Highlights
-
•
The manual epidemic intelligence system was quick and accurate.
-
•
All significant alerts were identified first through unofficial sources.
-
•
The system was adaptable and allowed for monitoring of events as they evolved.
Keywords: Epidemic intelligence, Disease surveillance, Early detection, Surveillance systems, Epidemiology
Abstract
Background
Epidemic intelligence (EI) for emerging infections is the process of identifying key information on emerging infectious diseases and specific incidents. Automated web-based infectious disease surveillance technologies are available; however, human input is still needed to review, validate, and interpret these sources. In this study, entries captured by Public Health England’s (PHE) manual event-based EI system were examined to inform future intelligence gathering activities.
Methods
A descriptive analysis of unique events captured in a database between 2013 and 2017 was conducted. The top five diseases in terms of the number of entries were described in depth to determine the effectiveness of PHE’s EI surveillance system compared to other sources.
Results
Between 2013 and 2017, a total of 22 847 unique entries were added to the database. The top three initial and definitive information sources varied considerably by disease. Ebola entries dominated the database, making up 23.7% of the total, followed by Zika (11.8%), Middle East respiratory syndrome (6.7%), cholera (5.5%), and yellow fever and undiagnosed morbidity (both 3.3%). Initial reports of major outbreaks due to the top five disease agents were picked up through the manual system prior to being publicly reported by official sources.
Conclusions
PHE’s manual EI process quickly and accurately detected global public health threats at the earliest stages and allowed for monitoring of events as they evolved.
Introduction
Emerging infections, either newly recognized or diseases increasing in a specific place or population, are increasingly affecting global human morbidity and mortality (Morens and Fauci, 2013). Epidemic intelligence (EI) for emerging infections is the process of identifying key information on emerging infectious diseases, specific incidents, and outbreaks. The importance of EI is recognized globally, and it is central to the surveillance component of the International Health Regulations (2005) (IHR) to protect the global community from public health risks and emergencies that cross international borders (WHO, 2005a). EI generally integrates both indicator-based (collection of structured data through surveillance systems) and event-based (unstructured data from formal and informal sources) components in a single surveillance system (Paquet et al., 2006). It can be based solely on event-based data, which is usually more valued as it has the ability to detect the earliest reports of events and undiagnosed diseases.
Few ministries of health publicly report real-time data due to logistical, financial, and political constraints. However, a significant amount of real-time information about disease outbreaks is available in various forms on web-based platforms. Several public and restricted access web-based surveillance technologies are available (e.g., the Global Public Health Intelligence Network (GPHIN) and the Global Health Security Initiative Early Alerting and Reporting project, both of which are restricted, and HealthMap). These are automated systems that continuously retrieve and post articles related to infectious diseases (Riccardo et al., 2014, Mykhalovskiy and Weir, 2006, Brownstein and Freifeld, 2007).
Including multiple sources has been shown to improve event detection and accuracy (Yan et al., 2017). However, aggregate reports still require human input for review, validation, and interpretation, which can prove time-consuming and challenging due to duplication, ‘information overload’, and substantial background noise. Web-accessible informal information sources (e.g., social media or blogs) are also playing an increasingly important role in early event detection, particularly in low-resource countries, where official diagnoses are either delayed or not attainable (Wilson and Brownstein, 2009). The utility of local, national, and international media and social media for early intelligence is well-recognized and is relied upon for the detection of first reports about diseases (Yan et al., 2017).
More manual forms of internet-based surveillance exist, where the information is either actively searched for and aggregated by individuals (e.g., FluTrackers https://flutrackers.com/forum/) or submitted by individuals for manual curation by moderators, as in the Program for Monitoring Emerging Diseases (ProMED, https://www.promedmail.org/) (Yu and Madoff, 2004, Madoff, 2004). Both track a wide range of infectious diseases and have proven utility in the early detection of major emerging infectious disease outbreaks (Yu and Madoff, 2004, Lau et al., 2014). They are free to access, retrieve information from multiple languages, use public sources of information, and specifically for ProMED, can include first-hand reports.
Within Public Health England (PHE), the Emerging Infections and Zoonoses (EIZ) Section started conducting epidemic intelligence for new and emerging infectious threats to the UK population in 2003. In the 16 years since its implementation, significant developmental changes have been made to the way EI has been undertaken by PHE, but the purpose remains the same. This article focuses on the manual event-based component of EI activities conducted by PHE, with special emphasis on the detection and assessment of potentially significant alerts detected by the system.
Methods
EI activities are conducted by a team of three epidemiologists in the EIZ team within PHE. More than 100 web-based resources are systematically reviewed by one of the epidemiologists from 07.00 to 17.00, 5 days per week. Certain sources are checked for urgent reports out of hours and at weekends. The on-duty epidemiologist manually navigates through each of the sources, reviewing them for new information. Standard operating procedures (SOP) are followed to ensure consistency in the performance of duties and optimal efficiency (for a summary of the SOP, see https://www.gov.uk/government/publications/emerging-infections-and-zoonoses-epidemic-intelligence-scanning-procedures/epidemic-intelligence-scanning-process).
Sources include both automated and manual aggregated surveillance systems (e.g., HealthMap and ProMED); informal information sources (e.g., Twitter); Google search terms, Google alerts, and Really Simple Syndication (RSS) feeds (a type of web feed that consolidates information sources in one place and provides updates when a site adds new content). Official sources, such as the World Health Organization (WHO) and US Centers for Disease Control and Prevention (CDC), and National Official Resources (NOR), such as ministries of health or other government resources, are searched for updates on specific events being monitored. International and local news media (collectively termed ‘media’) are reviewed for updates on specific events, as well as informal reports of disease outbreaks. The number and types of sources reviewed change reactively. Peer-reviewed journals that publish articles on relevant emerging infections, such as the New England Journal of Medicine, the International Journal of Infectious Diseases, and The Lancet, are monitored using RSS feeds and Table of Contents alerts. Information is occasionally obtained from restricted resources, where the information is not yet in the public domain. When this occurs, a targeted web search is conducted (including use of Twitter and Google search terms) to see if any of the information is publicly available to allow dissemination of the information without compromising the restricted resources.
Results from the daily systematic scan are discussed with other EI colleagues as necessary and logged in a Microsoft Access database (Microsoft, 2013). If an event is reported that appears unusual or worrying (such as an outbreak of an infectious disease with high morbidity or mortality), an informal risk assessment is also conducted to determine whether the event has the potential to quickly become a public health threat to the UK and whether any immediate action is required. The public health significance of incidents is determined using similar questions to the IHR framework for assessment and notification of events that may constitute a public health emergency of international concern (WHO, 2005a). For example, a determination is made as to whether there are unexpectedly high rates of illness or death, and whether there is the potential for spread beyond national borders.
Due to the inevitable delay in confirming a diagnosis, disease outbreaks are often initially reported as undiagnosed disease events, thus particular attention is paid to these types of reports. When an initial report of an emerging situation or undiagnosed disease/outbreak is found, a more targeted scan is conducted to determine the validity and significance of the report.
Results from the daily scan are used for many purposes, including providing daily updates of ongoing and emerging incidents to senior managers and government stakeholders; advising on current outbreaks and emerging risks, improving situational awareness of decision-makers across government, facilitating risk assessments, and generating briefings on international incidents. A public facing report summarizing ongoing and emerging incidents is produced on a monthly basis (https://www.gov.uk/government/publications/emerging-infections-monthly-summaries).
In this study, a descriptive analysis of unique emerging infection events captured by PHE’s EI system between January 2013 and December 2017 was conducted, focusing on sources of data (initial and definitive) and the geographic location of events. Initial sources are defined as those that are actively monitored for information and are the source of the first report of an event that is found by the EI system (e.g., a Tweet from a journalist or a Google Alert to a local news site). Initial sources are often neither the original nor conclusive source of intelligence, and information contained may require validation before further reporting. In these cases a definitive source of the quoted information or intelligence is searched for. A definitive source is defined as either the original place the information was published (e.g., ministry of health press release or a media report of a ministry of health press conference where an official release has not been published) or the most authoritative or conclusive resource (e.g., the WHO, ministry of health website, or US CDC). Initial sources of information are recorded to determine the utility of actively monitoring certain resources, and to allow the SOP to be revised as appropriate following review. For the purposes of this study, multiple entries with the same information on the same event/topic were removed. The top five infections in terms of number of entries were analysed to determine the timeliness and effectiveness of PHE’s EI surveillance system.
Role of the funding source
The funding source had no role in the writing of the manuscript or the decision to submit for publication.
Results
Between January 2013 and December 2017, a total of 22 847 unique entries were recorded in the database. These included incidents and research related to emerging infections or issues that could affect the emergence or distribution of various infectious diseases. The number of entries varied per year, with an average of 4569.4 (range 2256–5545) per year. Approximately 17–18 entries were added every working day. There was a doubling in entries in 2014 compared to 2013, with 4995 and 2256 entries, respectively, following the onset of the West Africa Ebola outbreak. The number of entries reached a peak in 2015, and then stabilized in 2016 and 2017.
A total of 17 234 (75.4%) entries related to a specific country; the remainder were associated with regions or were research articles. Over the study period, 37.4% of records were related to incidents in Africa, followed by Asia (23.9%), North America (16.1%), and Europe (14.5%).
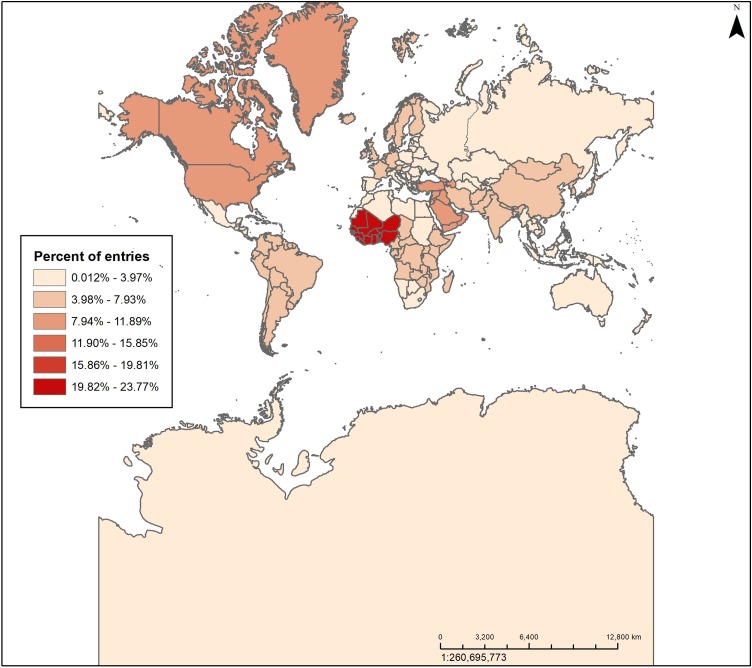
The heat map in Figure 1 shows the distribution of database entries by world region where the disease threat occurred using the United Nations geoscheme (United Nations, 2019). Most entries were related to events in West Africa (24%), reflecting the increase in surveillance activities in response to the West Africa Ebola outbreak (2014–2016). Eleven percent of the entries were related to Northern America; a result of updates and travel guidance issued by the US CDC and other official sources, as well as reports of locally acquired Ebola virus and Zika virus disease.
Figure 1.
Heat map of database entries by world region. The heat map created in ArcGIS shows the distribution of database entries by world region using the United Nations geoscheme.
The top 10 diseases in terms of number of entries (63.9% overall) are listed in Table 1 . Almost a quarter of all entries were related to Ebola virus. This was followed by Zika virus, Middle East respiratory syndrome coronavirus (MERS-CoV), Vibrio cholerae, and yellow fever virus and undiagnosed morbidity. The top three initial and definitive sources varied considerably by disease.
Table 1.
Top 10 diseases by the number of entries, 2013–2017.
| Disease | Number of entries | (% of all entries) | Top 3 initial sources and proportion (%) | Top 3 definitive sources and proportion (%) | ||
|---|---|---|---|---|---|---|
| Ebola virus | 5423 | 23.7 | Media | 15.2 | Media | 39.5 |
| WHO | 15.1 | WHO | 16.9 | |||
| Journals | 11.8 | Journals | 15.5 | |||
| Zika | 2685 | 11.8 | Journals | 27.1 | Journals | 34.1 |
| Media | 15.6 | Media | 27.9 | |||
| NOR | 12.7 | NOR | 17.7 | |||
| MERS | 1526 | 6.7 | WHO | 17.3 | NOR | 26.3 |
| ProMED | 17.0 | Media | 22.3 | |||
| NOR | 14.6 | WHO | 20.6 | |||
| Cholera | 1254 | 5.5 | WHO | 31.1 | WHO | 36.3 |
| ReliefWeb | 23.2 | Media | 27.7 | |||
| ProMED | 10.8 | Humanitarian | 18.4 | |||
| Yellow fever | 760 | 3.3 | WHO | 28.4 | WHO | 32.5 |
| ProMED | 22.9 | Media | 32.0 | |||
| NOR | 17.8 | NOR | 20.5 | |||
| Undiagnosed | 762 | 3.3 | ProMED | 36.4 | Media | 76.2 |
| FluTrackers | 19.6 | ProMED | 7.6 | |||
| Media | 15.6 | NOR | 4.9 | |||
| Avian influenza H7N9 | 660 | 2.9 | CIDRAP | 16.2 | Journals | 24.6 |
| ProMED | 15.2 | Media | 22.2 | |||
| FluTrackers | 14.7 | WHO | 16.7 | |||
| Polio | 585 | 2.6 | GPEI | 26.5 | GPEI | 29.2 |
| WHO | 15.4 | WHO | 20.9 | |||
| ProMED | 11.3 | Media | 20.5 | |||
| Chikungunya | 513 | 2.2 | ProMED | 23.6 | Media | 31.8 |
| Journals | 17.7 | Journals | 23.0 | |||
| CIDRAP | 15.6 | WHO | 16.0 | |||
| Dengue | 442 | 1.9 | Journals | 25.6 | Journals | 30.8 |
| ProMED | 18.1 | Media | 26.2 | |||
| WHO | 12.0 | WHO | 14.5 | |||
WHO, World Health Organization; NOR, National Official Resource; MERS, Middle East respiratory syndrome coronavirus; CIDRAP, Center for Infectious Disease Research and Policy; GPEI, Global Polio Eradication Initiative.
Top five specific infections
Ebola
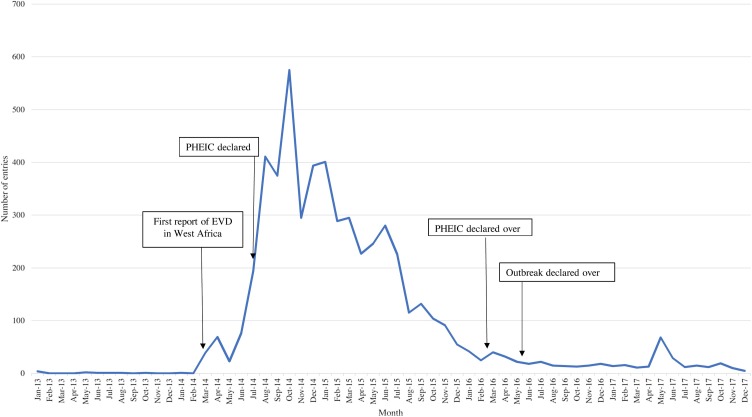
Most entries occurred during the 2014–2016 West Africa Ebola outbreak (Figure 2 ). Most journal articles were captured in 2016 and 2017, where they comprised over half of the Ebola entries in those years: 53.1% in 2016 and 55.4% in 2017. EI activities first picked up local media reports describing an undiagnosed disease in Guinea on March 15, 2014, 8 days before the outbreak was officially acknowledged by the WHO on March 23. For the duration of the outbreak, the EI system was used to detect suspected or confirmed cases of Ebola in new areas of previously affected countries, or countries that had not reported a case associated with the outbreak. During the 2014–2016 outbreak, 30.7% of initial sources came from NOR and other official resources. Media was the top definitive source (40.6%), as NOR communications were often limited or poor/slow. These media reports usually contained validated information from authorities that was not published elsewhere. Information on the latest epidemiology was closely monitored and used to inform assessments of the risk the outbreak presented to the UK population.
Figure 2.
Distribution of Ebola entries by month and year. A graphical depiction of the distribution of Ebola entries captured by PHE’s epidemic intelligence system over the study period. Important events are noted through the use of text boxes and arrows.
An increase in entries was also detected in May 2017, corresponding to an Ebola outbreak reported in Bas Uele Province, Democratic Republic of Congo (WHO, 2017). The outbreak was quickly contained and declared over on July 2, 2017.
Zika
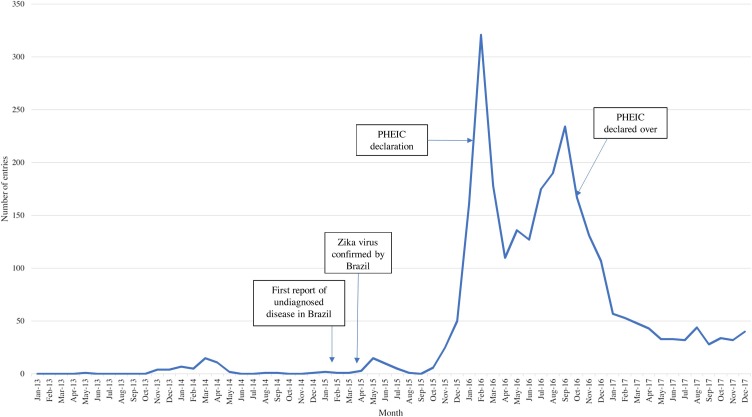
In late October 2013, PHE received notification from a restricted resource about a Zika virus outbreak in French Polynesia. A media report was found the same day, followed by a small, but steady, increase in media reports from French Polynesia and New Caledonia (Figure 3 ). Over the following months, there was an increase in reports from those countries and ensuing spread to the Cook Islands and Easter Island (Delatorre et al., 2018, Musso et al., 2018).
Figure 3.
Distribution of Zika entries by month and year. A graphical depiction of the distribution of Zika entries captured by PHE’s epidemic intelligence system over the study period. Important events are noted through the use of text boxes and arrows.
On February 10, 2015, EI activities detected local media reports of undiagnosed disease in Brazil (Figure 3). In March 2015, the Brazil Ministry of Health reported a large rash outbreak that was later officially acknowledged as being caused by Zika virus at the end of April. Outbreaks and evidence of transmission quickly appeared in other countries throughout the Americas, as well as Africa and Asia (Hennessey et al., 2016). During the outbreak (2015–2017), journals (32.7%) comprised the top definitive source due to the large volume of new research. Similar to the West African Ebola outbreak, the timeliness of official communications varied by country and was moderate at best. A significant spike in entries occurred in February 2016, corresponding to the clusters of microcephaly and other neurological disorders and their possible association with Zika virus being declared a Public Health Emergency of International Concern (PHEIC) by the WHO, (2005b). In October 2016, Zika virus was reported in Singapore for the first time, which led to an increase in media reports about cases in South East Asian countries. Information on disease epidemiology was used to inform the development of guidance and policy for the UK (https://www.gov.uk/government/collections/zika-virus-zikv-clinical-and-travel-guidance).
MERS
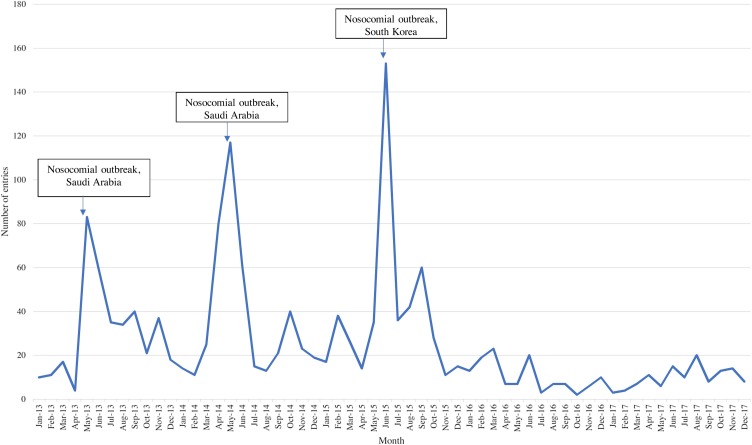
MERS-CoV was first identified as a cause of respiratory illness in September 2012 (Raj et al., 2014). The spikes seen in Figure 4 correspond to two nosocomial outbreaks in Saudi Arabia that took place between April and May 2013 and January through May 2014 (including an outbreak among camels), and a large nosocomial outbreak in South Korea in May through July 2015 (Figure 4) (Assiri et al., 2013, Oboho et al., 2015, Ki, 2015). EI activities detected local media reports about the first confirmed case in South Korea 4 days before the WHO officially acknowledged the situation.
Figure 4.
Distribution of Middle East respiratory syndrome (MERS) entries by month and year. A graphical depiction of the distribution of MERS entries captured by PHE’s epidemic intelligence system over the study period. Important events are noted through the use of text boxes and arrows.
Cholera
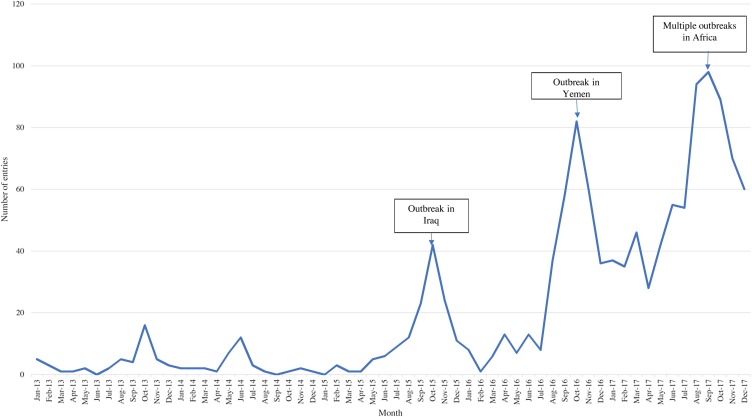
During the study period, a spike in cholera entries was reported in mid-2015, corresponding to an unusually large cholera outbreak reported in Iraq (Figure 5 ) (Mukhopadhyay et al., 2016). Media reports were detected at the beginning of August regarding an impending outbreak of cholera following rising water levels, although the outbreak was not officially declared until September 15, 2015. An overall increase in cholera entries occurred in August 2016, corresponding with the beginning of the cholera outbreak in Yemen, as well as a change in the EI protocol to monitor ongoing cholera outbreaks (Camacho et al., 2018). A spike in entries in late 2017 corresponded to multiple outbreaks being reported across Africa as well as the start of the second wave of cholera in Yemen.
Figure 5.
Distribution of cholera entries by month and year. A graphical depiction of the distribution of cholera entries captured by PHE’s epidemic intelligence system over the study period. Important events are noted through the use of text boxes and arrows.
During the study period, the cholera outbreak that began in Haiti in 2010 (and peaked with over 350 000 cases in 2011) was ongoing, although the number of cases had reduced. As the situation was ongoing and no longer emergent, reports of cholera in Haiti did not greatly contribute to a spike in entries.
Yellow fever
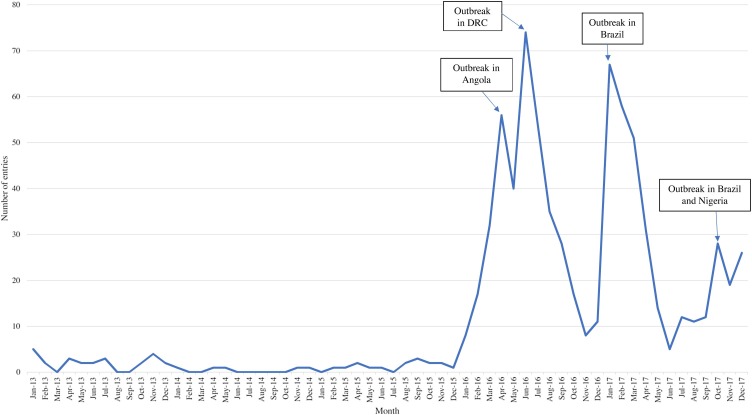
Most yellow fever entries occurred in 2016 and early 2017, corresponding to the outbreaks in Angola and the Democratic Republic of the Congo, and Brazil, respectively (Figure 6 ) (Kraemer et al., 2017, Goldani, 2017). EI activities first picked up a ProMED report about yellow fever in Angola on January 22, 2016, followed by a report from a NOR the same day. On March 11, 2016, a local media report about suspected cases of yellow fever in the Democratic Republic of the Congo was picked up through FluTrackers. The outbreak in the Democratic Republic of the Congo, associated with the outbreak in Angola, was officially acknowledged by the WHO on April 11 (WHO, 2019).
Figure 6.
Distribution of yellow fever entries by month and year. A graphical depiction of the distribution of yellow fever entries captured by PHE’s epidemic intelligence system over the study period. Important events are noted through the use of text boxes and arrows.
Starting in September 2016, EI activities registered an unseasonal increase in media reports regarding yellow fever among non-human primates in Brazil. This was followed by a media report about an increase in human cases on January 5, 2017, which was supported by a NOR report on January 9. Another spike in entries occurred in late 2017, corresponding to a large outbreak in Nigeria as well as an unseasonably early start to the yellow fever season in Brazil (WHO, 2019).
Discussion
EI is being increasingly recognized as an important component of global preparedness activities to identify and respond to new and emerging infectious events. It is important when developing such systems to be clear about the risk group, and PHE’s EI system has been developed to search for events that have the potential to impact the UK public health and UK interests. The standardized process of EI implemented by PHE promptly detected major emerging global public health events over this study period, for example the West African Ebola outbreak and the Zika virus outbreak of the Americas and Caribbean, prior to official confirmation. Other significant disease outbreaks that occurred during the study period were also quickly detected first through the media or aggregated surveillance websites. Such early detections have also been shown by other internet-based surveillance systems that monitor open source resources when compared to official reporting (Yan et al., 2017, Anema et al., 2014).
Early detection and alerting of emerging infectious events is crucial for a country’s preparedness and response activities. It also permits continuous data gathering from the earliest stages through the evolution of an event, which informs dynamic risk assessments, guidance, and policy. A major benefit of the manual EI system used by PHE is its responsiveness during any significant incidents, while remaining effective at detecting other ongoing situations. The use of open source resources and a systematic approach allows for reproducibility in other countries, including low-resource settings, and may help countries achieve IHR requirements for disease surveillance (WHO, 2005a).
PHE’s EI system has been continuously evaluated to improve event detection (e.g., through regular audits of recipients and their views as to its utility). Flexibility in scope of sources is a fundamental aspect of the system and allows for a degree of adaptation when responding to incidents, e.g. enhanced monitoring of journals (particularly those with rapid publication capabilities) for diseases that are poorly understood and for which case reports and early laboratory studies can provide essential information and evidence to inform guidance and policy. The fact that the top three sources varied considerably by disease demonstrates the importance of monitoring a variety of different sources. The increased demand for epidemic intelligence during major incidents by risk managers and other policy-makers can put a large strain on resources; however these systems are an essential component of evidence gathering to inform risk assessments, preparedness, guidance, and policy. Although EI systems need to be expansive in their scope to detect emerging incidents, the end use of the data may allow for rationalization of that scope during periods of increased demand.
Web-based social media are increasingly being used across different settings in public health, including EI (Milinovich et al., 2014). Over time, EI systems have had to become less reliant on official sources for first reports and more reliant on social media platforms. For example ministries of health and official organizations such as the WHO now use social media to distribute important event information quickly and to a wider audience, before the information is published elsewhere. In an increasingly digital world, Twitter has become an increasingly useful social media source for first information and is used frequently to support PHE’s EI system.
Despite the systematic processes used, the current system requires human input and so there might be some subjectivity in deciding which entries will go into the database. There are automated systems that use ‘webcrawling’ software to search for official reports, news, and rumours about infectious diseases, including outbreaks, around the world. They search in many languages and pull in information from specified sites. Although these ‘collection’ systems are used by large organizations such as the WHO and US CDC, what this type of automation struggles to do is turn the information gathered into usable intelligence. A well-known example of the limitations of automated ‘big data analysis’ systems to provide accurate, usable information is Google Flu Trends (Lazer et al., 2014). These automated systems generate listings of hundreds/thousands of reports every day, even when specific search terms are utilized. This volume of information will contain a high degree of ‘noise’ (Khan et al., 2012, Barboza et al., 2014). Generating usable intelligence requires additional processing and input including deduplication, translation, interpretation, and verification (Barboza et al., 2014, Fisichella et al., 2011). This requires human analysts who, in addition, can compare the latest data with historical reports, monitor trends, determine what is worth reporting, and provide additional information, expert scientific opinion, and context.
PHE’s EI system utilizes publically available sources of information for the early detection of incidents of potential public health significance. As such, this system would be useful for countries and organizations without access to restricted systems, such as GPHIN and the Global Health Security Initiative Early Alerting and Reporting project. An internal analysis of PHE’s system found that it is comparable to such systems in terms of timeliness and efficiency. PHE’s EI system detected some large-scale outbreaks prior to public acknowledgement of the outbreaks from the WHO, or at the same time, which led to improved governmental preparedness and response.
Manual EI using open source resources improves early detection and therefore response to emerging infectious disease events, including those that become public health emergencies of international concern, and should be viewed as an essential component of national surveillance systems. Open sources of information that are publicly available through the internet are important sources to quickly detect and monitor events. The encouragement of countries with limited technological infrastructure but high infectious disease burden to use such low-cost and effective forms of surveillance to monitor for global emerging infectious disease threats should be a global public health priority.
Author contributions
JW and CO’C conceived and designed the study. JW and CO’C conducted the data analyses. JW prepared drafts of the article. All authors contributed to the interpretation of the data, to editing and revising all drafts, and have approved the final submitted manuscript.
Ethical approval
Ethics approval was not required for this work.
Funding
This study was supported by Public Health England.
Conflict of interest
None.
Acknowledgements
The authors alone are responsible for the views expressed in this publication and they do not necessarily represent the decisions or policies of Public Health England. This work was supported by Public Health England. All authors are employed by Public Health England. The corresponding author had full access to all of the data in the study and had final responsibility for the decision to submit for publication.
Corresponding Editor: Eskild Petersen, Aarhus, Denmark.
References
- Anema A., Kluberg S., Wilson K., Hogg R., Khan K., Hay S. Digital surveillance for enhanced detection and response to outbreaks. Lancet Infect Dis. 2014;14(11):1035–1037. doi: 10.1016/S1473-3099(14)70953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., McGeer A., Perl T., Price C., Al Rabeeah A., Cummings D. Hospital outbreak of middle east respiratory syndrome coronavirus. N Engl J Med. 2013;369(5):407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza P., Vaillant L., Le Strat Y., Hartley D., Nelson N., Mawudeku A. Factors influencing performance of internet-based biosurveillance systems used in epidemic intelligence for early detection of infectious diseases outbreaks. PLoS ONE. 2014;9(3) doi: 10.1371/journal.pone.0090536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein J., Freifeld C. HealthMap: the development of automated real-time internet surveillance for epidemic intelligence. Euro Surveill. 2007;12 doi: 10.2807/esw.12.48.03322-en. 5. [DOI] [PubMed] [Google Scholar]
- Camacho, Bouhenia M., Alyusfi R., Alkohlani A., Naji M., de Radigues X. Cholera epidemic in Yemen, 2016–18: an analysis of surveillance data. Lancet Glob Health. 2018;6(6) doi: 10.1016/S2214-109X(18)30230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatorre E., Fernandez J., Bello G. Investigating the role of Easter Island in migration of Zika virus from South Pacific to Americas. Emerg Infect Dis. 2018;42(11):2119–2120. doi: 10.3201/eid2411.180586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisichella M., Stewart A., Cuzzocrea A., Denecke K. Detecting health events on the social web to enable epidemic intelligence. In: Grossi R., Sebastiani F., Silvestri F., editors. Lecture notes in computer science. Springer; Berlin, Heidelberg: 2011. pp. 87–103. [Google Scholar]
- Goldani L. Yellow fever outbreak in Brazil, 2017. Braz J Infect Dis. 2017;21(2) doi: 10.1016/j.bjid.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessey M., Fischer M., Staples J. Zika virus spreads to new areas — region of the Americas, May 2015 – January 2016. MMWR Morb Mortal Wkly Rep. 2016;65(3):55–58. doi: 10.15585/mmwr.mm6503e1. [DOI] [PubMed] [Google Scholar]
- Khan K., mcNabb S., Memish Z., Eckhardt R., Hu W., Kossowsky D. Infectious disease surveillance and modelling across geographic frontiers and scientific specialties. Lancet Infect Dis. 2012;12(3):222–230. doi: 10.1016/S1473-3099(11)70313-9. [DOI] [PubMed] [Google Scholar]
- Ki M. 2015 MERS outbreak in Korea: hospital-to-hospital transmission. Epidemiol Health. 2015;37:e2015033. doi: 10.4178/epih/e2015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer M., Faria N., Reiner R., Golding N., Nikolay B., Stasse S. Spread of yellow fever virus outbreak in Angola and the Democratic Republic of the Congo 2015–16: a modelling study. Lancet Infect Dis. 2017;17(3):330–338. doi: 10.1016/S1473-3099(16)30513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau E., Zheng J., Tsang T., Liao Q., Lewis B., Brownstein J. Accuracy of epidemiological inferences based on publicly available information: retrospective comparative analysis of line lists of human cases infected with influenza A(H7N9) in China. BMC Med. 2014;12:88. doi: 10.1186/1741-7015-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazer D., Kennedy R., King G., Vespignani A. The parable of Google Flu: Traps in big data analysis. Science. 2014;343(6176):1203–1205. doi: 10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- Madoff L. ProMED-mail: an early warning system for emerging diseases. Clin Infect Dis. 2004;39(2):227–232. doi: 10.1086/422003. [DOI] [PubMed] [Google Scholar]
- Milinovich G., Williams G., Clements A., Hi W. Internet-based surveillance systems for monitoring emerging infectious diseases. Lancet Infect Dis. 2014;14(2):160–168. doi: 10.1016/S1473-3099(13)70244-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D., Fauci A. Emerging infectious diseases: threats to human health and global stability. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A., Benwan K., Samanta P., Chowdhury G., Albert M.J. Vibrio cholerae O1 imported from Iraq to Kuwait, 2015. Emerg Infect Dis. 2016;22(9):1693–1694. doi: 10.3201/eid2209.160811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso D., Bossin H., Mallet H.P., Besnard M., Broult J., Baudouin L. Zika virus in French Polynesia 2013-14: anatomy of a completed outbreak. Lancet Infect Dis. 2018;18(5):PE172–182. doi: 10.1016/S1473-3099(17)30446-2. [DOI] [PubMed] [Google Scholar]
- Mykhalovskiy E., Weir L. The Global Public Health Intelligence Network and early warning outbreak detection: a Canadian contribution to global public health. Can J Public Health. 2006;97:42–44. doi: 10.1007/BF03405213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oboho I., Tomczyk S., Al-Asmari A., Banjar A., Al-Mugti H., Aloraini M. 2014 MERS-CoV outbreak in Jeddah — a link to health care facilities. N Engl J Med. 2015;372:846–854. doi: 10.1056/NEJMoa1408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet C., Coulombier D., Kaiser R., Ciotti M. Epidemic intelligence: a new framework for strengthening disease surveillance in Europe. EuroSurv. 2006;11(12):212–214. doi: 10.2807/esm.11.12.00665-en. [DOI] [PubMed] [Google Scholar]
- Raj V., Osterhaus A., Fouchier R., Haagmans B. MERS: emergence of a novel human coronavirus. Curr Op Virol. 2014;5:58–62. doi: 10.1016/j.coviro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardo F., Shigematsu M., Chow C., McKnight J., Linge J., Doherty B. Interfacing a Biosurveillance Portal and an International Network of Institutional Analysts to Detect Biological Threats. Biosecur Bioterror. 2014;12:325–336. doi: 10.1089/bsp.2014.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations . 2019. Methodology: Standard country or area codes for statistical use.https://unstats.un.org/unsd/methodology/m49/ , (Accessed 9 May 2019) [Google Scholar]
- WHO . 2005. Strengthening health security by implementing the International Health Regulations (2005)https://www.who.int/ihr/surveillance/en/ , (Accessed 6 June 2019) [Google Scholar]
- WHO . 2005. Statement on the first meeting of the International Health Regulations (2005) (IHR 2005) Emergency Committee on Zika virus and observed increase in neurological disorders and neonatal malformations.https://www.who.int/news-room/detail/01-02-2016-who-statement-on-the-first-meeting-of-the-international-health-regulations-(2005)-(ihr-2005)-emergency-committee-on-zika-virus-and-observed-increase-in-neurological-disorders-and-neonatal-malformations , (Accessed 6 June 2019) [Google Scholar]
- WHO . 2017. Ebola virus disease - Democratic Republic of Congo.https://www.who.int/csr/don/13-may-2017-ebola-drc/en/ , (Accessed 6 June 2019) [Google Scholar]
- WHO . 2019. Yellow Fever.https://www.who.int/csr/don/archive/disease/yellow_fever/en/ , (Accessed 7 June 2019) [Google Scholar]
- Wilson K., Brownstein J. Early detection of disease outbreaks using the Internet. CMAJ. 2009;180:829–831. doi: 10.1503/cmaj.090215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Chughati A., Macintyre C. Utility and potential of rapid epidemic intelligence from internet-based sources. Int J Infect Dis. 2017;63:77–87. doi: 10.1016/j.ijid.2017.07.020. [DOI] [PubMed] [Google Scholar]
- Yu V., Madoff L. ProMED-mail: an early warning system for emerging diseases. Clin Infect Dis. 2004;29(2):227–232. doi: 10.1086/422003. [DOI] [PubMed] [Google Scholar]