Abstract
Background
Antimicrobial resistance among nosocomial Gram-negative pathogens is a cause for concern in the Asia–Pacific region. The aims of this study were to measure the rates of resistance among clinical isolates collected in Asia–Pacific countries, and to determine the in vitro antimicrobial activities of ceftazidime–avibactam and comparators against these isolates.
Methods
CLSI broth microdilution methodology was used to determine antimicrobial activity and EUCAST breakpoints version 9.0 were used to determine rates of susceptibility and resistance. Isolates were also screened for the genes encoding extended-spectrum β-lactamases (ESBLs) or carbapenemases (including metallo-β-lactamases [MBLs]).
Results
Between 2015 and 2017, this study collected a total of 7051 Enterobacterales isolates and 2032 Pseudomonas aeruginosa isolates from hospitalized patients in Australia, Japan, South Korea, Malaysia, the Philippines, Taiwan, and Thailand. In the Asia–Pacific region, Enterobacterales isolates that were ESBL-positive, carbapenemase-negative (17.9%) were more frequently identified than isolates that were carbapenemase-positive, MBL-negative (0.7%) or carbapenemase-positive, MBL-positive (1.7%). Multidrug-resistant (MDR) isolates of P. aeruginosa were more commonly identified (23.4%) than isolates that were ESBL-positive, carbapenemase-negative (0.4%), or carbapenemase-positive, MBL-negative (0.3%), or carbapenemase-positive, MBL-positive (3.7%). More than 90% of all Enterobacterales isolates, including the ESBL-positive, carbapenemase-negative subset and the carbapenemase-positive, MBL-negative subset, were susceptible to amikacin and ceftazidime–avibactam. Among the carbapenemase-positive, MBL-positive subset of Enterobacterales, susceptibility to the majority of agents was reduced, with the exception of colistin (93.4%). Tigecycline was active against all resistant subsets of the Enterobacterales (MIC90, 1–4 mg/L) and among Escherichia coli isolates, > 90% from each resistant subset were susceptible to tigecycline. More than 99% of all P. aeruginosa isolates, including MDR isolates and the carbapenemase-positive, MBL-positive subset, were susceptible to colistin.
Conclusions
In this study, amikacin, ceftazidime–avibactam, colistin and tigecycline appear to be potential treatment options for infections caused by Gram-negative pathogens in the Asia–Pacific region.
Keywords: Ceftazidime–avibactam, Gram-negative, Antimicrobial susceptibility, Antimicrobial resistance, Antimicrobial surveillance, Asia–Pacific region
Background
Antimicrobial resistance is recognized to be a healthcare issue in the Asian continent [1], where there are high levels of resistant Gram-negative organisms, such as extended-spectrum β-lactamase (ESBL)-positive Enterobacterales and multidrug-resistant (MDR) Pseudomonas aeruginosa [2]. Of particular concern is the presence of metallo-β-lactamase (MBL)-positive Enterobacterales and newly-emerging resistance to the polymyxins (colistin or polymyxin B) [3–5]. Recent publications have shown that further afield in the Asia–Pacific region, Australia has relatively low levels of resistant Gram-negative organisms, although carbapenem-resistant Enterobacterales and P. aeruginosa are emerging as health risks to the Australian public [6, 7].
Ceftazidime–avibactam, a combination of a third-generation cephalosporin and a non-β-lactam β-lactamase inhibitor, has a spectrum of activity that covers MDR Enterobacterales and P. aeruginosa and is able to inhibit ESBLs and Klebsiella pneumoniae carbapenemases (KPCs) [8]. In Australia, Thailand and Taiwan, as well as in the United States and in Europe, ceftazidime–avibactam has been approved for the treatment of complicated intra-abdominal infection (in combination with metronidazole), complicated urinary tract infection (including pyelonephritis), and hospital-acquired and ventilator-associated bacterial pneumonia [9, 10]. In Europe, ceftazidime–avibactam is also indicated for the treatment of infections due to aerobic Gram-negative organisms in adult patients with limited treatment options [10].
The aims of this study were to determine the rates of resistance among clinical isolates of Enterobacterales and P. aeruginosa collected in Asia–Pacific countries between 2015 and 2017, and to show the in vitro antimicrobial activities of ceftazidime–avibactam and comparators against these isolates. The countries included are Australia, Japan, South Korea, Malaysia, the Philippines, Taiwan, and Thailand. Isolates collected in 2015 are also included in an INFORM publication on the Asia–Pacific region between 2012 and 2015 [11], and isolates collected in 2015 and 2016 are also included in INFORM global publications [12, 13].
Method
Each participating study center was required to collect a predefined number of Enterobacterales and P. aeruginosa isolates every year from adult and pediatric patients with specific bacterial infections. Only patient-derived, non-duplicate isolates suspected as the cause of each infection were included in the study. Isolates from external sources, such as drainage bottles or environmental samples, were excluded from the study. Demographic information was recorded for each isolate, including source of isolate and patient information, for example, age, sex and referring ward.
Isolates were identified at the local center of collection and shipped to a central laboratory (International Health Management Associates [IHMA] Inc., Schaumburg, IL, USA) for further testing. The central laboratory confirmed species identity and determined antimicrobial minimum inhibitory concentrations (MICs) using the Clinical Laboratory Standards Institute (CLSI) broth microdilution method [14]. MICs were interpreted using version 9.0 of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoint tables [15]. Enterobacterales and P. aeruginosa isolates were screened for the presence of the genes encoding ESBLs or carbapenemases (including MBLs), if they met the MIC criteria described below.
In 2015, Escherichia coli, K. pneumoniae, Klebsiella oxytoca, and Proteus mirabilis isolates with an MIC to ceftazidime or aztreonam of ≥ 2 mg/L were evaluated for ESBL activity using the CLSI phenotypic clavulanic acid combination test [14]. Isolates confirmed as having an ESBL-positive phenotype, or those that were found to be phenotypically ESBL-negative but with an MIC to ceftazidime of ≥ 16 mg/L, were screened for the genes encoding clinically-relevant β-lactamases (ESBLs: SHV, TEM, CTX-M, VEB, PER and GES; plasmid-mediated AmpC β-lactamases: ACC, ACT, CMY, DHA, FOX, MIR and MOX; serine carbapenemases: GES, KPC and OXA-48-like; and MBLs: NDM, IMP, VIM, SPM and GIM) using multiplex PCR assays, as previously described [16]. In 2016 and 2017, E. coli, K. pneumoniae, K. oxytoca, and P. mirabilis isolates with an MIC to meropenem, ceftazidime or aztreonam of ≥ 2 mg/L were screened for the presence of the above β-lactamase genes as previously described [16]. In addition, all other Enterobacterales isolates with MICs ≥ 2 mg/L to meropenem were also screened for β-lactamase genes using published multiplex PCR assays, as previously described [16]. All P. aeruginosa isolates with a meropenem MIC of ≥ 4 mg/L were screened for clinically-relevant β-lactamase genes (ESBLs: SHV, TEM, VEB, PER and GES; serine carbapenemases: GES, KPC and OXA-24; and MBLs: NDM, IMP, VIM, SPM and GIM) using multiplex PCR assays, as previously described [17].
Original-spectrum β-lactamases (TEM-type β-lactamases without substitutions at amino acid positions 104, 164 or 238, or SHV-type β-lactamases without substitutions at amino acid positions 146, 238 or 240) were not included. All detected genes were amplified using flanking primers and sequenced. Sequences were compared with publically available databases.
An MDR phenotype among isolates of P. aeruginosa was defined as resistance to one or more antimicrobial agents (given in parentheses) from three or more of the following anti-pseudomonal antimicrobial classes: aminoglycosides (amikacin), carbapenems (imipenem or meropenem), cephalosporins (ceftazidime or cefepime), fluoroquinolones (levofloxacin), β-lactam/β-lactamase inhibitor combinations (piperacillin–tazobactam), monobactams (aztreonam), and polymyxins (colistin) [18].
Results
Study centers and infection sources of isolates
Twenty-nine centers in the Asia–Pacific region collected Gram-negative isolates between 2015 and 2017: Australia, 7; Japan, 5; South Korea, 3; Malaysia, 2; the Philippines, 5; Taiwan, 4; and Thailand, 3. A total of 7051 Enterobacterales isolates and 2032 P. aeruginosa isolates were collected. Enterobacterales isolates were collected from the following sources of infection: urinary tract, 27.5% (n = 1941); lower respiratory tract, 27.1% (n = 1913); skin and skin structure, 21.1% (n = 1485); intra-abdominal, 14.5% (n = 1020) and blood, 9.8% (n = 692). P. aeruginosa isolate infection sources were: lower respiratory tract, 50.8% (n = 1032); skin and skin structure, 23.9% (n = 486); urinary tract, 14.0% (n = 285); intra-abdominal, 7.0% (n = 143); blood, 4.1% (n = 84); and other (skeletal), 0.1% (n = 2).
Resistance in the Asia–Pacific region and by country among all Enterobacterales
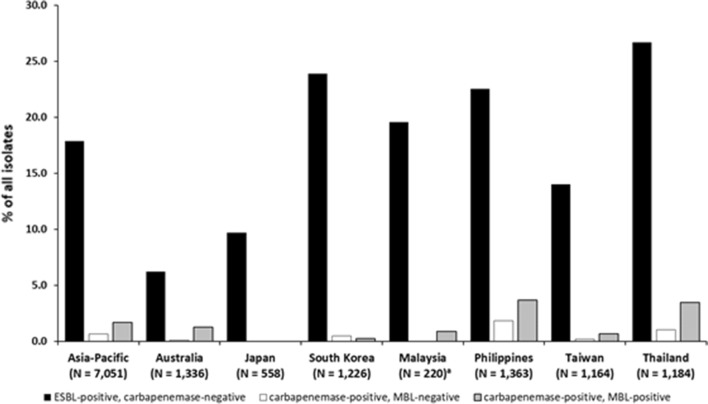
Figure 1 shows the rates of resistant subsets among Enterobacterales isolates from the Asia–Pacific region and by country. In the Asia–Pacific region as a whole, 17.9% of the Enterobacterales were ESBL-positive and carbapenemase-negative, 0.7% were carbapenemase-positive and MBL-negative, and 1.7% were carbapenemase-positive and MBL-positive. The country rates of ESBL-positive, carbapenemase-negative Enterobacterales were highest in Thailand (26.7%), South Korea (23.9%) and the Philippines (22.5%), and were lowest in Australia (6.2%) and Japan (9.7%). In each country, the rate of carbapenemase-positive, MBL-negative Enterobacterales was < 2% and the rate of carbapenemase-positive, MBL-positive isolates was < 4%. No carbapenemase-positive isolates (MBL-negative or MBL-positive) were identified among Enterobacterales from Japan, and no Enterobacterales isolates from Malaysia were carbapenemase-positive, MBL-negative.
Fig. 1.
Rates of resistant subsets (ESBL-positive, carbapenemase-negative; carbapenemase-positive, MBL-negative; and carbapenemase-positive, MBL-positive) among Enterobacterales isolates collected in the Asia–Pacific region (INFORM program, 2015–2017). ESBL, extended-spectrum β-lactamase; metallo-β-lactamase. Enterobacterales isolates (N = 7051) comprised: Citrobacter amalonaticus, 9; Citrobacter braakii, 22; Citrobacter farmeri, 8; Citrobacter freundii, 311; Citrobacter koseri, 231; Citrobacter murliniae, 1; Citrobacter sedlakii, 2; Enterobacter asburiae, 63; Enterobacter cloacae, 439; Enterobacter kobei, 22; Enterobacter ludwigii, 6; Enterobacter, non-speciated, 1; Escherichia coli, 2218; Klebsiella aerogenes, 287; Klebsiella oxytoca, 308; Klebsiella pneumoniae, 2082; Klebsiella variicola, 47; Morganella morganii, 164; Pluralibacter gergoviae, 6; Proteus hauseri, 56; Proteus mirabilis, 357; Proteus penneri, 7; Proteus vulgaris, 151; Providencia rettgeri, 61; Providencia stuartii, 65; Raoultella ornithinolytica, 5; Raoultella planticola, 1; Serratia liquefaciens, 2; Serratia marcescens, 118 and Serratia, non-speciated, 1. aCenters in Malaysia collected isolates in 2015 only
Resistance in the Asia–Pacific region and by country among E. coli and K. pneumoniae
For all E. coli and K. pneumoniae isolates collected in the Asia–Pacific region, the rate of ESBL-positive, carbapenemase-negative E. coli was greater than for K. pneumoniae (30.1% and 25.6%, respectively; Table 1). Rates of carbapenemase-positive, MBL-negative and carbapenemase-positive, MBL-positive K. pneumoniae in the Asia–Pacific region (1.9% and 2.5%, respectively) were greater than for E. coli (0.2% and 0.6%, respectively). By country, the highest rate of ESBL-positive, carbapenemase-negative E. coli was collected in Thailand (51.8%) and the highest rates of ESBL-positive, carbapenemase-negative K. pneumoniae were collected in South Korea, Malaysia, the Philippines and Thailand (33.5–38.8%). The rates of ESBL-positive, carbapenemase-negative E. coli and K. pneumoniae were both lowest in Australia (12.2% and 7.6%, respectively). Country rates of carbapenemase-positive, MBL-negative and carbapenemase-positive, MBL-positive E. coli were ≤ 0.3%, with the exception of the Philippines (carbapenemase-positive, MBL-positive E. coli, 0.9%) and Thailand (0.8% and 2.2%, respectively). The rates of carbapenemase-positive, MBL-negative and carbapenemase-positive, MBL-positive K. pneumoniae were highest in the Philippines (5.2% and 4.3%, respectively) and Thailand (2.5% and 7.3%, respectively).
Table 1.
Rates of resistant subsets among Escherichia coli and Klebsiella pneumoniae isolates collected in the Asia–Pacific region (INFORM program, 2015–2017)
| Organism/region/country | Total isolates | ESBL-positive, carbapenemase-negative | Carbapenemase-positive, MBL-negative | Carbapenemase-positive, MBL-positive | |||
|---|---|---|---|---|---|---|---|
| N | N | % | N | % | N | % | |
| Escherichia coli | |||||||
| Asia–Pacific | 2218 | 668 | 30.1 | 4 | 0.2 | 14 | 0.6 |
| Australia | 402 | 49 | 12.2 | 0 | 0.0 | 1 | 0.2 |
| Japan | 167 | 33 | 19.8 | 0 | 0.0 | 0 | 0.0 |
| South Korea | 449 | 160 | 35.6 | 0 | 0.0 | 1 | 0.2 |
| Malaysiaa | 72 | 12 | 16.7 | 0 | 0.0 | 0 | 0.0 |
| Philippines | 431 | 142 | 32.9 | 0 | 0.0 | 4 | 0.9 |
| Taiwan | 340 | 87 | 25.6 | 1 | 0.3 | 0 | 0.0 |
| Thailand | 357 | 185 | 51.8 | 3 | 0.8 | 8 | 2.2 |
| Klebsiella pneumoniae | |||||||
| Asia–Pacific | 2082 | 532 | 25.6 | 39 | 1.9 | 52 | 2.5 |
| Australia | 381 | 29 | 7.6 | 1 | 0.3 | 6 | 1.6 |
| Japan | 155 | 18 | 11.6 | 0 | 0.0 | 0 | 0.0 |
| South Korea | 335 | 123 | 36.7 | 5 | 1.5 | 0 | 0.0 |
| Malaysiaa | 80 | 31 | 38.8 | 0 | 0.0 | 1 | 1.3 |
| Philippines | 439 | 147 | 33.5 | 23 | 5.2 | 19 | 4.3 |
| Taiwan | 334 | 58 | 17.4 | 1 | 0.3 | 0 | 0.0 |
| Thailand | 358 | 126 | 35.2 | 9 | 2.5 | 26 | 7.3 |
aCenters in Malaysia collected isolates in 2015 only
Resistance in the Asia–Pacific region and by country among P. aeruginosa
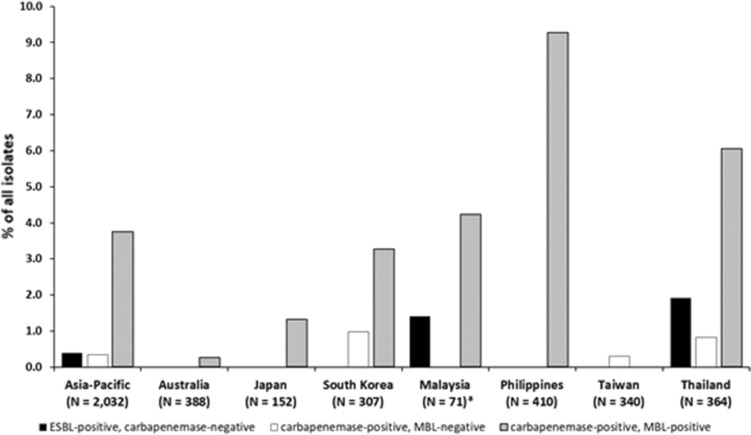
Figure 2 shows the rates of resistant subsets among P. aeruginosa isolates from the Asia–Pacific region and by country. Among all P. aeruginosa isolates collected in the Asia–Pacific region, the rate of carbapenemase-positive, MBL-positive isolates (3.7%) was greater than for ESBL-positive, carbapenemase-negative or carbapenemase-positive, MBL-negative isolates (0.4% and 0.3%, respectively). No ESBL-positive, carbapenemase-negative P. aeruginosa isolates were collected in Australia, Japan, South Korea, the Philippines, or Taiwan and no carbapenemase-positive, MBL-negative P. aeruginosa isolates were collected in Australia, Japan, Malaysia, or the Philippines. The rates of carbapenemase-positive, MBL-positive P. aeruginosa were highest in the Philippines (9.3%) and lowest in Taiwan and Australia (0.0% and 0.3%, respectively). The rate of MDR phenotypes among all P. aeruginosa isolates in the Asia–Pacific region was 23.4% and the country rates were: Australia, 14.9%; Malaysia, 18.3%; Taiwan, 19.1%; Japan, 21.1%; the Philippines, 24.1%; South Korea, 30.9%; and Thailand, 31.0%.
Fig. 2.
Rates of resistant subsets (ESBL-positive, carbapenemase-negative; carbapenemase-positive, MBL-negative; and carbapenemase-positive, MBL-positive) among Pseudomonas aeruginosa isolates collected in the Asia–Pacific region (INFORM program, 2015–2017). ESBL, extended-spectrum β-lactamase; MBL, metallo-β-lactamase. aCenters in Malaysia collected isolates in 2015 only
In vitro antimicrobial activity of 12 agents in the Asia–Pacific region against all Enterobacterales
The in vitro antimicrobial activities of a panel of agents against the pooled collection of Enterobacterales are presented in Table 2. Among all Enterobacterales isolates, the highest susceptibility rates of 96.8% to 98.1% were observed for amikacin, ceftazidime–avibactam, and meropenem. The activity of ceftazidime alone (MIC90, 64 mg/L) was lower than that of ceftazidime–avibactam (MIC90, 0.5 mg/L). Susceptibility to colistin was 83.9% and a colistin MIC90 value of ≥ 16 mg/L was observed against the Enterobacterales overall; however, colistin was active against E. coli and K. pneumoniae isolates (MIC90, 0.5 and 1 mg/L, respectively). Tigecycline was active against the collection of Enterobacterales (MIC90, 1 mg/L).
Table 2.
In vitro antimicrobial activity of ceftazidime–avibactam and comparators against Enterobacterales, Escherichia coli, and Klebsiella pneumoniae isolates collected in the Asia–Pacific region (INFORM program, 2015–2017)
| Organism/antimicrobial | MIC50 (mg/L) | MIC90 (mg/L) | MIC range (mg/L) | % S | % R |
|---|---|---|---|---|---|
| Enterobacterales (N = 7051)a | |||||
| Amikacin | 2 | 4 | ≤ 0.25–≥ 64 | 96.8 | 1.6 |
| Amoxicillin–clavulanic acid | 8 | ≥ 64 | ≤ 0.12–≥ 64 | 51.3 | 48.7 |
| Aztreonam | 0.12 | 64 | ≤ 0.015–≥ 256 | 69.4 | 26.6 |
| Cefepime | ≤ 0.12 | ≥ 32 | ≤ 0.12–≥ 32 | 75.5 | 20.3 |
| Ceftazidime | 0.25 | 64 | ≤ 0.015–≥ 256 | 69.0 | 26.7 |
| Ceftazidime–avibactam | 0.12 | 0.5 | ≤ 0.015–≥ 256 | 98.1 | 1.9 |
| Colistin | 0.25 | ≥ 16 | ≤ 0.06–≥ 16 | 83.9 | 16.1 |
| Imipenem | 0.25 | 2 | ≤ 0.03–≥ 16 | 85.3 | 2.3 |
| Levofloxacin | 0.12 | ≥ 16 | ≤ 0.004–≥ 16 | 66.2 | 27.8 |
| Meropenem | 0.03 | 0.12 | ≤ 0.004–≥ 16 | 97.7 | 1.6 |
| Piperacillin–tazobactam | 2 | 64 | ≤ 0.25–≥ 256 | 82.5 | 13.4 |
| Tigecyclineb | 0.5 | 1 | ≤ 0.015–≥ 16 | N/A | N/A |
| Enterobacterales, ESBL-positive, carbapenemase-negative (N = 1259) | |||||
| Amikacin | 2 | 8 | ≤ 0.25–≥ 64 | 91.0 | 3.7 |
| Amoxicillin–clavulanic acid | 16 | 32 | 1–≥ 64 | 26.7 | 73.3 |
| Aztreonam | 64 | ≥ 256 | 0.5–≥ 256 | 0.5 | 91.2 |
| Cefepime | ≥ 32 | ≥ 32 | ≤ 0.12–≥ 32 | 3.2 | 85.5 |
| Ceftazidime | 32 | ≥ 256 | 0.25–≥ 256 | 5.9 | 79.7 |
| Ceftazidime–avibactam | 0.25 | 0.5 | ≤ 0.015–≥ 256 | 99.9 | 0.1 |
| Colistin | 0.25 | 1 | 0.12–≥ 16 | 96.6 | 3.4 |
| Imipenem | 0.25 | 0.5 | 0.06–≥ 16 | 98.1 | 0.6 |
| Levofloxacin | ≥ 16 | ≥ 16 | 0.03–≥ 16 | 18.0 | 71.7 |
| Meropenem | 0.03 | 0.12 | 0.008–8 | 98.6 | 0.0 |
| Piperacillin–tazobactam | 8 | ≥ 256 | ≤ 0.25–≥ 256 | 62.0 | 26.6 |
| Tigecyclineb | 0.25 | 1 | ≤ 0.015–≥ 16 | N/A | N/A |
| Enterobacterales, carbapenemase-positive, MBL-negative (N = 46) | |||||
| Amikacin | 2 | 8 | ≤ 0.25–≥ 64 | 93.5 | 2.2 |
| Amoxicillin–clavulanic acid | ≥ 64 | ≥ 64 | ≥ 64–≥ 64 | 0.0 | 100 |
| Aztreonam | ≥ 256 | ≥ 256 | 8–≥ 256 | 0.0 | 100 |
| Cefepime | ≥ 32 | ≥ 32 | ≤ 0.12–≥ 32 | 4.3 | 91.3 |
| Ceftazidime | 128 | ≥ 256 | 8–≥ 256 | 0.0 | 100 |
| Ceftazidime–avibactam | 1 | 2 | 0.12–4 | 100 | 0.0 |
| Colistin | 0.25 | ≥ 16 | 0.12–≥ 16 | 87.0 | 13.0 |
| Imipenem | 8 | ≥ 16 | 0.5–≥ 16 | 23.9 | 67.4 |
| Levofloxacin | ≥ 16 | ≥ 16 | 0.06–≥ 16 | 10.9 | 82.6 |
| Meropenem | ≥ 16 | ≥ 16 | 0.5–≥ 16 | 26.1 | 58.7 |
| Piperacillin–tazobactam | ≥ 256 | ≥ 256 | 128–≥ 256 | 0.0 | 100 |
| Tigecyclineb | 0.5 | 1 | 0.12–≥ 16 | N/A | N/A |
| Enterobacterales, carbapenemase-positive, MBL-positive (N = 121) | |||||
| Amikacin | 4 | ≥ 64 | 0.5–≥ 64 | 75.2 | 16.5 |
| Amoxicillin–clavulanic acid | ≥ 64 | ≥ 64 | 16–≥ 64 | 0.0 | 100 |
| Aztreonam | 128 | ≥ 256 | ≤ 0.015–≥ 256 | 12.4 | 81.8 |
| Cefepime | ≥ 32 | ≥ 32 | 2–≥ 32 | 0.0 | 94.2 |
| Ceftazidime | ≥ 256 | ≥ 256 | 32–≥ 256 | 0.0 | 100 |
| Ceftazidime–avibactam | ≥ 256 | ≥ 256 | 32–≥ 256 | 0.0 | 100 |
| Colistin | 0.5 | 1 | ≤ 0.06–≥ 16 | 93.4 | 6.6 |
| Imipenem | ≥ 16 | ≥ 16 | 1–≥ 16 | 13.2 | 77.7 |
| Levofloxacin | ≥ 16 | ≥ 16 | 0.06–≥ 16 | 11.6 | 81.8 |
| Meropenem | ≥ 16 | ≥ 16 | 0.5–≥ 16 | 9.9 | 71.1 |
| Piperacillin–tazobactam | ≥ 256 | ≥ 256 | 1–≥ 256 | 7.4 | 90.9 |
| Tigecyclineb | 0.5 | 4 | 0.12–8 | N/A | N/A |
| E. coli (N = 2218) | |||||
| Amikacin | 2 | 8 | ≤ 0.25–≥ 64 | 97.1 | 0.6 |
| Amoxicillin–clavulanic acid | 8 | 32 | ≤ 0.12–≥ 64 | 55.7 | 44.3 |
| Aztreonam | 0.12 | 64 | ≤ 0.015–≥ 256 | 62.4 | 32 |
| Cefepime | ≤ 0.12 | ≥ 32 | ≤ 0.12–≥ 32 | 67.6 | 27.6 |
| Ceftazidime | 0.25 | 32 | ≤ 0.015–≥ 256 | 64.0 | 28.4 |
| Ceftazidime–avibactam | 0.12 | 0.25 | ≤ 0.015–≥ 256 | 99.4 | 0.6 |
| Colistin | 0.25 | 0.5 | ≤ 0.06–≥ 16 | 99.3 | 0.7 |
| Imipenem | 0.12 | 0.25 | ≤ 0.03–≥ 16 | 99.2 | 0.7 |
| Levofloxacin | 0.5 | ≥ 16 | ≤ 0.004–≥ 16 | 50.6 | 45.4 |
| Meropenem | 0.03 | 0.06 | 0.008–≥ 16 | 99.1 | 0.5 |
| Piperacillin–tazobactam | 2 | 16 | ≤ 0.25–≥ 256 | 89.0 | 7.3 |
| Tigecyclineb | 0.25 | 0.5 | 0.03–≥ 16 | 97.7 | 2.3 |
| E. coli, ESBL-positive, carbapenemase-negative (N = 668) | |||||
| Amikacin | 4 | 8 | 0.5–≥ 64 | 92.8 | 1.2 |
| Amoxicillin–clavulanic acid | 16 | 32 | 1–≥ 64 | 36.4 | 63.6 |
| Aztreonam | 32 | 128 | 0.5–≥ 256 | 0.1 | 88.9 |
| Cefepime | ≥ 32 | ≥ 32 | ≤ 0.12–≥ 32 | 1.6 | 86.8 |
| Ceftazidime | 16 | 64 | 0.25–≥ 256 | 8.4 | 70.1 |
| Ceftazidime–avibactam | 0.12 | 0.25 | ≤ 0.015–4 | 100 | 0.0 |
| Colistin | 0.25 | 0.5 | 0.12–8 | 98.4 | 1.6 |
| Imipenem | 0.12 | 0.25 | 0.06–≥ 16 | 99.7 | 0.3 |
| Levofloxacin | ≥ 16 | ≥ 16 | 0.03–≥ 16 | 14.2 | 81.9 |
| Meropenem | 0.03 | 0.06 | 0.008–8 | 99.6 | 0.0 |
| Piperacillin–tazobactam | 4 | 32 | ≤ 0.25–≥ 256 | 82.2 | 10.0 |
| Tigecyclineb | 0.25 | 0.5 | 0.03–≥ 16 | 96.9 | 3.1 |
| E. coli, carbapenemase-positive, MBL-positive (N = 14) | |||||
| Amikacin | 4 | ≥ 64 | 2–≥ 64 | 71.4 | 21.4 |
| Amoxy/clav | ≥ 64 | ≥ 64 | 32–≥ 64 | 0.0 | 100 |
| Aztreonam | ≥ 256 | ≥ 256 | 0.12–≥ 256 | 7.1 | 92.9 |
| Cefepime | ≥ 32 | ≥ 32 | ≥ 32–≥ 32 | 0.0 | 100 |
| Ceftazidime | ≥ 256 | ≥ 256 | 128–≥ 256 | 0.0 | 100 |
| Ceftazidime–avibactam | ≥ 256 | ≥ 256 | 64–≥ 256 | 0.0 | 100 |
| Colistin | 0.25 | 1 | 0.12–1 | 100 | 0.0 |
| Imipenem | ≥ 16 | ≥ 16 | 2–≥ 16 | 7.1 | 92.9 |
| Levofloxacin | ≥ 16 | ≥ 16 | 1–≥ 16 | 0.0 | 92.9 |
| Meropenem | ≥ 16 | ≥ 16 | 4–≥ 16 | 0.0 | 78.6 |
| Pip/taz | ≥ 256 | ≥ 256 | 64–≥ 256 | 0.0 | 100 |
| Tigecyclineb | 0.25 | 0.5 | 0.12–2 | 92.9 | 7.1 |
| K. pneumoniae (N = 2082) | |||||
| Amikacin | 1 | 4 | ≤ 0.25–≥ 64 | 95.6 | 2.5 |
| Amoxicillin–clavulanic acid | 4 | ≥ 64 | ≤ 0.12–≥ 64 | 64.0 | 36.0 |
| Aztreonam | 0.12 | ≥ 256 | ≤ 0.015–≥ 256 | 66.1 | 31.3 |
| Cefepime | ≤ 0.12 | ≥ 32 | ≤ 0.12–≥ 32 | 69.7 | 26.7 |
| Ceftazidime | 0.25 | ≥ 256 | ≤ 0.015–≥ 256 | 65.0 | 32.4 |
| Ceftazidime–avibactam | 0.12 | 0.5 | ≤ 0.015–≥ 256 | 97.5 | 2.5 |
| Colistin | 0.25 | 1 | ≤ 0.06–≥ 16 | 98.1 | 1.9 |
| Imipenem | 0.25 | 1 | ≤ 0.03–≥ 16 | 95.5 | 3.8 |
| Levofloxacin | 0.12 | ≥ 16 | 0.008–≥ 16 | 65.4 | 25.9 |
| Meropenem | 0.06 | 0.12 | 0.008–≥ 16 | 95.4 | 3.3 |
| Piperacillin–tazobactam | 4 | ≥ 256 | ≤ 0.25–≥ 256 | 73.0 | 21.0 |
| Tigecyclineb | 0.5 | 1 | ≤ 0.015–≥ 16 | N/A | N/A |
| K. pneumoniae, ESBL-positive, carbapenemase-negative (N = 532) | |||||
| Amikacin | 2 | 16 | ≤ 0.25–≥ 64 | 89.1 | 6.2 |
| Amoxicillin–clavulanic acid | 16 | ≥ 64 | 2–≥ 64 | 11.8 | 88.2 |
| Aztreonam | 128 | ≥ 256 | 0.5–≥ 256 | 0.6 | 95.1 |
| Cefepime | ≥ 32 | ≥ 32 | ≤ 0.12–≥ 32 | 3.4 | 86.1 |
| Ceftazidime | 64 | ≥ 256 | 0.25–≥ 256 | 2.4 | 91.9 |
| Ceftazidime–avibactam | 0.25 | 1 | ≤ 0.015–≥ 256 | 99.8 | 0.2 |
| Colistin | 0.25 | 1 | 0.12–≥ 16 | 96.8 | 3.2 |
| Imipenem | 0.25 | 1 | 0.06–≥ 16 | 98.7 | 0.8 |
| Levofloxacin | 8 | ≥ 16 | 0.03–≥ 16 | 20.3 | 62.8 |
| Meropenem | 0.06 | 0.12 | 0.015–8 | 97.6 | 0.0 |
| Piperacillin–tazobactam | 16 | ≥ 256 | 1–≥ 256 | 35.5 | 48.5 |
| Tigecyclineb | 0.5 | 2 | ≤ 0.015–≥ 16 | N/A | N/A |
| K. pneumoniae, carbapenemase-positive, MBL-negative (N = 39) | |||||
| Amikacin | 2 | 4 | ≤ 0.25–≥ 64 | 94.9 | 2.6 |
| Amoxicillin–clavulanic acid | ≥ 64 | ≥ 64 | ≥ 64–≥ 64 | 0.0 | 100 |
| Aztreonam | ≥ 256 | ≥ 256 | 8–≥ 256 | 0.0 | 100 |
| Cefepime | ≥ 32 | ≥ 32 | ≤ 0.12–≥ 32 | 5.1 | 92.3 |
| Ceftazidime | 128 | ≥ 256 | 32–≥ 256 | 0.0 | 100 |
| Ceftazidime–avibactam | 1 | 2 | 0.12–4 | 100 | 0.0 |
| Colistin | 0.25 | ≥ 16 | 0.25–≥ 16 | 87.2 | 12.8 |
| Imipenem | 8 | ≥ 16 | 0.5–≥ 16 | 20.5 | 69.2 |
| Levofloxacin | ≥ 16 | ≥ 16 | 0.06–≥ 16 | 5.1 | 87.2 |
| Meropenem | ≥ 16 | ≥ 16 | 0.5–≥ 16 | 23.1 | 61.5 |
| Piperacillin–tazobactam | ≥ 256 | ≥ 256 | 128–≥ 256 | 0.0 | 100 |
| Tigecyclineb | 0.5 | 2 | 0.25–≥ 16 | N/A | N/A |
| K. pneumoniae, carbapenemase-positive, MBL-positive (N = 52) | |||||
| Amikacin | 4 | ≥ 64 | 0.5–≥ 64 | 67.3 | 17.3 |
| Amoxicillin–clavulanic acid | ≥ 64 | ≥ 64 | 16–≥ 64 | 0.0 | 100 |
| Aztreonam | 128 | ≥ 256 | 0.06–≥ 256 | 3.8 | 94.2 |
| Cefepime | ≥ 32 | ≥ 32 | 4–≥ 32 | 0.0 | 94.2 |
| Ceftazidime | ≥ 256 | ≥ 256 | 128–≥ 256 | 0.0 | 100 |
| Ceftazidime– avibactam | ≥ 256 | ≥ 256 | 32–≥ 256 | 0.0 | 100 |
| Colistin | 0.5 | 1 | 0.12–≥ 16 | 94.2 | 5.8 |
| Imipenem | ≥ 16 | ≥ 16 | 1–≥ 16 | 5.8 | 88.5 |
| Levofloxacin | ≥ 16 | ≥ 16 | 0.25–≥ 16 | 3.8 | 86.5 |
| Meropenem | ≥ 16 | ≥ 16 | 2–≥ 16 | 1.9 | 84.6 |
| Piperacillin–tazobactam | ≥ 256 | ≥ 256 | 16–≥ 256 | 0.0 | 96.2 |
| Tigecyclineb | 1 | 4 | 0.12–8 | N/A | N/A |
aEnterobacterales isolates (N = 7051) comprised: Citrobacter amalonaticus, 9; Citrobacter braakii, 22; Citrobacter farmeri, 8; Citrobacter freundii, 311; Citrobacter koseri, 231; Citrobacter murliniae, 1; Citrobacter sedlakii, 2; Enterobacter asburiae, 63; Enterobacter cloacae, 439; Enterobacter kobei, 22; Enterobacter ludwigii, 6; Enterobacter, non-speciated, 1; Escherichia coli, 2218; Klebsiella aerogenes, 287; Klebsiella oxytoca, 308; Klebsiella pneumoniae, 2082; Klebsiella variicola, 47; Morganella morganii, 164; Pluralibacter gergoviae, 6; Proteus hauseri, 56; Proteus mirabilis, 357; Proteus penneri, 7; Proteus vulgaris, 151; Providencia rettgeri, 61; Providencia stuartii, 65; Raoultella ornithinolytica, 5; Raoultella planticola, 1; Serratia liquefaciens, 2; Serratia marcescens, 118 and Serratia, non-speciated, 1
bEUCAST breakpoints for tigecycline are only available with E. coli
Among ESBL-positive, carbapenemase-negative Enterobacterales isolates, susceptibility to amikacin, ceftazidime–avibactam, colistin, imipenem and meropenem was ≥ 91% (Table 2). The MIC90 value for tigecycline was 1 mg/L. Fewer than 6% of ESBL-positive, carbapenemase-negative isolates were susceptible to aztreonam, cefepime or ceftazidime. Among the carbapenemase-positive, MBL-negative subset, susceptibility was highest to ceftazidime–avibactam (100%), amikacin (93.5%), and colistin (87.0%). For the majority of agents with EUCAST breakpoints, susceptibility was reduced among carbapenemase-positive, MBL-positive isolates, compared with the Enterobacterales overall. Colistin was the only agent to which susceptibility was maintained for the carbapenemase-positive, MBL-positive subset (93.4%). No carbapenemase-positive, MBL-positive isolates were susceptible to ceftazidime–avibactam, compared with 100% of carbapenemase-positive, MBL-negative isolates. No carbapenemase-positive, MBL-negative or carbapenemase-positive, MBL-positive isolates were susceptible to amoxicillin–clavulanic acid or ceftazidime.
In vitro antimicrobial activity of 12 agents in the Asia–Pacific region against E. coli and K. pneumoniae
More than 95% of all E. coli and K. pneumoniae isolates were susceptible to amikacin, ceftazidime–avibactam, colistin, imipenem and meropenem (Table 2). Among E. coli isolates, 97.7% were susceptible to tigecycline and a tigecycline MIC90 value of 0.5 mg/L was observed. No EUCAST breakpoints are available for tigecycline against K. pneumoniae. Susceptibility rates to amikacin, ceftazidime–avibactam, colistin, imipenem and meropenem were > 89% among ESBL-positive, carbapenemase-negative E. coli and K. pneumoniae isolates. Among ESBL-positive, carbapenemase-negative E. coli isolates, 96.9% were susceptible to tigecycline.
Fewer carbapenemase-positive, MBL-negative and carbapenemase-positive, MBL-positive E. coli isolates (N = 4 and N = 14, respectively) were collected than for those of K. pneumoniae (N = 39 and N = 52, respectively; Table 2). Data for the carbapenemase-positive, MBL-negative subset of E. coli are not presented in Table 2 because of the small isolate numbers. Against this subset, the lowest MIC ranges were observed for colistin and tigecycline (both agents, 0.12–0.5 mg/L), followed by ceftazidime–avibactam (0.25–2 mg/L), whereas amoxicillin–clavulanic acid, aztreonam, ceftazidime, and piperacillin–tazobactam were not active. For the carbapenemase-positive, MBL-positive subset of E. coli and the carbapenemase-positive, MBL-negative and carbapenemase-positive, MBL-positive subsets of K. pneumoniae, the susceptibility rates to the majority of agents were < 24%, with the exception of amikacin (67.3–94.9%) and colistin (87.2–100%; Table 2). Susceptibility to tigecycline among carbapenemase-positive, MBL-positive E. coli isolates was 92.9%. All carbapenemase-positive, MBL-negative K. pneumoniae isolates were susceptible to ceftazidime–avibactam; however, no carbapenemase-positive, MBL-positive E. coli or K. pneumoniae isolates were susceptible to ceftazidime–avibactam.
In vitro antimicrobial activity of 12 agents in the Asia–Pacific region against P. aeruginosa
The highest rate of susceptibility among P. aeruginosa isolates was to colistin (99.7%), followed by ceftazidime–avibactam and amikacin (92.7% and 92.3%, respectively; Table 3). The activity of ceftazidime alone (MIC90, 64 mg/L) was less potent than that of ceftazidime–avibactam (MIC90, 8 mg/L). Among the resistant subsets of P. aeruginosa, eight isolates were identified as ESBL-positive, carbapenemase-negative and seven as carbapenemase-positive, MBL-negative. These data are not presented in Table 3 because of the small isolate numbers; however, the lowest MIC range was observed for colistin (0.5–2 mg/L against both subsets). Susceptibility to the majority of agents was reduced among the carbapenemase-positive, MBL-positive subset, when compared with the susceptibility among all P. aeruginosa isolates, with the exception of colistin (100% susceptible; Table 3). Amoxicillin–clavulanic acid (MIC90, ≥ 64 mg/L) and tigecycline (MIC90, ≥ 16 mg/L) were inactive against all MDR, or carbapenemase-positive, MBL-positive P. aeruginosa isolates.
Table 3.
In vitro antimicrobial activity of ceftazidime–avibactam and comparators against Pseudomonas aeruginosa isolates collected in the Asia–Pacific region (INFORM program, 2015–2017)
| Organism/antimicrobial | MIC50 (mg/L) | MIC90 (mg/L) | MIC range (mg/L) | % S | % R |
|---|---|---|---|---|---|
| Pseudomonas aeruginosa (N = 2032) | |||||
| Amikacin | 4 | 8 | ≤ 0.25–≥ 64 | 92.3 | 5.2 |
| Amoxicillin–clavulanic acida | ≥ 64 | ≥ 64 | 0.5–≥ 64 | N/A | N/A |
| Aztreonam | 8 | 32 | ≤ 0.015–≥ 256 | 80.6 | 19.4 |
| Cefepime | 4 | ≥ 32 | ≤ 0.12–≥ 32 | 81.9 | 18.1 |
| Ceftazidime | 2 | 64 | 0.06–≥ 256 | 78.4 | 21.6 |
| Ceftazidime–avibactam | 2 | 8 | ≤ 0.015–≥ 256 | 92.7 | 7.3 |
| Colistin | 1 | 2 | ≤ 0.06–8 | 99.7 | 0.3 |
| Imipenem | 2 | ≥ 16 | 0.12–≥ 16 | 80.0 | 20.0 |
| Levofloxacin | 0.5 | ≥ 16 | 0.008–≥ 16 | 68.5 | 31.5 |
| Meropenem | 0.5 | ≥ 16 | 0.008–≥ 16 | 80.3 | 10.8 |
| Piperacillin–tazobactam | 8 | ≥ 256 | ≤ 0.25–≥ 256 | 73.7 | 26.3 |
| Tigecyclinea | 8 | ≥ 16 | ≤ 0.015–≥ 16 | N/A | N/A |
| P. aeruginosa, MDR (N = 475) | |||||
| Amikacin | 4 | ≥ 64 | ≤ 0.25–≥ 64 | 71.6 | 21.5 |
| Amoxicillin–clavulanic acida | ≥ 64 | ≥ 64 | 32–≥ 64 | N/A | N/A |
| Aztreonam | 32 | 128 | 0.25–≥ 256 | 25.9 | 74.1 |
| Cefepime | 16 | ≥ 32 | 1–≥ 32 | 26.3 | 73.7 |
| Ceftazidime | 64 | ≥ 256 | 0.5–≥ 256 | 20.2 | 79.8 |
| Ceftazidime–avibactam | 8 | 128 | 0.25–≥ 256 | 68.6 | 31.4 |
| Colistin | 1 | 2 | 0.12–2 | 100 | 0.0 |
| Imipenem | ≥ 16 | ≥ 16 | 0.25–≥ 16 | 41.1 | 58.9 |
| Levofloxacin | 8 | ≥ 16 | 0.06–≥ 16 | 25.1 | 74.9 |
| Meropenem | 8 | ≥ 16 | 0.12–≥ 16 | 38.3 | 43.8 |
| Piperacillin–tazobactam | 128 | ≥ 256 | ≤ 0.25–≥ 256 | 5.1 | 94.9 |
| Tigecyclinea | ≥ 16 | ≥ 16 | 0.03–≥ 16 | N/A | N/A |
| P. aeruginosa, carbapenemase-positive, MBL-positive (N = 76) | |||||
| Amikacin | 32 | ≥ 64 | 2–≥ 64 | 10.5 | 76.3 |
| Amoxicillin–clavulanic acida | ≥ 64 | ≥ 64 | ≥ 64–≥ 64 | N/A | N/A |
| Aztreonam | 16 | ≥ 256 | 0.25–≥ 256 | 52.6 | 47.4 |
| Cefepime | ≥ 32 | ≥ 32 | 16–≥ 32 | 0.0 | 100 |
| Ceftazidime | 128 | ≥ 256 | 16–≥ 256 | 0.0 | 100 |
| Ceftazidime–avibactam | 128 | ≥ 256 | 2–≥ 256 | 3.9 | 96.1 |
| Colistin | 1 | 1 | 0.5–2 | 100 | 0.0 |
| Imipenem | ≥ 16 | ≥ 16 | 4–≥ 16 | 1.3 | 98.7 |
| Levofloxacin | ≥ 16 | ≥ 16 | 0.5–≥ 16 | 3.9 | 96.1 |
| Meropenem | ≥ 16 | ≥ 16 | 8–≥ 16 | 0.0 | 96.1 |
| Piperacillin–tazobactam | 128 | ≥ 256 | 8–≥ 256 | 6.6 | 93.4 |
| Tigecyclinea | ≥ 16 | ≥ 16 | 4–≥ 16 | N/A | N/A |
aEUCAST breakpoints for amoxicillin–clavulanic acid and tigecycline are not available with P. aeruginosa
Discussion
This study reports the in vitro antimicrobial susceptibilities and the rates of resistant subsets for clinical isolates of Enterobacterales and P. aeruginosa from the Asia–Pacific region. For the pooled collection of Enterobacterales, susceptibility was highest to amikacin, ceftazidime–avibactam and meropenem, and among P. aeruginosa was highest to amikacin, ceftazidime–avibactam and colistin. Enterobacterales isolates that were ESBL-positive, carbapenemase-negative were more common than carbapenemase-positive, MBL-negative or carbapenemase-positive, MBL-positive isolates. MDR isolates of P. aeruginosa were more frequently observed than the other resistant subsets.
This study is an update to the 2012–2015 INFORM study, which reported similar susceptibility rates to amikacin, ceftazidime–avibactam, colistin and meropenem among a pooled collection of Enterobacteriaceae [11]. Any comparisons should be treated with caution, however, as the isolates collected in 2015 were included in both studies and the 2012–2015 INFORM study also included isolates from China and Hong Kong. Yet, the findings of these studies together would indicate a level of stability in antimicrobial susceptibility among the Enterobacterales in the Asia–Pacific region.
In the current study, susceptibility to colistin and its antimicrobial activity was lower for the pooled collection of Enterobacterales, when compared with E. coli and K. pneumoniae isolates individually. This reduced susceptibility to colistin among the pooled group of Enterobacterales is likely to be due to the presence of intrinsically colistin-resistant species, such as P. mirabilis and Serratia marcescens [19].
Among the resistant subsets presented in this study, > 98% of ESBL-positive, carbapenemase-negative Enterobacterales isolates remained susceptible to amikacin, ceftazidime–avibactam, colistin, imipenem and meropenem. The range of antimicrobial susceptibilities to these agents was similar to the 2012–2015 INFORM study for the isolates of Enterobacteriaceae molecularly-characterized as ESBL-positive (95.1–99.6%) [11]. In the present study, susceptibility to colistin, imipenem and meropenem was reduced among the carbapenemase-positive, MBL-negative subset of Enterobacterales, compared with the ESBL-positive, carbapenemase-negative subset; however, susceptibility to amikacin and ceftazidime–avibactam among this subset remained > 93%. Similar trends were observed among K. pneumoniae isolates, although there were relatively low isolate numbers in the resistant subsets for this species. Studies of antimicrobial resistance in South Korea and Taiwan, on isolates collected between 2012 and 2017, have reported the rates of amikacin resistance to be < 10% among E. coli and K. pneumoniae isolates [20–22], and among carbapenem-nonsusceptible E. coli isolates (Taiwan only) [23].
In the current study, the group of carbapenemase-positive, MBL-positive Enterobacterales remained susceptible to colistin (93.4%) and similar results were seen for these resistant subsets of K. pneumoniae (94.2%) and E. coli (100%) isolates. Furthermore, nearly 100% of P. aeruginosa isolates, including all MDR or carbapenemase-positive, MBL-positive isolates, were susceptible to colistin. Colistin has been described in the literature as a ‘last-resort’ antimicrobial agent due to its clinical efficacy against antimicrobial-resistant isolates of Enterobacterales and P. aeruginosa [19]. However, resistance to colistin, along with the carbapenems, has been emerging in Southeast Asia, leading to increased hospital mortality rates in the case of the carbapenems [5]. A clinical trial involving patients with infections predominantly caused by carbapenem-resistant K. pneumoniae found that ceftazidime–avibactam was superior to colistin and may be an alternative treatment option [24]. Avibactam is inactive against MBL-positive isolates, therefore it is presumed that carbapenem resistance among K. pneumoniae from these clinical studies was mediated by the serine carbapenemases, such as KPC and OXA-48 [8, 25], which are susceptible to ceftazidime–avibactam.
For P. aeruginosa isolates in the current study, the MDR subset accounted for 23.4%. Lower rates of MDR P. aeruginosa were recently reported in the literature for the Asia–Pacific region (15.0% [2013–2016] and 14.8% [2012–2015]) [11, 26]. Each study used the MDR definition proposed by Magiorakos et al. [18]; however, variations in the participating countries and time periods for each study may account for the differences in the overall regional rates.
One of the limitations of this antimicrobial surveillance study is that a predefined number of isolates were collected at each center, so the findings cannot describe the epidemiology of resistance. Additionally, the lower numbers of isolates collected from Japan and Malaysia than elsewhere may have been insufficient to allow the detection of resistant subsets, resulting in relatively low resistant rates being reported for these countries. Furthermore, this study did not include data from China and India where antimicrobial resistance is prevalent, exemplified by the discovery of the first colistin-resistant Enterobacteriaceae isolate [27] and the first blaNDM-1 gene in K. pneumoniae [28] in these two countries, respectively. Nevertheless, reporting the local antimicrobial activities of contemporary agents in this region, through programs such as INFORM, is a key to guide physicians toward appropriate use of antimicrobials.
Conclusions
In the Asia–Pacific region, ESBL-positive, carbapenemase-negative Enterobacterales (17.9%) were identified more frequently than carbapenemase-positive, MBL-negative (0.7%) or carbapenemase-positive, MBL-positive isolates (1.7%). The rates of ceftazidime–avibactam, meropenem and amikacin susceptibility were highest (> 90%) among all Enterobacterales, including ESBL-positive, carbapenemase-negative, and carbapenemase-positive, MBL-negative isolates. Among the carbapenemase-positive, MBL-positive Enterobacterales, the rate of meropenem susceptibility (9.9%) was notably reduced, and no isolates in this subset were susceptible to ceftazidime–avibactam. For P. aeruginosa, carbapenemase-positive, MBL-positive isolates (3.7%) were more common than ESBL-positive, carbapenemase-negative (0.4%), or carbapenemase-positive, MBL-negative isolates (0.3%).
Among all P. aeruginosa isolates, including the resistant subsets, the rate of colistin susceptibility (> 99%) was higher than rates for ceftazidime–avibactam and amikacin, particularly among carbapenemase-positive, MBL-positive isolates (3.9% and 10.5%, respectively). Data from this study have shown the variability in antimicrobial activity depending on the β-lactamase profile of isolates, and multicenter surveillance of antimicrobial resistance remains essential for public health and clinical management of bacterial infections.
Acknowledgements
The authors would like to thank all participating investigators and laboratories and would also like to thank the staff at IHMA Inc. for their coordination of the study.
Abbreviations
- CLSI
Clinical Laboratory Standards Institute
- ESBL
Extended-spectrum β-lactamase
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- INFORM
International Network for Optimal Resistance Monitoring
- KPC
Klebsiella pneumoniae carbapenemase
- MBL
Metallo-β-lactamase
- MDR
Multidrug-resistant
- MIC
Minimum inhibitory concentration (mg/L)
- MIC50
MIC required to inhibit growth of 50% of isolates (mg/L)
- MIC90
MIC required to inhibit growth of 90% of isolates (mg/L)
- N/A
Not available
- PCR
Polymerase chain reaction
- R
Resistant
- S
Susceptible (standard dosing)
Authors’ contributions
WCK participated in data collection and interpretation, as well as drafting and reviewing the manuscript. GGS was involved in the study design, participated in data interpretation, and drafted and reviewed the manuscript. Both authors read and approved the final manuscript.
Funding
This study is funded by Pfizer. Medical writing support was provided by Neera Hobson Ph.D. at Micron Research Ltd., Ely, UK, which was funded by Pfizer. Micron Research Ltd. also provided data management services which were funded by Pfizer.
Availability of data and materials
Data from the study can be accessed at https://atlas-surveillance.com. The datasets used and/or analyzed during the current study of isolates collected in the Asia–Pacific region, 2015 to 2017, are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
WCK has no competing interests to declare. GGS is an employee and a shareholder of Pfizer Inc., and is also a shareholder of AstraZeneca.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zellweger RM, Carrique-Mas J, Limmathurotsakul D, Day NPJ, Thwaites GE, Baker S, Southeast Asia Antimicrobial Resistance Network A current perspective on antimicrobial resistance in Southeast Asia. J Antimicrob Chemother. 2017 doi: 10.1093/jac/dkx260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suwantarat N, Carroll KC. Epidemiology and molecular characterization of multidrug-resistant Gram-negative bacteria in Southeast Asia. Antimicrob Resist Infect Control. 2016 doi: 10.1186/s13756-016-0115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kazmierczak KM, Rabine S, Hackel M, McLaughlin RE, Biedenbach DJ, Bouchillon SK, et al. Multiyear, multinational survey of the incidence and global distribution of metallo-β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2015 doi: 10.1128/AAC.02379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu LY, Apisarnthanarak A, Khan E, Suwantarat N, Ghafur A, Tambyah PA. Carbapenem-resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin Microbiol Rev. 2017 doi: 10.1128/CMR.00042-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malchione MD, Torres LM, Hartley DM, Koch M, Goodman J. Carbapenem and colistin resistance in Enterobacteriaceae in Southeast Asia: review and mapping of emerging and overlapping challenges. Int J Antimicrob Agents. 2019 doi: 10.1016/j.ijantimicag.2019.07.019. [DOI] [PubMed] [Google Scholar]
- 6.Pfaller MA, Shortridge D, Sader HS, Flamm RK, Castanheira M. Ceftolozane–tazobactam activity against drug-resistant Enterobacteriaceae and Pseudomonas aeruginosa causing healthcare-associated infections in Australia and New Zealand: report from an Antimicrobial Surveillance Program (2013–2015) J Glob Antimicrob Resist. 2017 doi: 10.1016/j.jgar.2017.05.025. [DOI] [PubMed] [Google Scholar]
- 7.Williamson DA, Howden BP, Paterson DL. The risk of resistance: what are the major antimicrobial resistance threats facing Australia? Med J Aust. 2019 doi: 10.5694/mja2.50249. [DOI] [PubMed] [Google Scholar]
- 8.Falcone M, Paterson D. Spotlight on ceftazidime/avibactam: a new option for MDR Gram-negative infections. J Antimicrob Chemother. 2016 doi: 10.1093/jac/dkw239. [DOI] [PubMed] [Google Scholar]
- 9.Allergan. Avycaz (ceftazidime–avibactam) for injection, for intravenous use: prescribing information. Madison: Allergan. 2017. https://www.allergan.com/assets/pdf/avycaz_pi. Accessed 29 Aug 2019.
- 10.Pfizer. Zavicefta: summary of product characteristics. Groton: Pfizer. 2016. https://www.ema.europa.eu/en/documents/product-information/zavicefta-epar-product-information_en.pdf. Accessed 29 Aug 2019.
- 11.Karlowsky JA, Kazmierczak KM, Bouchillon SK, de Jonge BLM, Stone GG, Sahm DF. In vitro activity of ceftazidime–avibactam against clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa collected in Asia-Pacific countries: results from the INFORM global surveillance program, 2012 to 2015. Antimicrob Agents Chemother. 2018 doi: 10.1128/AAC.02569-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramalheira E, Stone GG. Longitudinal analysis of the in vitro activity of ceftazidime/avibactam versus Enterobacteriaceae, 2012–2016. J Glob Antimicrob Resist. 2019 doi: 10.1016/j.jgar.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Stone GG, Smayevsky J, Kazmierczak K. Longitudinal analysis of the in vitro activity of ceftazidime–avibactam vs. Pseudomonas aeruginosa, 2012–2016. Diagn Microbiol Infect Dis. 2020 doi: 10.1016/j.diagmicrobio.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute (CLSI) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI standard M07. 10. Wayne: CLSI; 2015. [Google Scholar]
- 15.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0, 2019. http://www.eucast.org. Accessed 21 Aug 2019.
- 16.Lob SH, Kazmierczak KM, Badal RE, Hackel MA, Bouchillon SK, Biedenbach DJ, et al. Trends in susceptibility of Escherichia coli from intra-abdominal infections to ertapenem and comparators in the United States according to data from the SMART program, 2009 to 2013. Antimicrob Agents Chemother. 2015 doi: 10.1128/AAC.05186-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols WW, de Jonge BL, Kazmierczak KM, Karlowsky JA, Sahm DF. In vitro susceptibility of global surveillance isolates of Pseudomonas aeruginosa to ceftazidime–avibactam (INFORM 2012 to 2014) Antimicrob Agents Chemother. 2016 doi: 10.1128/AAC.00220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012 doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 19.Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014 doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YA, Park YS. Epidemiology and treatment of antimicrobial-resistant Gram-negative bacteria in Korea. Korean J Intern Med. 2018 doi: 10.3904/kjim.2018.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H, Yoon EJ, Kim D, Jeong SH, Won EJ, Shin JH, et al. Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 to April 2017: first one-year report from Kor-GLASS. Euro Surveillance. 2016 doi: 10.2807/1560-7917.ES.2018.23.42.1800047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao CH, Lee NY, Tang HJ, Lee SS, Lin CF, Lu PL, et al. Antimicrobial activities of ceftazidime–avibactam, ceftolozane–tazobactam, and other agents against Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa isolated from intensive care units in Taiwan: results from the Surveillance of Multicenter Antimicrobial Resistance in Taiwan in 2016. Infect Drug Resist. 2019 doi: 10.2147/IDR.S193638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang YT, Siu LK, Wang JT, Wu TL, Chen YH, Chuang YC, et al. Resistance mechanisms and molecular epidemiology of carbapenem-nonsusceptible Escherichia coli in Taiwan, 2012–2015. Infect Drug Resist. 2019 doi: 10.2147/IDR.S208231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Duin D, Lok JJ, Earley M, Cober E, Richter SS, Perez F, et al. Colistin versus ceftazidime–avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin Infect Dis. 2018 doi: 10.1093/cid/cix783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheu CC, Chang YT, Lin SY, Chen YH, Hsueh PR. Infections caused by carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Front Microbiol. 2019 doi: 10.3389/fmicb.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shortridge D, Gales AC, Streit JM, Huband MD, Tsakris A, Jones RN. Geographic and temporal patterns of antimicrobial resistance in Pseudomonas aeruginosa over 20 years from the SENTRY antimicrobial surveillance program, 1997–2016. Open Forum Infect Dis. 2019 doi: 10.1093/ofid/ofy343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016 doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 28.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009 doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data from the study can be accessed at https://atlas-surveillance.com. The datasets used and/or analyzed during the current study of isolates collected in the Asia–Pacific region, 2015 to 2017, are available from the corresponding author on reasonable request.