Summary
Introduction
Severe acute respiratory syndrome (SARS) is an emerging infectious disease that first occurred in humans in the People's Republic of China in November 2002 and has subsequently spread worldwide. A novel virus belonging to the Coronaviridae family has been identified as the cause of this pulmonary disease. The severity of the disease combined with its rapid spread requires the development of fast and sensitive diagnostic assays.
Results
A real-time quantitative RT-PCR was designed in the nsp11 region of the replicase 1B domain of the SARS-coronavirus (SARS-CoV) genome. To evaluate this quantitative RT-PCR, cRNA standards were constructed by in vitro transcription of SARS-CoV Frankfurt 1 RNA using T7 RNA polymerase, followed by real-time RT-PCR. The assay allowed quantitation over a range of 102 to 108 RNA copies per reaction.
Conclusions
Extrapolated to clinical samples, this novel assay has a detection range of 104 to 1010 copies of viral genome equivalents per millilitre. In comparison to the current de facto cRNA Artus Biotech standard, the in-house cRNA standard gives a 100-fold higher absolute quantity, suggesting a possible underestimation of the viral load when using the Artus Biotech standard.
Keywords: TaqMan PCR, Quantitative PCR, Real-time PCR, SARS-CoV, cRNA standard, RT-PCR
Introduction
Severe acute respiratory syndrome (SARS) has recently emerged as a new severe human disease that resulted in 774 deaths and more than 8000 reported probable cases worldwide.1 The SARS epidemic began in the Guangdong province in southern China, where several cases of atypical pneumonia of unknown etiology were reported in November 2002.
A novel member of the Coronaviridae family has been identified as the causative agent of this pulmonary disease.2, 3, 4, 5, 6 The SARS-coronavirus (SARS-CoV) is a new coronavirus type, of which the morphology and genome organization are very similar to those of other coronaviruses. Phylogenetic analyses and sequence comparisons showed that SARS-CoV is not closely related to any of the previously characterized coronaviruses.6, 7
The clinical case definition of SARS is essentially one of fever and pneumonia, with or without a contact history. In the absence of a definite history of contact with other SARS patients, it is not easy to differentiate SARS from other causes of atypical pneumonia. Laboratory SARS-CoV confirmation tests are therefore a critical clinical need. Detection of specific SARS-CoV antibodies has been a reliable way to confirm the diagnosis. Due to the fact that the antibody response appears only around day ten of the illness, IgM or other subtype assays have not been useful for closing the diagnostic window within the first week of illness.
Virus culture is relatively insensitive, and takes too much time to be clinically relevant. During the epidemic, a variety of reverse transcription-polymerase chain reaction (RT-PCR) assays were developed3, 8 including a commercial ready-to-use RT-PCR kit (Artus Biotech, Hamburg, Germany). Detection of viral RNA by RT-PCR in clinical samples offers the opportunity of diagnosis in an earlier stage of the disease.9
In contrast to many other acute respiratory infections, viral loads in clinical samples are low in the first few days of illness and peak around day ten of the disease. Recently Ng and colleagues demonstrated the role of serum SARS-CoV viral load measurement as a prognostic marker, even on the first day of hospital admission. A high systemic viral load may directly lead to severe tissue damage, or indirectly through the activation of a potentially damaging immune reaction.10 For SARS-CoV diagnosis, plasma or serum RT-PCR can be performed with high sensitivity during the first week of the disease. Stool and respiratory samples can be used during the second week, and serologic testing for antibodies against SARS-CoV can be used from day 21 onward.11 A major advantage of real-time PCR is that amplification and analysis are completed in a closed system. The risk of contamination, which can confound conventional (nested) RT-PCR protocols, is markedly reduced. This paper presents a very sensitive and specific quantitative RT-PCR assay, which can contribute to a rapid and efficient diagnosis of SARS in case of a new outbreak.
Materials and methods
Viral propagation
The SARS-CoV Frankfurt 1 (FFM-1) strain was kindly provided by Professor Dr H.F. Rabenau and Professor Dr H.W. Doerr from the Johann Wolfgang Goethe University, Frankfurt, Germany. SARS-CoV was propagated in African green monkey kidney cells (Vero E6 cells), obtained from the American Type Culture Collection (ATCC C1008). Supernatant was harvested after four days incubation at 37 °C in a 5% CO2 atmosphere. Human coronaviruses OC43 and 229E were also obtained from the American Type Culture Collection (VR-759 and VR-740). Viral RNA was isolated using the QIAamp viral RNA kit (Qiagen, Westburg, Leusden, The Netherlands).
Primer and probe design
The nsp11 region of the replicase 1B domain of the SARS-CoV genome was screened for primer and probe target sites that would be compatible with TaqMan PCR requirements (ABI 7700 Users Manual), using the Primer express 2.0 software. Primers spanning a target region of 68 bp were selected, with matched dissociation temperatures and a minimal likelihood for duplex or hairpin formation: forward primer SARS-FP (5′-CACCCGCGAAG-AAGCTATTC-3′), MGB probe SARS-TP (FAM 5′-TGCGTGGATTGGCTT-3′NFQ-MGB), and reverse primer SARS-RP (5′-TTGCATGACAGCCCTCTACATC-3′).
Construction of cRNA standards
The TaqMan SARS-CoV forward primer was modified with a T7-promoter sequence at the 5′-end (SARS-FPT7 5′-TAATACGACTCACTATAGGGAGGCACCCGCGAAGAAGCTA-TTC-3′). PCR products amplified with the modified primer pairs were quantified spectrophotometrically at 260 nm. Two hundred nanograms of PCR product were used for in vitro transcription (MEGAshortscript T7 kit, Ambion, Austin, TX, USA) performed at 37 °C overnight in a 20 μl reaction mix containing 2 μl of reaction buffer, 2 μl of each dNTP and 2 μl enzyme mix. The cDNA was removed by digestion with two units of RNase-free DNase I for 15 min at 37 °C. The cRNA was precipitated by adding 3 μl 3 M NaOAc and 60 μl of 96% ethanol and a subsequent incubation at –20 °C for 30 min. After centrifugation at 13000 rpm for 15 min, the supernatant was removed and 500 μl 70% ethanol was added. After another 5 min of centrifugation at 13000 rpm, the supernatant was removed and the pellet was dissolved in 200 μl RNase free H2O (Sigma-Aldrich NV, Bornem, Belgium) and stored at –80 °C. Quantitation of cRNA was performed spectrophotometrically at 260 nm.12
Real-time quantitative RT-PCR
A 25 μl RT-PCR was carried out using 5 μl of extracted RNA or standard cRNA, 12.5 μl of a real-time One-Step RT-PCR Master Mix containing ROX as a passive reference (Applied Biosystems, Foster City, CA, USA), 900 nM forward and reverse primer, and 150 nM MGB probe. Amplification and detection were performed in a ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) under the following conditions: an initial reverse transcription at 48 °C for 30 min, followed by PCR activation at 95 °C for 10 min and 45 cycles of amplification (15 seconds at 95 °C and 1 min at 60 °C). During amplification, the ABI PRISM sequence detector monitored real-time PCR amplification by quantitative analysis of fluorescence emissions. The reporter dye (FAM) signal was measured against the internal reference dye (ROX) signal to normalize for non-PCR-related fluorescence fluctuations occurring from well to well. The threshold cycle represented the refraction cycle number at which a positive amplification was measured, and was set at ten times the standard deviation of the mean baseline emission calculated for PCR cycles 3 to 15. Analysis of the copy numbers and linear regression curve was performed using the Sequence Detector v1.9 software (Applied Biosystems, Foster City, CA, USA).
One-step RT-PCR
To compare the sensitivity of the real-time RT-PCR with a conventional RT-PCR, a cRNA sample was used (kindly provided by Professor Dr C. Drosten from the Bernard Nocht Institute for Tropical Medicine, Hamburg, Germany). A 195 bp fragment was amplified by a one-step RT-PCR using the BNI outer primers (5′-ATGAATTACCAAGTCAATGGTTAC-3′ as the sense primer and 5′-CATAACCAGTCGGTACAGCTAC-3′ as the antisense primer). The RT-PCR was carried out on a Geneamp PCR System 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA). Each reaction contained 0.6 μM of sense and antisense primers, 0.4 mM of each dNTP and 2 μl Qiagen OneStep RT-PCR enzyme mix. RT-PCR conditions were as follows: an initial reverse transcription for 30 min at 50 °C followed by a PCR activation for 15 min at 95 °C, 45 cycles of amplification (30 sec at 94 °C, 30 seconds at 50 °C and 1 min at 72 °C) and a final extension step at 72 °C for 10 min.
Results
Absolute viral RNA load quantitation
To determine the amount of genome equivalents/ml, i.e. the viral RNA load, in cell culture samples, SARS-CoV cRNA standards were used for the construction of a standard curve (Figure 1 ). Ten-fold serial dilutions of cRNA transcripts were used corresponding to copy numbers of 102 to 108. Equal volumes of standard and sample were used for PCR amplification. SARS-CoV RNA, extracted from the supernatant of infected Vero E6 cells, which contained a virus titer of 103 CCID50/ml was analysed and was found to correspond to a viral RNA load of 1.7 × 1012 RNA copies/ml.
Figure 1.
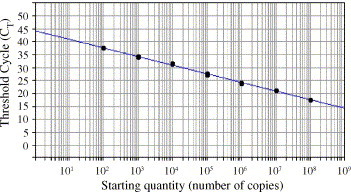
Ten-fold serial dilutions ranging from 108 to 102 copies of cRNA were tested in duplicate in real-time RT-PCR. A standard curve graph was generated by plotting the CT values on the y-axis and the log of the input amounts on the x-axis. The slope of this standard curve was −3.43 and the correlation coefficient was 0.997.
Dynamic range
To investigate the dynamic range of the assay, 10-fold serial dilutions of the cRNA standard, ranging from 10 to 1012 molecules in the reaction, were tested. The results were analysed in terms of CT value (the cycle in which a target sequence is detected). As shown in Figure 2 , the dynamic range of the assay spanned 6 logs ranging from 102 to 108 molecules SARS-CoV per reaction, corresponding to a range in CT values from 37.45 ± 0.04 to 17.40 ± 0.04.
Figure 2.
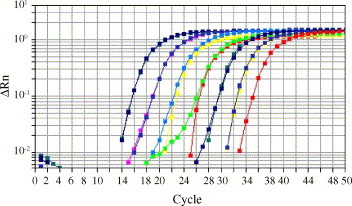
The dynamic range of the assay was tested running 10-fold serial dilutions of the authors’ cRNA standard ranging from 10 to 1012 molecules in the reaction. The amplification plot shows delta Rn on the y-axis (Rn: fluorescence emission intensity of the reporter dye normalized to a passive reference, delta Rn: Rn of an unreacted sample – Rn value of the reaction) against the cycle number displayed on the x-axis. The dynamic range of the assay spans 6 logs ranging from 102 to 108 log molecules of SARS-CoV RNA per reaction, corresponding to CT values ranging from 37.45 ± 0.04 for 102 copies to 17.40 ± 0.04 for 108 copies. cRNA standards with a copy number above or below the dynamic range are not shown.
Specificity, sensitivity and reproducibility
Experiments were undertaken to assess diagnostic criteria such as specificity, sensitivity and reproducibility. Specificity of the primers and probe was tested using other human coronaviruses OC43 and 229E. No detectable non-specific amplification of these coronaviruses took place. The sensitivity of the real-time RT-PCR was investigated by assaying ten replicates of samples containing 10, 102, 103 or 104 copies of the cRNA standard per reaction. One hundred copies of the standard cRNA were detected in all ten replicates. For ten genome equivalents per reaction, only one out of ten replicates was found to be positive. The standard deviation of the mean threshold cycle varied between 0.34 and 0.98 cycles, resulting in coefficients of variation ranging from 0.97 to 2.57%. The intra-assay reproducibility of the SARS-CoV real-time RT-PCR was assessed by running dilutions of the cRNA standards ranging from 107 to 102 copies per reaction. Each of these dilutions was assayed four times per run. The mean values of CT, the standard deviations and the coefficients of variation were determined. Table 1 shows that the SARS-CoV cRNA standards can be detected with a coefficient of variation of less than 0.7%. The interassay reproducibility of the SARS-CoV real-time RT-PCR was assessed by running dilutions of the cRNA standards ranging from 108 to 105 copies per reaction in several assay runs. The mean values of the CT, the standard deviations and the coefficients of variation were determined (Table 2 ). The results show that the SARS-CoV cRNA standard can be detected with a coefficient of variation of less than 4%.
Table 1.
Intra-assay reproducibility of the SARS-CoV quantitative RT-PCR.
| Sample | Dilution | cRNA copies | Mean CT | S.D. | CV (%) |
|---|---|---|---|---|---|
| cRNA 1 | 1 × 10−6 | 1.86 × 107 | 18.50 | 0.07 | 0.36 |
| 1 × 10−7 | 1.86 × 106 | 21.66 | 0.08 | 0.39 | |
| 1 × 10−8 | 1.86 × 105 | 25.33 | 0.12 | 0.48 | |
| 1 × 10−9 | 1.86 × 104 | 28.28 | 0.12 | 0.41 | |
| 1 × 10−10 | 1.86 × 103 | 32.61 | 0.22 | 0.66 | |
| 1 × 10−11 | 1.86 × 102 | 35.31 | 0.17 | 0.48 |
The intra-assay reproducibility was assessed by running dilutions of the cRNA standards ranging from 102 to 107 copies per reaction. Each of these dilutions was assayed four times per run. The mean values of the CT, the standard deviations (S.D.) and the coefficients of variation (CV) were determined.
Table 2.
Interassay reproducibility of the SARS-CoV quantitative RT-PCR.
| Sample | Dilution | cRNA copies | Mean CT | S.D. | CV (%) |
|---|---|---|---|---|---|
| cRNA 1 | 1 × 10−5 | 1.86 × 108 | 14.80 | 0.53 | 3.61 |
| 1 × 10−6 | 1.86 × 107 | 17.62 | 0.23 | 1.32 | |
| 1 × 10−7 | 1.86 × 106 | 21.16 | 0.18 | 0.87 | |
| 1 × 10−8 | 1.86 × 105 | 24.22 | 0.06 | 0.23 | |
| cRNA 2 | 1 × 10−5 | 1.94 × 108 | 15.39 | 0.36 | 2.34 |
| 1 × 10−6 | 1.94 × 107 | 18.89 | 0.40 | 2.10 | |
| 1 × 10−7 | 1.94 × 106 | 21.77 | 0.15 | 0.71 | |
| 1 × 10−8 | 1.94 × 105 | 25.00 | 0.09 | 0.38 | |
| cRNA 3 | 1 × 10−5 | 2.08 × 108 | 15.43 | 0.41 | 2.69 |
| 1 × 10−6 | 2.08 × 107 | 18.45 | 0.27 | 1.49 | |
| 1 × 10−7 | 2.08 × 106 | 21.57 | 0.15 | 0.68 | |
| 1 × 10−8 | 2.08 × 105 | 25.23 | 0.10 | 0.38 | |
| cRNA 4 | 1 × 10−5 | 1.85 × 108 | 16.22 | 0.19 | 1.15 |
| 1 × 10−6 | 1.85 × 107 | 19.49 | 0.09 | 0.46 | |
| 1 × 10−7 | 1.85 × 106 | 23.18 | 0.08 | 0.36 | |
| 1 × 10−8 | 1.85 × 105 | 26.95 | 0.10 | 0.39 | |
The interassay reproducibility was assessed by running dilutions of the cRNA standards ranging from 105 to 108 copies per reaction in three assay runs. The mean values of the CT, the standard deviations (S.D.) and the coefficients of variation (CV) were determined.
Comparison of the in-house cRNA standard with the cRNA standard of Artus Biotech
To validate the in-house cRNA standard, serial ten-fold dilutions of the in-house standard together with equivalent dilutions of the commercially available Artus Biotech standard were assessed in the same run. Analysing a specific CT value with the two standard curves resulted in a 100-fold difference in copy number.
Comparison of real-time RT-PCR to RT-PCR
To demonstrate the feasibility of broad usage of real-time RT-PCR for both research and clinical studies, the sensitivity of the real-time RT-PCR was compared to that of the RT-PCR using the BNI-outer primer pair. For both assays, ten-fold dilutions of a sample containing 109 copies per μl of the BNI cRNA fragment were used. Whereas the conventional RT-PCR could detect up to 104 copies of cRNA per reaction, the detection rate of this real-time RT-PCR assay was 100-fold higher, with a sensitivity up to 102 cRNA copies per reaction.
Discussion
Although the initial global outbreak of SARS has been successfully contained, SARS will remain a serious concern. The severity of the disease combined with its rapid spread requires a diagnostic test that can detect the SARS-CoV infection early in the course of illness. The SARS-CoV can be detected by conventional and real-time RT-PCR in specimens obtained from the upper and lower respiratory tracts, in plasma or serum, and in feces.9, 10, 13, 14, 15, 16, 17
A conventional RT-PCR, although performed as a one-step reaction in a thermocycler, still requires the use of PAGE analysis for detection and it takes four to five hours to complete the test. This real-time RT-PCR, on the other hand, can generate complete results within a period of three hours. The speed of this assay combined with a minimum of manipulations that need to be performed, make this real-time RT-PCR an ideal routine protocol for high-throughput screening of possible SARS-CoV-infected samples.
The assay described here has been shown to be highly sensitive, with a detection limit of 102 copies of SARS-CoV per reaction, which is 2 logs more sensitive than a conventional SARS-CoV RT-PCR. Furthermore, this assay has a sensitivity that is similar or even better than that reported for other existing real-time SARS-CoV assays. This sensitivity is achieved without the need for a nested step in the RT-PCR reaction.18
During the first week of illness, the viral load in the upper respiratory tract specimens is very low, but will reach a peak around day ten of illness. Since early diagnosis is important, highly sensitive diagnostic methods are necessary to avoid false-negative results in samples that contain only trace amounts of the virus in the early phase of the infection. This study presents a diagnostic real-time quantitative assay that is sensitive enough to detect the presence of SARS-CoV even at an early stage of the illness. The potential for quantitation over a wide dynamic range (at least 6 logs) was demonstrated with low intra- and inter-assay variation. Furthermore, this method was highly specific for the SARS-CoV and gave no false-positive results with HCoV-OC43 and HCoV-229E.
Comparison of the in-house cRNA standard with the existing cRNA standard of Artus Biotech2, 18 showed a 100-fold difference in absolute quantity. Using the in-house standard, this assay can detect 100 copies per reaction, whereas with the Artus standard, the detection limit of our assay is one RNA copy per reaction.
Drosten and co-workers were the first to develop a standard to quantify the SARS-CoV, and this standard is also used in the commercial ready-to-use RT-PCR kit (Artus Biotech). Even after repeating the cRNA construction several times, the same results were obtained. At the moment the correct absolute quantities cannot be predicted. The Artus Biotech standard is currently the de facto standard, and it is necessary to point out that there is a possible 100-fold underestimation of the copy numbers. This underestimation of the viral load can be important for in vitro and in vivo antiviral testing, where real-time RT-PCR is used for virus yield assays and time-of-drug-addition experiments.19, 20
Acknowledgements
The authors wish to thank their colleagues at the laboratory of Clinical and Epidemiological Virology, Department of Microbiology & Immunology, Rega Institute for Medical Research, University of Leuven, Belgium for their helpful comments and discussion. This work was supported by a fellowship from the Flemish Fonds voor Wetenschappelijk Onderzoek (FWO) to Leen Vijgen, and by FWO-grant G.0288.01.
Conflict of interest: No conflict of interest to declare.
Corresponding Editor: Jane Zuckerman, London, UK
References
- 1.Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., SARS study group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 7.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 8.Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., National Microbiology Laboratory, Canada, Canadian Severe Acute Respiratory Syndrome Study Team Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 9.Poon L.L., Chan K.H., Wong O.K., Yam W.C., Yuen K.Y., Guan Y. Early diagnosis of SARS-coronavirus infection by real-time RT-PCR. J Clin Virol. 2003;28:233–238. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng E.K., Hui D.S., Chan K.C., Hung E.C., Chiu R.W., Lee N. Quantitative analysis and prognostic implication of SARS-coronavirus RNA in the plasma and serum of patients with severe acute respiratory syndrome. Clin Chem. 2003;49:1976–1980. doi: 10.1373/clinchem.2003.024125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., HKU/UCH SARS Study Group. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fronhoffs S., Totzke G., Stier S., Wernert N., Rothe M., Bruning T. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol Cell Probes. 2002;16:99–110. doi: 10.1006/mcpr.2002.0405. [DOI] [PubMed] [Google Scholar]
- 13.Lau L.T., Fung Y.W., Wong F.P., Lin S.S., Wang C.R., Li H.L. A real-time PCR for SARS-coronavirus incorporating target gene pre-amplification. Biochem Biophys Res Commun. 2003;312:1290–1296. doi: 10.1016/j.bbrc.2003.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon L.L., Chan K.H., Wong O.K., Cheung T.K., Ng I., Zheng B. Detection of SARS-coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin Chem. 2004;50:67–72. doi: 10.1373/clinchem.2003.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant P.R., Garson J.A., Tedder R.S., Chan P.K., Tam J.S., Sung J.J. Detection of SARS-coronavirus in plasma by real-time RT-PCR. N Engl J Med. 2003;349:2468–2469. doi: 10.1056/NEJM200312183492522. [DOI] [PubMed] [Google Scholar]
- 16.Ng L.F., Wong M., Koh S., Ooi E.E., Tang K.F., Leong H.N. Detection of severe acute respiratory syndrome coronavirus in blood of infected patients. J Clin Microbiol. 2004;42:347–350. doi: 10.1128/JCM.42.1.347-350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhai J., Briese T., Dai E., Wang X., Pang X., Du Z. Real-time polymerase chain reaction for detecting SARS coronavirus, Beijing, 2003. Emerg Infect Dis. 2004;10:300–303. doi: 10.3201/eid1002.030799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang S.S., Chen T.C., Yang J.Y., Hsiung C.A., Su I.J., Liu Y.L. Sensitive and quantitative detection of severe acute respiratory syndrome coronavirus infection by real-time nested polymerase chain reaction. Clin Infect Dis. 2004;38:293–296. doi: 10.1086/380841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem Biophys Res Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto N., Yang R., Yoshinaka Y., Amari S., Nakano T., Cinatl J. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem Biophys Res Commun. 2004;318:719–725. doi: 10.1016/j.bbrc.2004.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]


