the number of susceptible individuals required for the measles virus to continue circulation in an area is 200,000 or more. In general, pathogen transmission is related to the population density of susceptible individuals. This relationship is reflected in the actual value of the basic reproductive rate (R 0) of an infectious disease: it is proportional to the current number of susceptible individuals ( Fig. 2).
Fig. 2.
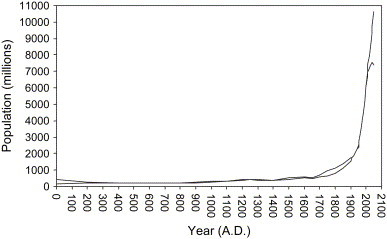
World human population growth curve. Two curves represent the human population in upper and lower medium values. (Source: U.S. Census Bureau International Database and U.N. Population Division Population Database.)
In fact, one of the critical human factors is the sheer increase in the number of Homo sapiens on the planet after the start of the Industrial Revolution in the 18th century and medical development in the 19th century. The dramatic increase in the human population means that more space for living is needed. Therefore, the expansion of the human population into virgin forests could disturb the virus reservoirs and increase the opportunities for viral transmission from animals to humans, ultimately causing EIDs.
3.2. Urbanization
Population growth is correlated with a dramatic increase in human density in a given area, in the process of urbanization. Urbanization brings with it the problems of housing, sanitation, pollution, drinking water, and health care facilities. Increasing population densities and urban poverty encourage the spread of viruses; under poor sanitary conditions, people are more susceptible to pulmonary and gastrointestinal infections. Moreover, the R 0 is also proportional to the number of contacts in a population. Throughout history, an increasing trend of urbanization has been observed; however, in the second half of the 20th century, the rate of urbanization multiplied dramatically. In 1000 A.D., densely populated cities such as Cordova, the capital city of the Empire of Cordova in Europe, and Kaifeng, the capital city of the Sung Dynasty in China, had populations of only 0.4 to 0.45 million ( Table 2). In 1800, the only city with a population over 1 million was Peking, the capital city of the Ching Dynasty in China. In 1900, however, there were at least 10 cities with a population over 1 million. In 2003, there were 37 “megacities”, most of them located in Asia, with over 5 million inhabitants (Table 2).
Table 2.
Metropolitan population
| Year | Urbanites (million) | Megacities (population in million) |
|---|---|---|
| 100 | ≥0.15 | Europe: Rome (0.45) |
| E. Asia: Loyang (0.42) | ||
| Middle East: Seleucia (0.25), Antioch (0.15) | ||
| Africa: Alexandria (0.25) | ||
| 1000 | ≥0.2 | Europe: Cordova (0.45) |
| E. Asia: Kaifeng (0.4), Angkor (0.2) | ||
| Middle East: Constantinople (0.3) | ||
| 1500 | ≥0.25 | E. Asia: Peking (0.672), Hangchow (0.25) |
| S. Asia: Vijayanagar (0.5) | ||
| Middle East: Tabriz (0.25) | ||
| Africa: Cairo (0.4) | ||
| 1800 | ≥0.5 | Europe: London (0.86), Paris (0.55) |
| E. Asia: Peking (1.1), Canton (0.8), Edo (0.69) | ||
| Middle East: Constantinople (0.57) | ||
| 1900 | ≥1 | Europe: London (6.48), Paris (3.33), Berlin (2.707), Vienna (1.7), St. Petersburg (1.4), Manchester (1.4) |
| E. Asia: Tokyo (1.5) | ||
| N. America: New York (4.242), Chicago (1.717), Philadelphia (1.4) | ||
| 2003 | ≥5 | Europe: Paris (9.3), Moskva (8.5), London (7.1) |
| E. Asia: Tokyo (12.1), Seoul (10.2), Shanghai (8.2), Beijing (7.4), Hong Kong (6.8), Tianjin (5.9) | ||
| S. Asia: Mumbai (Bombay) (16.4), Kolkata (Calcutta) (13.2), Delhi (12.8), Jakarta (9.4), Karãchi (9.3), Bangkok (7.5), Chennai (Madras) (6.4), Bandung (5.9), Bangalore (5.7), Hyderabad (5.5), Lahore (5.1), Bogor (5.0) | ||
| Middle East: Istanbul (8.5), Tehran (6.8) | ||
| Africa: Cairo (6.8) | ||
| N. America: New York (21.2), Los Angeles (16.4), Chicago (9.2), Washington DC (7.6), San Francisco (7.1), Boston (5.8), Detroit (5.5) | ||
| S. America: Mexico City (20.3), Buenos Aires (11.3), São Paulo (10.0), Bogota (6.4), Lima (5.7), Rio de Janeiro (5.6) | ||
Source: U.N. Population Division; World Bank.
3.3. Human population movements
Human beings are considered the most highly traveled animals on earth. Due to intense human traffic, the spread of infectious diseases can be led to new areas at any time. Viruses present in rural areas of the world, such as rural areas of Africa or Asia, may show up in more developed parts of the world, such as in Europe or the United States.
3.3.1. Planned migration
The commercial and industrial hubs of the megacities attract low-income citizens, who migrate from the countryside to the city to find work. This situation is occurring in sub-Saharan Africa, South America, Central and Southeast Asia. People from rural areas bring typically rural diseases into the high-density urban area. In addition, people migrating and living at the edge of the city create crowded living conditions and poor sanitation in areas surrounding the city. As more people migrate and reproduce, the strained infrastructure of public health faces increased pressure, and can ultimately break down.
3.3.2. Human travel
Global travel enables viremic travelers to reach any part of the world in less than 24 hours, possibly initiating a global pandemic. In past centuries, when a new disease spread from one continent to another continent, it took many months, years, or even decades to spread around the world. Presently, due to an increase in the amount and speed of international travel, new diseases can spread around the world faster than ever before. A person can now fly from Africa to the US in a day; before the invention of the airplane, it would have taken months or years to make this same journey.
A recent example of the effect of travel on global pandemics is the epidemic of SARS, demonstrating that virus can be spread around the world by international air travelers. Travelers from Singapore, Canada and Vietnam became infected visiting Hong Kong in March 2003 and carried SARS-CoV back to their countries. The SARS outbreaks in Singapore and Canada were related to exposure to the index case in Hong Kong. Within a month, almost twenty countries reported cases of SARS.
3.3.3. Vector travel
It is believed that aircraft can carry virus-infected animals or arthropods from country-to-country and continent-to-continent. It has been demonstrated that mosquitoes can survive in the wheel bays of international flights [13]. The West Nile virus (WNV) outbreak in New York in 1999 is an example. WNV is indigenous to the region of the West Nile River, hence its name, and is common in Middle Eastern countries such as Israel. This virus is transmitted to humans by mosquitoes (Culex pipiens) ( Fig. 3). A leading theory is that airplanes carried WNV-infected mosquitoes when they flew across the Atlantic Ocean from the Middle East.
Fig. 3.
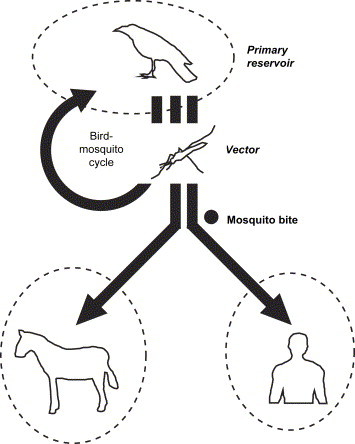
Schematic diagram of the principal routes of transmission of WNV. WNV is maintained in an enzootic bird-mosquito cycle, which involves wild birds as the primary reservoir and mosquitoes as the vector. The virus moves out of this bird-mosquito cycle when infected mosquitoes bite humans or other vertebrate animals (such as horses). However, humans and other mammals serve as “dead-end hosts”, which do not sufficiently amplify virus for mosquito transmission. Arrows represent the route of transmission. Circles with dotted lines represent a species. Arrows crossing between two circles represent cross-species transmission.
3.3.4. Unplanned migration
Unplanned migration due to war or natural disasters has played a large part in introducing infectious diseases into humans. Refugee camps, resettlement areas, and temporary shelters are often characterized by crowded living conditions, poor sanitation, and populations having poor nutrition status, limited access to clean water and medical care, as well as being poorly vaccinated, and lacking separation from insects and animals in the environment. Under these conditions, people become highly susceptible to infectious agents. For example, outbreaks of measles occurred in four refugee camps in Tanzania in mid-2000 [14] and in an Indonesian refugee camp two weeks after the massive earthquake and tsunami of December 2004 in the Indian Ocean [15]. Viral hepatitis has also been found to be prevalent in a large refugee population from Albania [16].
3.4. Human factor: hunting and pasture practices
Approximately 60% of known pathogens of humans have a zoonotic origin [17]. In addition, 75% of EIDs result from exposure to zoonotic pathogens [18], [19]. “Zoonosis” is a disease that is acquired from a nonhuman vertebrate. It is a trans-species transmission of an infectious agent between a human and nonhuman animal species. The close proximity of wildlife to humans is not uncommon; this situation breaks down species barriers and causes interspecies transmission. The viruses are not pathogenic in their natural host, but upon crossing the species barrier, they can lead to disease in the new host. Then, when the disease has adapted, it can be transmitted to another human being and lead to a pandemic outbreak of EIDs.
3.4.1. Hunting
In Africa, close contact between nonhuman primates and humans occurs frequently, due to the traditional habit of hunting animals for food [20], and sometimes hunters slaughter pathogen-infected animals. In Cameroon, more than 60% of the population is directly exposed to fresh nonhuman primate blood and bodily fluids from hunting, butchering, or petting [21], [22].
HIV originated in Africa between the late 1940s and the early 1950s. The earliest known case was in 1959, in a man from the Democratic Republic of Congo (DRC, formerly, Belgian Congo). His blood, stored since 1959, tested positive for HIV [23]. HIV is now not considered a zoonosis, since every case of human acquired immunodeficiency syndrome (AIDS) is acquired from another human being. However, HIV was zoonotic in origin. It was a cross-species transmission from the simian immunodeficiency virus (SIV), which is found in nonhuman primates, to human beings several decades earlier [20], [24]. In fact, HIV and SIV are transmitted the same way (i.e. blood-borne) and found in blood. The two viruses are very similar in genomic organization. Data from phylogenetic trees and geographic habitats show that HIV-1 is most closely related to SIVcpz, an SIV found in a chimpanzee (Pan troglodytes troglodytes) [25]. Human immunodeficiency virus type 2 (HIV-2) is most closely identified with SIVsm, an SIV found in the sooty mangabey (Cercocebus atys) [26]. Although HIV causes AIDS in humans, SIV does not cause any disease in its natural hosts. SIVs might enter man via monkey blood and could be transmitted from nonhuman primates to humans either by eating raw monkey meat, drinking monkey blood, or perhaps through another method of direct exposure to monkey bodily fluids ( Fig. 4).
Fig. 4.
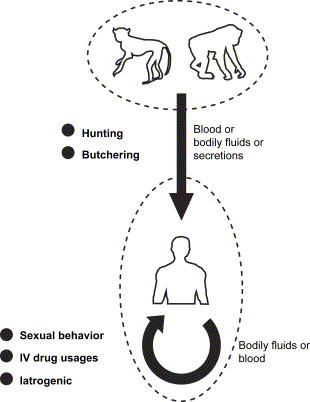
Schematic diagram of the principal route of the transmission of AIDS. HIV-1 and HIV-2 originated from SIV in chimpanzees and sooty mangabeys, respectively. The virus might enter humans during animal hunting and butchering. After that, HIV is transmitted from person-to-person through exchange of bodily fluids, including blood, semen, and breast milk.
3.4.2. Pasturage practices
Monocultures of genetically identical individuals for the purpose of raising productivity and facilitating management of livestock and poultry, promote susceptibility to infection. In addition, large artificially high-density livestock populations facilitate the rapid spread of pathogens throughout livestock populations.
In developing countries, pasturage practices are the cause of the proximity of animals to humans, which can cause viruses to merge. For example, in southern China, especially Guangdong Province and the Pearl River Delta, even though trade in wildlife is illegal, wild animal species (such as badgers, beavers, civet cats, domestic cats, hares, raccoon dogs, snakes, pangolin) are commonly sold in live-animal markets (also called “wet markets”). In the Chinese culture, fresh foods from wild animals are considered great delicacies, (called “wild taste”). The captured wild animals are bred in markets or restaurants for human consumption and kept in squalid animal-holding cages made of metal or wood. It has been thought that a zoonotic virus can be transmitted from animals to food handlers who handle, kill, or butcher animals. Direct contact of humans with animal blood, bodily fluids, or excreta during food handling appears to be the simplest and most plausible explanation for the cross-species transmission of viruses.
As mentioned above, the recombination and reassortment of viruses require the coexistence of two viral genomes in the same cell (Fig. 1A,B). In southern China, captured wild animals are bred and sold as livestock. Before being slaughtered or butchered, the captured animals are kept in the same cage ( Fig. 5A) or stacked cage upon cage (Fig. 5B). Animals may fight in the same cage and some die; blood, secretions, or excretions deposited in the animal-holding cage would provide a source of contamination to the cage beneath it. Usually, the cages are not washed and more animals are placed in them immediately. These practices can contribute to virus strain cross-contamination.
Fig. 5.
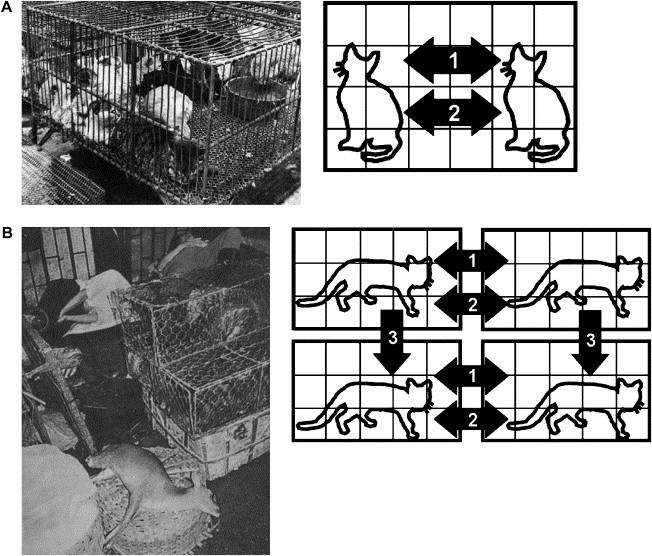
Virus transmission during pasture practices. (A) In southern China, captured animals were put in the same animal-holding cage before slaughtering or butchering. The animals were in close proximity and fought (arrow 1) or were bitten (arrow 2) inside the cage. Therefore, the virus was transmitted to the animals through bloody wounds or close contact. (B) To save storage space, the animal-holding cages are often stacked cage upon cage. Animals at the same level can transfer disease by fighting (arrow 1) or biting (arrow 2). The excreted droppings from the upper level wound increase the chance of infection in the animals held in the lower level (arrow 3). Cross-species transmission will occur if there are different animals in the cages that are staked together.
The SARS outbreak is a typical example. Sequence and phylogenetic analyses reveal that the SARS-CoV is the mutant or recombinant strain from mammalian-like and avian-like CoVs (see Section 2.2). The CoVs in one animal may cross-infect other animals during pasturage and animal management in southern China. Several of the SARS index patients worked as restaurant chefs, food handlers, or lived near the live-animal market [27]. Although scientists isolated SARS-CoV from caged Himalayan palm civets (Paguma larvata) and raccoon dog (Nyctereutes procyonoides) in wild animal markets in Guangdong [28], civets from the wild were free from SARS-CoV infection [29]. Recent study indicated that wild Chinese horseshoe bats (Rhinolophus sinicus) may be the reservoir animal of SARS-CoV [30]. It is probable that the virus could have spread to the human population from these animals.
The influenza virus is another notable example of cross-species transmission. The transmission route of influenza A virus is well understood ( Fig. 6). The primary natural reservoir of influenza A viruses is the aquatic wildfowl (waterfowl). Ponds, the resting site of those birds, are particularly involved in fecal-oral transmission. Occasionally, viruses infect domestic poultry via bird droppings on equipment, cages, feed, vehicles, shoes, or clothing. In Mainland China, proximity of pigpens and backyard poultry to human habitation is very common. Since pigs are susceptible to both avian and human influenza viruses [31], [32], they serve as “mixing vessels” to generate reassortants which are potential candidates for new pandemic strains.
Fig. 6.
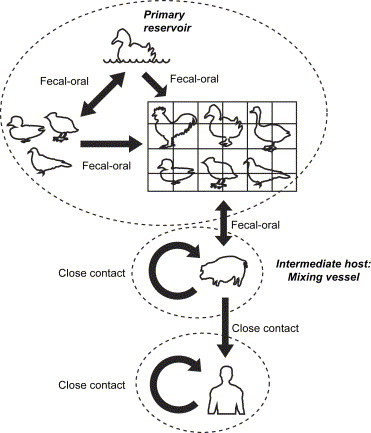
Schematic diagram of the principal routes of transmission of influenza A virus. Influenza A viruses have an extensive reservoir in aquatic wildfowl, in which they usually cause asymptomatic infections. Influenza viruses in birds replicate in the intestines of the avian and are transmitted primarily through the fecal-oral route. Since the tracheal epithelium of pigs contains both receptors for avian and human influenza viruses, pigs can be simultaneously infected with both avian and mammalian viruses. Therefore, pigs were proposed to be the intermediate hosts in which genetic reassortment would take place, giving rise to novel influenza A viruses. Humans can be infected by the viruses from infected pigs. Such infections are usually sporadic and tend to occur in individuals who are exposed to infected pigs. Influenza viruses are transmitted from person to person very easily through aerosolized particles from coughing and sneezing.
These environments, therefore, provide favorable conditions for genetic mixing and spreading of new virus strains. Moreover, the domestic wholesale and retail poultry markets provide the opportunity for the spread of viruses directly to humans. Recently, there were sudden outbreaks of avian influenza virus in a human population. Here again, pasture practice was the major factor that contributed to the outbreak of avian influenza virus in south China. Firstly, the Chinese desire “fresh” (i.e. recently slaughtered) poultry for food consumption and various cultural activities. Secondly, live poultry are housed in crowded and unhygienic conditions in the stalls of retail outlets and traditional markets, without physical separation. Thirdly, without central slaughtering facilities, the chicken slaughtering has to be performed at retail outlets in proximity to crowded residential complexes, with close proximity of poultry-to-poultry and poultry-to-human occurring in live-animal markets or poultry farms. Under such conditions, the influenza virus can easily spread from poultry to humans.
These examples suggest that co-infection by genetically distinct virus strains may be a more important aspect of viral epidemiology than previously realized, and it has the capacity to make new variants available for natural selection.
3.5. Human factor: agricultural practices and deforestation/land development
3.5.1. Expanding agriculture
The extension of agricultural cultivation into forests may alter the existing zoonotic transmission cycles. The removal of forests and development of new farmland expose farmers and workers to disease-carrying arthropods and rodents. As humans encroach upon the forests, it becomes more likely that they will be exposed to new infectious agents.
Agricultural workers in forest-farmland margins are at risk of infection by arenaviruses, which are often carried by rodents, causing hemorrhagic fever (HF) in infected persons. Argentine hemorrhagic fever (AHF), caused by the Junín virus, emerged through agricultural changes [33], [34]. As people cleared the pampas for agriculture and began to plant maize, the rodent Calomys musculinus, a natural host of Junín virus, began to flourish. The infected Calomys species shed virus in their urine. Agricultural machinery (such as excavators) can stir up dried rodent urine containing the virus. Agricultural workers can become infected by inhalation of virus-contaminated aerosols produced from rodent excreta. Agricultural machinery can also create aerosols of infective blood if the animals are accidentally ground by the machines. A similar situation existed with Bolivian HF and Lassa fever, caused by the Machupo virus and Lassa fever virus, respectively, via their natural respective host rodents Calomys callosus and Mastomys natalensis [35], [36]. Therefore, increased agriculture led to an increase of HF cases.
3.5.2. Deforestation/land development
Logging, dam construction, mining, and oil/gas exploitation are also direct causes of deforestation. Expanding human habitation in a region may put people in direct contact with new animal species and thus their viruses. An example can be observed in people in central and western Africa infected with monkeypox. When people develop settlements at the edge of a rain forest and then encroach upon the forest, they come in contact with infected rodents that inhabit the forest. Monkeypox is most often transmitted to humans through contact with the animal's blood or through a rodent bite.
Another example is the Nipah virus (NiV) outbreak in Malaysia between September 1998 and April 1999, also a result of deforestation. During 1997–8, slash-and-burn deforestation formed a severe haze in a few months that blanketed much of Southeast Asia. Lacking their food supply, the fruitbats (also known as flying-foxes; Pteropus species), the natural reservoir of NiV, migrated from the forest to cultivated fruit orchards. In pigsties located around the fruit orchards, pigs ingested bat saliva from half-eaten dropped fruit containing the NiV. The virus spread easily in the overcrowded farms and allowed the transmission of NiV from the fruitbat to the domestic pigs and ultimately to humans [37], [38] ( Fig. 7A). Similar conditions occurred with the HeV outbreak in 1994–5 in Queensland, Australia [39]. During that time, horses were infected with HeV from bat placental fluid. The HeV was then transmitted to humans by close contact with the horses (Fig. 7B).
Fig. 7.
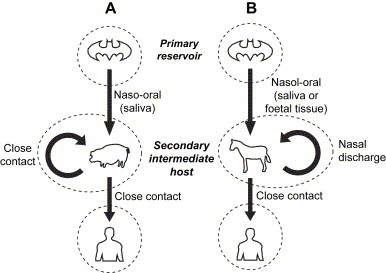
Schematic diagram of the principal routes of transmission of NiV and HeV. (A) The fruitbats of the genus Pteropus have been identified as a primary natural reservoir host for NiV. The natural transmission of the virus from fruitbats to pigs is possible via the naso-oral route. Viruses have been isolated from half-eaten fruit dropped near pig farms, and which may have enough virus to infect an animal that subsequently ingests them. The close proximity of pigs in many Malaysian pig farms probably contributed significantly to pig-to-pig transmissions. The lack of human-to-human transmission could be due to the lower virus load in human respiratory secretions and urine compared to that in pigs. (B) It is possible that the natural transmission of HeV from bats to horses is by the naso-oral route. It has also been hypothesized that transmission from bats to horses is affected by contact with infected fetal tissue or fluids, most probably via the ingestion of recently contaminated pasture. The possibility of contact with nasal discharge exists for horse-to-horse transmission.
3.5.3. Dam construction
In order to increase agricultural productivity, many dam construction projects for irrigation and flood control were undertaken in the 1930s. Two thirds of rivers on the earth have at least one dam. Dams alter free-flowing water systems by reducing river levels, blocking the flow of nutrients, changing water temperature and oxygen levels, and impeding or preventing fish and wildlife migration. The construction of dams created large artificial lakes that covered forests and open land. Thus, dams changed the chemical, physical, and biological processes of river ecosystems.
In 1987, following dam construction on the Senegal River, a Rift Valley fever (RVF) epidemic occurred for the first time in Mauritania, leading to 200 human deaths from HF [40]. In 1993, a RVF outbreak followed the opening of the Aswan dam in Egypt. Three mosquito species (Aedes cumminsil, Ae. Circumluteolus, and Ae. Mcintoshi) are known vectors for RVF virus. Floods and heavy rainfall are factors in the spread of the virus because mosquito populations increase dramatically during these periods.
3.6. Human factor: globalization of commerce
3.6.1. Global food production chain
Today, mass food processing combines large amounts of raw materials for worldwide distribution. Unfortunately, global trade expanded the markets for imported foods, which occasionally contain microbial contaminants due to uneven sanitary practices during manufacture and processing of food in different parts of the world. For example, a common life-threatening complication of the food-borne illnesses called hemolytic uremic syndrome (HUS) may have developed from Escherichia coli O157:H7. Food-borne viral infections are also increasingly recognized as an additional cause of illness in humans. Norwalk virus (NV), Sapporo virus, and hepatitis A virus (HAV) pose the greatest risk of food-borne transmission to humans.
3.6.2. Animal trade
Imported experimental monkeys, such as vervet monkeys (Cercopithecus aethiops) used in biomedical research, from tropical countries into Western countries, were responsible for an outbreak of Marburg virus in Marburg (Germany) and Ebola virus in Virginia (US) in 1969 and 1989, respectively [41], [42].
In addition, the pet trade and exotic animals are of concern. A recent example is the pet prairie dog (Cynomys species), a type of wild rodent from Africa: the prairie dog was responsible for a monkeypox outbreak in six Midwestern states in the US that infected 37 people in the summer of 2003 [43]. All the cases involved contact with prairie dogs, but no person-to-person transmission was reported. This is the first documented human infection from monkeypox in the Western hemisphere that is outside of the virus' geographic origin of Africa.
3.7. Human factor: human social behavior
3.7.1. Sexual behavior and intravenous drug abuse
Having multiple sex partners increases the risk of sexually transmitted diseases (STDs). From a computer simulation model, long-term simultaneous partnerships (or sex-relationships) in populations dramatically increased the size, speed, and variability of the HIV epidemic [44]. As mentioned earlier (Section 3.4.1), HIV underwent cross-species transmission from nonhuman primate to humans. Once HIV entered humans, it spread from person-to-person primarily through homosexual contact or by needle sharing. Today, AIDS is primarily spread by heterosexual contact.
3.7.2. Traditional funeral rites
In Africa, many Ebola virus outbreaks were linked to funeral practices. It is common for the women to prepare (i.e. wash and clean) the body of their deceased relatives for the ceremony. This traditional funeral rite allows the rapid spread of the virus from blood-to-blood contact.
3.7.3. Research study and bioterrorism
Keeping dangerous viral pathogens (such as SARS-CoV and smallpox) in research laboratories raises concern for lapses in biosafety. With SARS-CoV infection cases at three different institutions, in Singapore, Taipei, and Beijing in 2003 and 2004, it is feared that the next SARS epidemic may be more likely to emerge from a research laboratory than from a natural reservoir [45], [46]. In 2005, an H2N2 influenza A strain, which killed millions of people in a worldwide pandemic in 1957, was sent to thousands of laboratories across 19 countries [47]. This serious mistake poses a threat to public health if the virus escapes from any of the laboratories. Moreover, the increased threat of the use of viruses as biological weapons by bioterrorists is a great concern [48].
3.8. Human factor: modern medicine and unsafe practices
Healthcare-related infections affect millions of people worldwide every year. Injudicious use or misuse of antibiotics and anti-viral drugs often occur in poor countries, and can hasten the evolution of resistant strains. Medical devices for intracerebral surgery also carry a risk of transmitting Creutzfeld–Jakob disease (CJD), and so-called iatrogenic CJD (iCJD). Both antibiotic-resistant bacteria and CJD are not viral diseases and have been extensively reviewed elsewhere; they will not be discussed in this review.
3.8.1. Blood transfusions and selling blood
Transfusions can be a very efficient way of transmitting blood-borne viruses. An example was the large-scale HIV transmission through a contaminated blood transfusion in China in the 1990s. Because of the Chinese cultural belief that losing blood is unhealthy, there is often a shortage of blood at local blood banks; blood was therefore often collected from several people at the same time. In order to let the blood donors sell their blood more frequently without developing anemia, the blood centers pooled the blood, retained the plasma, and re-infused the remaining red blood cells back into the blood donors. Therefore, at least 250,000 paid blood donors in seven provinces in central China may have been infected with HIV through this procedure, as well as through the reuse of needles and non-sterilized equipment. Many patients in China received contaminated blood transfusions as well.
3.8.2. Hospital practices
Poor hospital practices inadvertently help the spread of disease. Almost no hospital in developing countries in rural Africa can afford single-use syringes and needles. Usually five or six syringes are used throughout the day with minimal sterilization between patients. This was one of the primary ways of transmission of EIDs such as Ebola and Marburg. Another example is SARS. During the early phase of the SARS epidemic in 2003, the exhaled droplets from SARS patients coming out of the side vents of their standard oxygen facemasks may have increased the risk for contracting SARS-CoV by medical staff, other patients, or hospital visitors. This helped the transmission of droplet-borne respiratory infection [49].
3.8.3. Immunosuppression
The use of immunosuppressive drugs during organ or bone marrow transplants, chemotherapy, renal dialysis, and chronic corticosteroid treatment decreases the capacities of the immune system and encourage opportunistic infections including human cytomegalovirus (HCMV), varicella-zoster virus (VZV), herpes simplex virus-1 (HSV-1), Epstein–Barr virus (EBV), human herpesvirus-6 (HHV-6) and -7 (HHV-7), human papillomavirus (HPV), adenovirus, respiratory syncytial virus (RSV), and viral hepatitis.
3.8.4. Xenotransplantation
The use of animal organs as a source of transplantation, xenotransplantation, increases the possibility of directly introducing animal viruses into human beings. Baboon cytomegalovirus (BCMV) DNA was detected in a baboon liver recipient after xenotransplantation [50].
3.9. Human factor: breakdown in public health measures
Public health measures, including basic sanitation and disease control activities, are not well maintained during war, civil strife, or natural disaster. For example, many refugee populations in African countries do not receive support from national or global AIDS strategy plans [51]. War is accompanied by economic collapse, famine, and homelessness as well as water-borne, rodent-related diseases, and STDs are associated with refugee campus (Section 3.3.4).
4. Ecological factors
Many EID outbreaks have occurred after extreme climatic conditions such as floods and droughts, demonstrating a clear relationship between environmental characteristics and virus emergence. In the 21st century, extreme climatic events are expected to increase, with changes in the global climate, and frequency of EID outbreaks.
4.1. El Niño Southern Oscillation (ENSO)
The El Niño Southern Oscillation (ENSO) is the most potent source of climate variability. The emergence of hantavirus pulmonary syndrome (HPS) in 1993, caused by Sin Nombre virus (SNV), in the Four Corners region of the US was linked to ENSO [52]. The dramatic increase in precipitation during the 1992–3 ENSO was associated with an increased production of piñon nuts, a food for rodents, and resulted in an increase in deer mouse (Peromyscus maniculatus) populations in the southwest United States. The virus was excreted in mouse droppings and contaminated floors and blankets. Another example is the NiV outbreak in 1997–8 (Section 3.5.2). A drought driven by the ENSO led to an acute reduction in the already scarce fruiting forest trees in the region. This forced the fruitbats to invade areas of fruit cultivation that were also used as pig farms.
4.2. Global warming
A slight rise in global temperature could trigger explosions of the earth's insect population. Computer simulation models have also predicted that global warming will cause mountain glacier recessions and lead to highland environment changes. The warm climate triggers the increase in plant and insect populations in mountain areas. In the past 15 years, mosquito-borne infections (including dengue fever and yellow fever) are being reported at high elevations in South and Central America, Asia, and East and Central Africa since the expansion of mosquito populations [53], [54].
The warming trend also helped to establish the WNV-mosquito cycle in New York. The mild winter in 1998–9 enabled virus-bearing mosquitoes to survive in hibernation. The subsequent dry spring and summer in 1999 decreased the number of predators for mosquitoes such as frogs, lacewings, and ladybugs. In the meantime, water for larvae breeding sites evaporated, and nourishing organic compounds became concentrated. The mosquito population grew very large. As more and more mosquitoes became infected, they easily spread the WNV to birds and ultimately to humans.
5. Conclusion
In recent years, we have seen the emergence and re-emergence of many viruses. EID outbreaks highlight the shortcomings in our understanding of the complexities of infectious diseases. The reasons for emergence and re-emergence are multiple and complex ( Table 3). Factors that contribute to the emergence of EIDs are (1) virus evolution and adaptation, (2) human factors, and (3) ecological changes. Clearly, human activities are a key factor for driving disease emergence, and the current knowledge of the various emerging viruses is far from complete. It could be said that the current global situation favors disease emergence, and we may be faced with more outbreaks or pandemics of EIDs in the future.
Table 3.
Reasons for emergence of new viral diseases
| Disease or outbreak | AIDS | HPS (1993) | Avian flu (1997) | NiV (1997–8) | WN (1999) | SARS (2003) |
|---|---|---|---|---|---|---|
| Viral pathogen | HIV | SNV | Influenza A virus | NiV | WNV | SARS-CoV |
| Viral factor | ||||||
| Mutation | ● | ● | ● | ● | ||
| Human factors | ||||||
| Population growth and urbanization | ● | ● | ● | |||
| Human population movements | ● | ● | ● | |||
| Hunting and pasture practices | ● | ● | ● | ● | ||
| Agricultural practices and deforestation/land development | ● | |||||
| Globalization commerce | ||||||
| Human social behavior | ● | |||||
| Modern medicine and unsafe practice | ● | ● | ||||
| Breakdown in public health measures | ● | |||||
| Ecological factor | ● | ● | ● | |||
Acknowledgments
The author would like to thank Dr. Debi P. Nayak, Dr. Brian J. Van Lenten, Alan Wagner, Dr. Anthony Lee, Ee Ming Yap, and Nancy Fong for critical reading of the manuscript and helpful suggestions.
References
- 1.Domingo E., Biebrichen C.K., Eigen M., Holland J.J. Landes Bioscience; Georgetown, Texas, USA: 2001. Quasispecies and RNA virus evolution: principles and consequences. <Eurekah.com>. [Google Scholar]
- 2.Mansky L.M. Forward mutation rate of human immunodeficiency virus type 1 in a T-lymphoid cell line. AIDS Res. Hum. Retroviruses. 1996;12:307–314. doi: 10.1089/aid.1996.12.307. [DOI] [PubMed] [Google Scholar]
- 3.Consortium TCSme: Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 4.Daar E.S., Richman D.D. Confronting the emergence of drug-resistant HIV type 1: impact of antiretroviral therapy on individual and population resistance. AIDS Res. Hum. Retroviruses. 2005;21:343–357. doi: 10.1089/aid.2005.21.343. [DOI] [PubMed] [Google Scholar]
- 5.Hatta M., Gao P., Halfmann P., Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 6.Worobey M., Holmes E.C. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- 7.Lai M.M.C. Recombination in large RNA viruses: coronaviruses. Semin. Virol. 1996;7:381–388. doi: 10.1006/smvy.1996.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai M.M.C., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strauss J.H. Recombination in the evolution of RNA viruses. In: Morse S.S., editor. Emerging Viruses. Oxford University Press; New York: 1993. pp. 241–251. [Google Scholar]
- 10.Magiorkinis G., Magiorkinis E., Paraskevis D., Vandamme A.M., Van Ranst M., Moulton V., Hatzakis A. Phylogenetic analysis of the full-length SARS-CoV sequences: evidence for phylogenetic discordance in three genomic regions. J. Med. Virol. 2004;74:369–372. doi: 10.1002/jmv.20187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stavrinides J., Guttman D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X.W., Yap Y.L., Danchin A. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch. Virol. 2004;150:1–20. doi: 10.1007/s00705-004-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell R.C. Survival of insects in the wheel bays of a Boeing 747B aircraft on flights between tropical and temperate airports. Bull. WHO. 1987;65:659–662. [PMC free article] [PubMed] [Google Scholar]
- 14.Kamugisha C., Cairns K.L., Akim C. An outbreak of measles in Tanzanian refugee camps. J. Infect. Dis. 2003;187:S58–S62. doi: 10.1086/368057. [DOI] [PubMed] [Google Scholar]
- 15.Cranmer H.H. The public health emergency in Indonesia: one patient at a time. N. Engl. J. Med. 2005;352:965. doi: 10.1056/NEJMp058045. [DOI] [PubMed] [Google Scholar]
- 16.Chironna M., Germinario C., Lopalco P.L., Carrozzini F., Barbuti S., Quarto M. Prevalence rates of viral hepatitis infections in refugee Kurds from Iraq and Turkey. Infection. 2003;31:70–74. doi: 10.1007/s15010-002-3100-3. [DOI] [PubMed] [Google Scholar]
- 17.Cleaveland S., Laurenson M.K., Taylor L.H. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Phil. Trans. R. Soc. Lond. B. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor L.H., Latham S.M., Woolhouse M.E.J. Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres-Vélez F., Brown C. Emerging infections in animals: potential new zoonoses? Clin. Lab. Med. 2004;24:825–838. doi: 10.1016/j.cll.2004.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn B.H., Shaw G.M., De Cock K.M., Sharp P.M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 21.Wolfe N.D., Switzer W.M., Carr J.K., Bhullar V.B., Shanmugam V., Tamoufe U., Prosser A.T., Torimiro J.N., Wright A., Mpoudi-Ngole E., McCutchan F.E., Birx D.L., Folks T.M., Buke D.S., Heneine W. Naturally acquired simian retrovirus infections in central African hunters. Lancet. 2004;363:932–937. doi: 10.1016/S0140-6736(04)15787-5. [DOI] [PubMed] [Google Scholar]
- 22.Wolfe N.D., Heneine W., Carr J.K., Garcia A.D., Shanmugam V., Tamoufe U., Torimiro J.N., Prosser A.T., LeBreton M., Mpoudi-Ngole E., McCutchan F.E., Birx D.L., Folks T.M., Burke D.S., Switzer W.M. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc. Natl. Acad. Sci. USA. 2005;102:7994–7999. doi: 10.1073/pnas.0501734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu T., Korber B.T., Nahmias A.J., Hooper E., Sharp P.M., Ho D.D. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 24.Korber B., Muldoon M., Theiler J., Gao F., Gupta R., Lapedes A., Hahn B.H., Wolinsky S., Bhattacharya T. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 25.Gao F., Bailes E., Robertson D.L., Chen Y., Rodenburg C.M., Michael S.F., Cummins L.B., Arthur L.O., Peeters M., Shaw G.M., Sharp P.M., Hahn B.H. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z., Telfier P., Gettie A., Reed P., Zhang L., Ho D.D., Marx P.A. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu R.-H., He J.-F., Evans M.R., Peng G.-W., Field H.E., Yu D.-W., Lee C.-K., Luo H.-M., Lin W.-S., Lin P., Li L.-H., Liang W.-J., Lin J.-Y., Schnur A. Epidemiologic clues to SARS origin in China. Emerg. Infect. Dis. 2004;10:1030–1037. doi: 10.3201/eid1006.030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan Y., Zheng B.J., He Q.Y., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S.M., Poon L.L.M. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 29.Poon L.L.M., Chu D.K.W., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H.C., Lau S.K.P., Woo P.C.Y., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S.M. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.-W., Wong B.H.L., Wong S.S.Y., Leung S.-Y., Chan K.-H., Yuen K.-Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito T., Couceiro N.S.S., Kelm S., Baum L.G., Krauss S., Castrucci M.R., Donatelli I., Kida H., Paulson J.C., Webster R.G., Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ninomiya A., Takada A., Okazaki K., Shortridge K.F., Kida H. Seroepidemiological evidence of avian H4, H5, and H9 influenza A virus transmission to pigs in southeastern China. Vet. Microbiol. 2002;88:107–114. doi: 10.1016/s0378-1135(02)00105-0. [DOI] [PubMed] [Google Scholar]
- 33.Maiztegui J.I. Clinical and epidemiological patterns of Argentine haemorrhagic fever. Bull. WHO. 1975;52:567–575. [PMC free article] [PubMed] [Google Scholar]
- 34.Enria D.A., Feuillade M.R. Argentine haemorrhagic fever (Junin virus, Arenaviridae): a review on clinical, epidemiological, ecological, treatment and preventive aspects of the disease. In: Travassos da Rosa A.P.A., Vasconcelos P.F.C., Travassos da Rosa J.F.S., editors. An Overiew of Arbovirology in Brazil and Neighboring Countries. Instituto Evandro Chaggas; Belem, Brazil: 1998. pp. 219–232. [Google Scholar]
- 35.Vainrub B., Salas P. Latin American hemorrhagic fever. Infect. Dis. Clin. North Am. 1994;8:47–59. [PubMed] [Google Scholar]
- 36.Richmond J.K., Baglole D. Lassa fever: epidemiology, clinical features, and social consequences. BMJ. 2003;327:1271–1275. doi: 10.1136/bmj.327.7426.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enserink M. Malaysian researchers trace Nipah virus outbreak to bats. Science. 2000;289:518–519. doi: 10.1126/science.289.5479.518. [DOI] [PubMed] [Google Scholar]
- 38.Chua K.B. Nipah virus outbreak in Malaysia. J. Clin. Virol. 2003;26:265–275. doi: 10.1016/s1386-6532(02)00268-8. [DOI] [PubMed] [Google Scholar]
- 39.Field H., Young P., Yob J.M., Mills J., Hall L., Mackenzie J. The natural history of Hendra and Nipah viruses. Microbes. Infect. 2001;3:307–314. doi: 10.1016/s1286-4579(01)01384-3. [DOI] [PubMed] [Google Scholar]
- 40.Nabeth P., Kane Y., Abdalahi M.O., Diallo M., Ndiaye K., Ba K., Schneegans F., Sall A.A., Mathiot C. Rift Valley fever outbreak, Mauritania, 1998: seroepidemiologic, virologic, entomologic, and zoologic investigations. Emerg. Infect. Dis. 2001;7:1052–1054. doi: 10.3201/eid0706.010627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters C.J., LeDuc J.W. An introduction to Ebola: the virus and the disease. J. Infect. Dis. 1999;179(Suppl. 1):ix–xvi. doi: 10.1086/514322. [DOI] [PubMed] [Google Scholar]
- 42.Slenczka W.G. The Marburg virus outbreak of 1967 and subsequent episodes. Curr. Top. Microbiol. Immunol. 1999;235:49–75. doi: 10.1007/978-3-642-59949-1_4. [DOI] [PubMed] [Google Scholar]
- 43.Di Giulio D.B., Eckburg P.B. Human monkeypox: an emerging zoonosis. Lancet Infect. Dis. 2004;4:15–25. doi: 10.1016/S1473-3099(03)00856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris M., Kretzschman M. Concurrent partnerships and the spread of HIV. AIDS. 1997;11:641–648. doi: 10.1097/00002030-199705000-00012. [DOI] [PubMed] [Google Scholar]
- 45.Senior K. Recent Singapore SARS case a laboratory accident. Lancet Infect. Dis. 2003;3:679. doi: 10.1016/S1473-3099(03)00815-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Normile D. Lab accidents prompt calls for new containment program. Science. 2004;304:1223–1225. doi: 10.1126/science.304.5675.1223a. [DOI] [PubMed] [Google Scholar]
- 47.Check E. Heightened security after flu scare sparks biosafety debate. Nature. 2005;434:943. doi: 10.1038/434943a. [DOI] [PubMed] [Google Scholar]
- 48.Rotz L.D., Khan A.S., Lillibridge S.R., Ostroff S.M., Hughes J.M. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 2002;8:225–230. doi: 10.3201/eid0802.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somogyi R., Vesely A.E., Azami T., Preiss D., Fisher J., Correia J., Fowler R.A. Dispersal of respiratory droplets with open vs closed oxygen delivery masks: implications for the transmission of severe acute respiratory syndrome. Chest. 2004;125:1155–1157. doi: 10.1378/chest.125.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michaels M.G., Jenkins F.J., George K.S., Nalesnil M.A., Starzl T.E., Rinaldo C.R.J. Detection of infectious baboon cytomegalovirus after baboon-to-human liver xenotransplantation. J. Virol. 2001;75:2825–2828. doi: 10.1128/JVI.75.6.2825-2828.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zarocostas J. Refugees are often left out of AIDS plans, report says. Lancet. 2004;363:713. doi: 10.1016/S0140-6736(04)15669-9. [DOI] [PubMed] [Google Scholar]
- 52.Hijelle B., Glass G.E. Outbreak of hantavirus infection in the Four Corners region of the United States in the wake of the 1997–1998 El Nino-southern oscillation. J. Infect. Dis. 2000;181:1569–1573. doi: 10.1086/315467. [DOI] [PubMed] [Google Scholar]
- 53.Patz J.A., Epstein P.R., Burke T.A., Balbus J.M. Global climate change and emerging infectious diseases. JAMA. 1996;275:217–223. [PubMed] [Google Scholar]
- 54.Patz J.A., Reisen W.K. Immunology, climate change and vector-borne diseases. Trends Immunol. 2001;22:171–172. doi: 10.1016/s1471-4906(01)01867-1. [DOI] [PubMed] [Google Scholar]


