Highlights
-
•
The bovine respiratory disease (BRD) complex is widespread in cattle production requiring high use of antimicrobials.
-
•
Quantifying bacterial and viral pathogens by qPCR provides good predictive value for disease diagnosis.
-
•
A commercially-available multiplex qPCR (Pneumo4) quantified relevant pathogens over a 5-log range.
-
•
The assay had good agreement with currently used methods for detection of relevant pathogens.
Keywords: Bovine respiratory disease, Calf pneumonia, Multiplex qPCR
Abstract
Bovine respiratory disease complex is the most common disease requiring the use of antimicrobials in industrial calf production worldwide. Pathogenic bacteria (Mannheimia haemolytica (Mh), Pasteurella multocida (Pm), Histophilus somni (Hs), and Mycoplasma bovis) and a range of viruses (bovine respiratory syncytial virus, bovine coronavirus, bovine parainfluenza virus type 3, bovine viral diarrhea virus and bovine herpesvirus type 1) are associated with this complex. As most of these pathogens can be present in healthy and diseased calves, simple detection of their presence in diseased calves carries low predictive value. In other multi-agent diseases of livestock, quantification of pathogens has added substantially to the predictive value of microbiological diagnosis. The aim of this study was to evaluate the ability of two recently developed quantitative PCR (qPCR) kits (Pneumo4B and Pneumo4V) to detect and quantify these bacterial and viral pathogens, respectively.
Test efficiencies of the qPCR assays, based on nucleic acid dilution series of target bacteria and viruses, were 93–106% and 91–104%, respectively, with assay detection limits of 10–50 copies of nucleic acids. All 44 strains of target bacteria were correctly identified, with no false positive reactions in 135 strains of non-target bacterial species. Based on standard curves of log10 CFU versus cycle threshold (Ct) values, quantification was possible over a 5-log range of bacteria. In 92 tracheal aspirate samples, the kappa values for agreement between Pneumo4B and bacterial culture were 0.64-0.84 for Mh, Pm and Hs. In an additional 84 tracheal aspirates, agreement between Pneumo4B or Pneumo 4V and certified diagnostic qPCR assays was moderate (0.57) for M. bovis and high (0.71–0.90) for viral pathogens. Thus Pneumo4 kits specifically detected and quantified the relevant pathogens.
Introduction
Bovine respiratory disease (BRD) is common in calves between two weeks and six months of age and is the major cause of livestock mortality in beef calves and feedlots (Johnson et al., 2011, Larsen et al., 1999, Virtala et al., 1999). A wide range of pathogens are involved in BRD, including bovine respiratory syncytial virus (BRSV), bovine coronavirus (BCoV), bovine parainfluenza virus type 3 (BPI3), Mannheimia haemolytica, Pasteurella multocida, Histophilus somni and Mycoplasma bovis (Angen et al., 2009, Friis and Krogh, 1983, Kusiluka et al., 2000, Larsen et al., 1999, Tegtmeier et al., 1999). Bovine herpesvirus 1 (BoHV-1) and bovine viral diarrhoea virus (BVDV) can also contribute, but these pathogens have been eradicated in several countries, including Denmark1 .
In the United States, more than 90% of feedlots are affected by BRD after arrival,2 and a BRD incidence of 18.2% in medium to large Australian feedlots has been reported (Hay et al., 2014). The estimated mortality rate in Danish dairy calves in 2014 was 7–10% and BRD was considered responsible for 10–35 % of deaths (Grønbæk et al., 2016). In recent investigations, 10% of calf mortalities in an Irish study were attributed to BRD, and a prevalence of 45.9% and incidence of 10.1% was found for BRD in the United Kingdom (Johnson et al., 2017). BRD can be controlled by several strategies such as correct use of colostrum (Pardon et al., 2015) or vaccination programs (Richeson et al., 2008). The widespread use of antimicrobials for BRD treatment and control (De Briyne et al., 2014) represents one of the major uses of antimicrobials in food producing animals (Fulton, 2009, Nickell and White, 2010).
BRD is a multifactorial disease associated with host susceptibility, environmental conditions, management and presence and load of pathogens. Infection results in inflammation of and damage to the respiratory tract, with severe cases resulting in death. Pathogenesis has been attributed to a primary viral infection and subsequent bacterial colonization of compromised epithelium causing a secondary infection (Angen et al., 2009, Sudaryatma et al., 2018). Viral interaction with the immune system can lead to immune suppression, which impairs host responses to secondary bacterial infections (Czuprynski et al., 2004).
The field diagnosis of BRD is based on clinical signs which can be supported by microbiology of clinical specimens such as tracheal aspirate fluid, nasal swabs and lung tissue samples. However, false negative results are commonly encountered as the causal bacteria may be fastidious or slow growing, and possibly overgrown by other organisms (Bell et al., 2014, Kugadas et al., 2014, Shanthalingam et al., 2014, Tegtmeier et al., 2000). While serological assays such as ELISA are frequently used to indicate prior infection with M. bovis, false negative results in early infection occur as seroconversion normally takes two weeks or more (Howard et al., 1986, Petersen et al., 2018). A wide range of polymerase chain reaction (PCR) assays have been developed to test individually for each of the pathogens involved in BRD (Amer et al., 2013, Bell et al., 2014, Klem et al., 2019, Kishimoto et al., 2017, Rahpaya et al., 2018). However, testing for multiple BRD pathogens in separate assays is prohibitively expensive for livestock producers. To overcome this problem, a multiplex PCR assay has recently been developed (Zhang et al., 2017).
Although individual bacterial or viral pathogens can cause disease alone, mixed infections are most common and problematic in BRD. A rapid diagnosis of these pathogens is also essential in the implementation of appropriate treatment and control measures. Conventional PCR is problematic because pathogens can be present in both healthy and diseased calves. In other situations with multi-agent diseases with opportunistic pathogens, quantification of the pathogens in clinical specimens has added substantially to the value of microbiological diagnosis (Lima et al., 2016, Pedersen et al., 2014, Pedersen et al., 2013). Recently, a multiplex quantitative PCR (qPCR) method to detect relevant bacterial pathogens of BRD was published, demonstrating better performance than culture in co-infections with two or more pathogens (Loy et al., 2018). To offer a quick and complete laboratory result of multiple BRD pathogens, a set of two commercially available multiplex qPCR assays has been developed under the name Pneumo4 (DNA Diagnostic A/S, Risskov, Denmark). These multiplex qPCR kits (Pneumo4B and Pneumo4V) can specifically detect and quantitate four bacterial (M. haemolytica, P. multocida, H. somni, M. bovis) and five viral (BPI3, BCoV, BRSV, BoHV-1 and BVDV) pathogens, respectively, which are most frequently associated with BRD. In this study, the performance of the Pneumo4 kits was evaluated by comparing the results of test of field samples with bacterial culture and PCR tests for M. bovis and BPI3, BCoV and BRSV routinely used in Denmark as reference standard methods.
Materials and methods
Sample collection and processing
Two sets of 92 and 84 tracheal aspirate (TA) samples from calves with and without signs of pneumonia were used. These samples were collected in 2017 from 11 and 14 Danish calf farms, respectively. Two sets of samples were used, because Pneumo4B was developed prior to Pneumo4V. The first set were used only for validation of the Pneumo4B (bacterial) assay, as we did not want to risk false negative results on subsequent viral tests due to prolonged sample storage. For viral testing, the second set of samples was obtained. Ethical approval of the study was not required since samples were taken as part of the routine work performed by herd veterinarians.
All TA samples were collected as previously described (Doyle et al., 2017) by authorized veterinarians or veterinary students under their supervision. Each TA sample (approx. 15−30 mL) was divided into multiple aliquots. For the first set of 92 samples, one aliquot was used for plating for detection of M. haemolytica, P. multocida and H. somni by culture which was carried out immediately after sampling; a second aliquot was used for detecting the pathogens of BRD by Pneumo4B, and the last aliquot was stored as a reserve sample at −20 °C. For the second set of 84 samples, one aliquot without preservation was used for the detection of BPI3, BCoV, BRSV and M. bovis by real-time RT-PCR at the Danish National Veterinary Institute (NVI), Lyngby, Denmark; a second was used for detecting M. bovis by Pneumo4B, a third for detecting viral pathogens by Pneumo4V, and the last aliquot was stored as a reserve sample at −20 °C.
Bacterial identification by culture and propagation of cultures for quantification
An aliquot of TA sample (5 mL) was centrifuged for 5 min at 4000 g. The pellet was suspended in 500 μL of phosphate buffered saline (PBS, pH 7.4). One, 10 and 100 μL of each sample were spread with a Drigalski spatula on three blood agar plates (Oxoid CM0055 containing 5% calf blood) supplemented with 1% amphotericin B (Sigma-Aldrich) and incubated at 37 °C in 10% CO2. After incubation for 24 h–72 h, colonies displaying typical morphology of P. multocida, M. haemolytica or H. somni were subjected to MALDI-TOF MS identification using a VITEK® MS RUO instrument (bioMérieux, Marcy l’Etoile, France) and CHCA matrix solution (Vitek® MS−CHCA, bioMérieux SA) according to the manufacturer's standard procedure. Spectra data was analysed with the SARAMIS database. All purified cultures were suspended in Brain Heart Infusion broth (Oxoid, CM1135) containing 30% glycerol and stored at −80 °C to allow re-identification.
In vitro transcription of constructed viral RNA genes
To estimate the analytical sensitivity of the Pneumo4V assays, cloned viral RNA genes of the target sequences were used. Without revealing the target genes and sequences to the author consortium, the manufacturer of Pneumo4V supplied obtained plasmid DNA with the cloned viral target gene sequences from a commercial supplier (GenScript Biotech). Plasmids were transfected into E. coli DH5α (Life Technologies Europe BV) and purified from E. coli using the QIAGEN Plasmid Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA was then transcribed in vitro using the MEGAscript T7 Transcription Kit (Life Technologies Europe BV) according to the manufacturer's protocol. Residual DNA was removed with Turbo DNase (Life Technologies Europe BV), and the RNA was further purified using the MEGAclear Transcription Clean-Up Kit (Life Technologies Europe BV). RNA quality and integrity were measured in a NanoDrop spectrophotometer (Saveen and Werner ApS).
Nucleic acid extractions
RNA and DNA from TA samples were extracted using a Pneumo4 extraction kit (DNA Diagnostic A/S) according to the manufacturer’s instructions. Briefly, RNA and DNA were extracted from 500 μL of sample using pre-lysis buffer at 37 °C for 10 min. Then cells were harvested by centrifugation at 5000 g and washed with washing buffer. The RNA and DNA were recovered at 37 °C for 20 min followed by incubation at 95 °C for 15 min in 60 μL of lysis mix buffers, then debris was pelleted by centrifugation. The lysate supernatant contained both RNA and/or DNA which was either used immediately or stored at −20 °C for bacterial qPCR detection and at −80 °C for viral RT-qPCR detection.
Multiplex qPCR and RT-qPCR protocols
Each Pneumo4 kit consists of two sets of qPCR assays. The Pneumo4B kit (DNA Diagnostic A/S) contains primers and TaqMan probes designed to detect DNA of M. haemolytica, P. multocida, H. somni and M. bovis. The primer sequences have not been released by the company. The qPCR was performed using an Mx3005 P qPCR System with the following cycling parameters: 95 °C for 1 min, followed by 40 cycles of 95 °C for 5 s and 60 °C for 25 s. The machine was set to acquire fluorescence on the CY5, ROX, HEX, FAM and ATTO channels for M. haemolytica, P. multocida, H. somni, M. bovis amplicons and Internal Amplification Control (IAC), respectively.
The Pneumo4V kit (DNA Diagnostic A/S) contains primers and TaqMan probes to detect sequences of BPI3, BCoV, BRSV, BoHV-1 and BVDV. Viral RNA extracted from the samples was tested using a two-step RT-qPCR method. First the RT reaction for the cDNA synthesis in Pneumo4V was incubated at 37 °C for 60 min and then incubated at 70 °C for 15 min. The second step for the qPCR stage was performed with the same cycling parameters as for the Pneumo4B assay. The machine was set to acquire fluorescence on the CY5, ROX, HEX, FAM and ATTO channels for BPI3, BCoV, BRSV, BoHV-1/BVDV and Internal Amplification Control (IAC), respectively. For both the Pneumo4B and Pneumo4V testing, results were analyzed using MxPro qPCR Software and samples with a cycle threshold (Ct) of 37 cycles or less were considered positive.
Viral pathogens and M. bovis detection by standard qPCR
To evaluate the performance of Pneumo4V for viral pathogen detection and Pneumo4B with respect to M. bovis detection, comparisons with standard singleplex qPCR methods used by NVI, Denmark were undertaken.
Determination of Pneumo4 efficiency and analytical sensitivity
The efficiency and sensitivity of Pneumo4 kits were evaluated by amplification of 10-fold serial dilutions of copy numbers of bacterial or viral nucleic acids. For the four bacterial targets, the genomic DNA of each was used, while for the five viral targets, the RNAs generated from plasmids containing target gene sequences of each virus were used. Dilutions of each target nucleic acids were performed in PBS. Each dilution was run in triplicate. The mean Ct values were plotted against the log10 of nucleic acid copy numbers of targets. The PCR efficiency (E) for each target (as a percentage) was calculated based on the slope of each curve using the formula -
E = [10−(1/slope) -1] ×100 (Svec et al., 2015).
Quantification of bacterial numbers by Pneumo4B PCR
To quantify the bacterial load, Ct values of the qPCR test were compared with bacterial colony forming units (CFU) of target bacteria. For each of four bacteria, three different isolates were tested and 10-fold serial dilutions in 0.9% NaCl were made from overnight cultures in serum bouillon broth (SSI Diagnostica A/S). DNA was extracted from 0.5 mL of all dilutions and tested by Pneumo4B PCR. Simultaneously, the number of bacterial cells was enumerated by plating the dilutions onto blood agar plates (Eurofins Steins Laboratorium A/S) for M. haemolytica, P. multocida and H. somni, and on mycoplasma agar plates (Oxoid CM0401B) containing supplement G (Oxoid SR0059C) for M. bovis. Blood agar plates were incubated at 37 °C in 10% CO2 for 24 h while mycoplasma agar plates were incubated for 14 days. The CFU of the target bacteria was calculated from the plates containing between 10 and 300 colonies and bacterial loads (log10 CFU/0.5 mL) were plotted against the corresponding Ct values.
Determination of Pneumo4B analytical specificity
Pneumo4B assay specificity was evaluated on pure cultures of M. haemolytica (n = 15), P. multocida (n = 16), H. somni (n = 10) and M. bovis (n = 3) and 135 strains of non-target bacteria (Refer Supplementary Table 1 for strain details).
Determination of diagnostic sensitivity and specificity of Pneumo4B and Pneumo4V
The diagnostic sensitivity and specificity of the Pneumo4 kits were evaluated by testing the nucleic acids of targets extracted from clinical samples. The first set of 92 TA was examined to compare results of Pneumo4B with bacterial culture as a reference standard method for detecting M. haemolytica, P. multocida and H. somni. The second set of 84 TA samples were analyzed by the Pneumo4 V for viral pathogens and Pneumo4B for M. bovis and compared to the results of the standard qPCR for viral pathogens and M. bovis described above.
Statistics
The diagnostic sensitivity and specificity with 95% confidence intervals were calculated using a DAG Stat spreadsheet (Mackinnon, 2000). Cohen’s kappa coefficient was used to describe agreement between results of Pneumo4 and bacterial culture or PCRs for viruses and M. bovis. The kappa values were interpreted as: 0.00–0.20, slight agreement; 0.21–0.40, fair agreement; 0.41–0.60, moderate agreement; 0.61–0.80, substantial agreement; 0.81–1.00, almost perfect agreement (Landis and Koch, 1977).
Results
Pneumo4 efficiency, limit of detection and analytical sensitivity
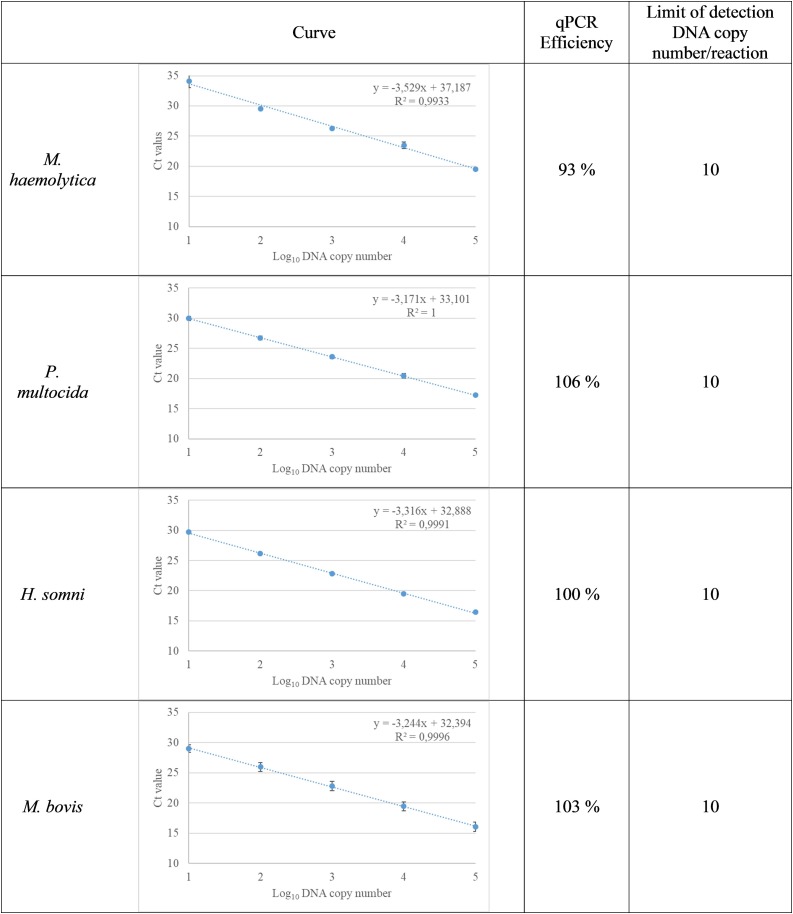
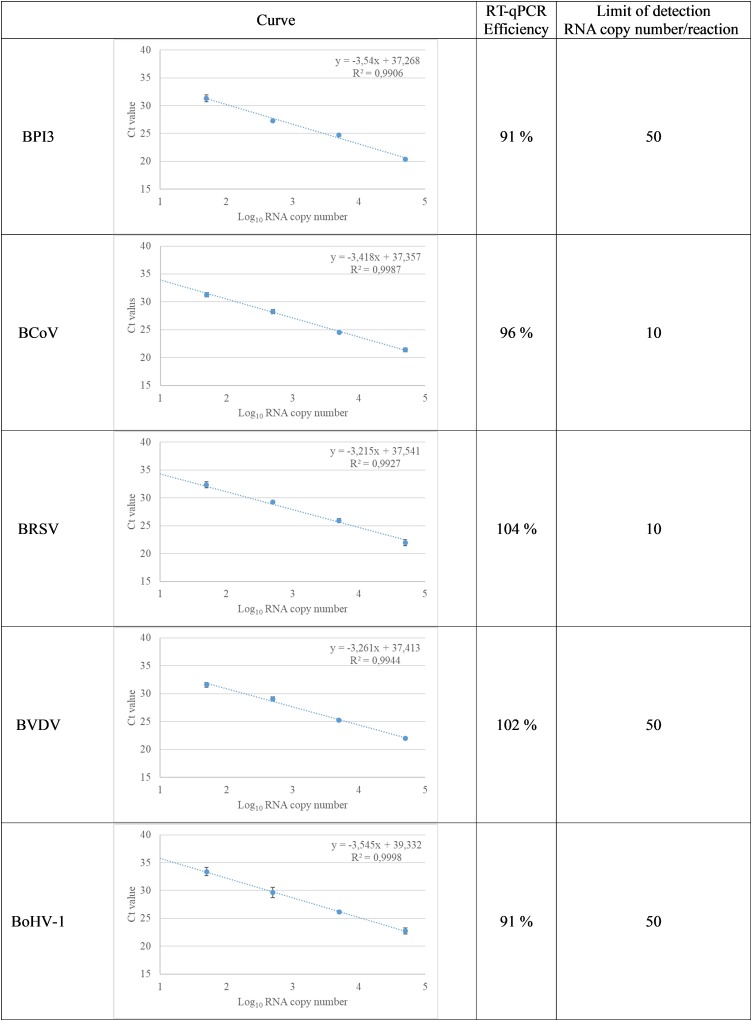
Based on the 10-fold serial dilutions of genomic DNA or plasmid RNA, the limits of detection (Ct ≤ 37) were between 10 and 50 copies of nucleic acid per reaction depending on the assay. The coefficient of determination ( r2) between the Ct values and the copies of the nucleic acids per assay was above 0.99 for all assays. The qPCR efficiencies (E) ranged from 93% to 106% for Pneumo4B (Fig. 1 ) and 91%–104% for Pneumo4 V (Fig. 2 ).
Fig. 1.
Standard curves showing the linear relationship between cycle threshold (Ct) values and log10 DNA copy numbers of bacterial targets. The results of 10-fold dilution series of bacterial DNA copy numbers are shown with each dilution run in triplicate. The mean values with standard deviation are shown, as well as equations (y) and determination coefficients ( r2).
Fig. 2.
Standard curves showing the linear relationship between cycle threshold (Ct) values and log10 RNA copy numbers of viral targets. The results of 10-fold dilution series of viral RNA copy numbers are shown with each dilution run in triplicate. The mean values with standard deviation are shown, as well as equations (y) and determination coefficients ( r2). BPI3, bovine parainfluenza virus type 3; BCoV, bovine corona virus; BRSV, bovine respiratory syncytial virus; BVDV, bovine viral diarrhoea virus; BoHV-1, bovine herpesvirus 1.
Quantification of bacterial pathogens using bacterial cells from pure culture
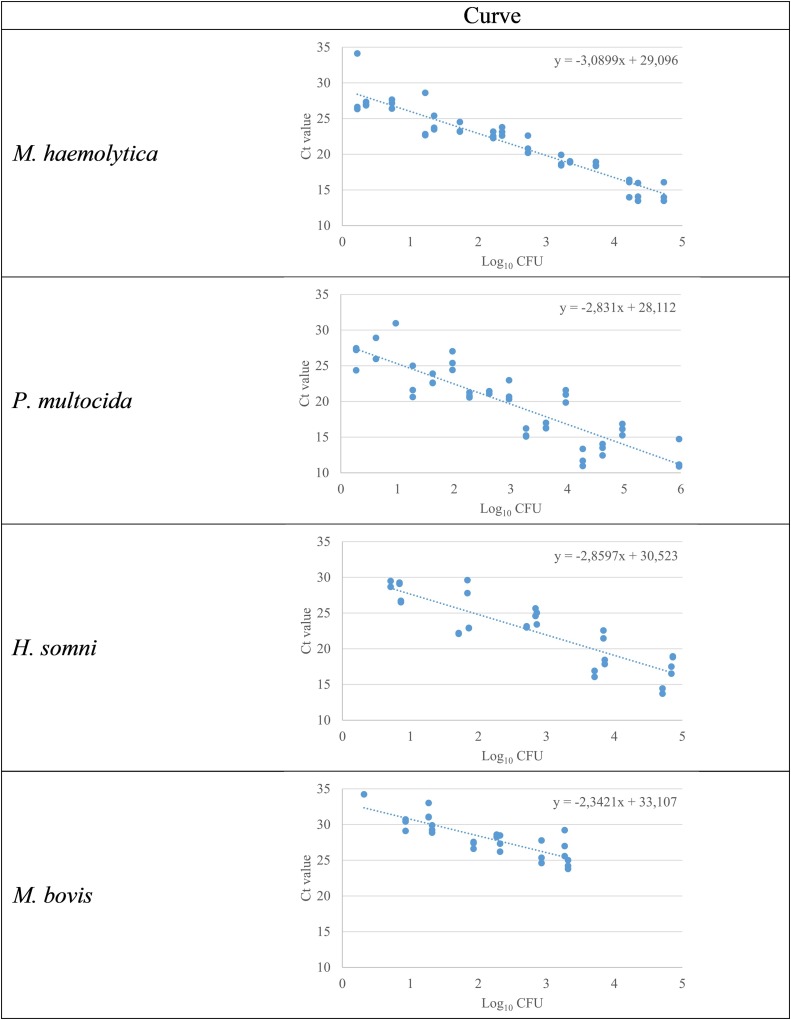
The quantification of bacterial targets was in the range of 0.4–4.7 log10 CFU/0.5 mL for M. haemolytica, 0.4–5.9 log10 CFU/0.5 mL for P. multocida, 1.1–4.8 log10 CFU/0.5 mL for H. somni and 1.1–3.3 log10 CFU/0.5 mL for M. bovis (Fig. 3 ).
Fig. 3.
Scatter plot showing the relationship between bacterial counts (log10 CFU/0.5 mL) and cycle threshold (Ct) values in the Pneumo4B qPCR. Tenfold serial dilution of each target bacteria cells were subjected to DNA extraction and qPCR. Three different isolation of each target were tested.
Pneumo4 analytical specificity
The Pneumo4B kit detected all strains of M. haemolytica, P. multocida, H. somni and M. bovis tested. No positive reactions were observed with 135 non-target bovine bacteria.
Pneumo4 diagnostic sensitivity and specificity
For the first set of TA samples (n = 92) tested using Pneumo4B and bacterial culture, results and Ct values per sample appear in Supplementary Table 2. Ct values in positive samples ranged from 22 to 37 for M. haemolytica, 19–34 for P. multocida and 22–31 for H. somni. Using a standard curve (Fig. 3), this corresponded to a bacterial load of (log10 values) 4.1 to <1, 4.3 to <1 and 3.1 to <1 in the positive samples for the three pathogens, respectively.
Results of the comparison of Pneumo4B PCR and bacterial culture for M. haemolytica, P. multocida, and H. somni based on 92 TA samples is shown in Table 1 . With bacterial culture as a reference standard, the corresponding diagnostic sensitivities and specificities of the PCR were 0.72 and 0.91 for M. haemolytica, 0.85 and 0.88 for P. multocida, 0.85 and 0.96 for H. somni (Table 1). Agreement between the Pneumo4B PCR and bacterial culture were moderate to high for all bacterial targets (kappa values of 0.64, 0.69, 0.84 for M. haemolytica, P. multocida and H. somni, respectively).
Table 1.
Comparison of Pneumo4B multiplex qPCR and bacterial culture for Mannheimia haemolytica, Pasteurella multocida, Histophilus somni in 92 tracheal aspirate (TA) samples.
| n (%) TA samples | ||||||||
|---|---|---|---|---|---|---|---|---|
| Pathogen | C + PCR+ | C + PCR- | C-PCR+ | C- PCR- | Diagnostic sensitivity | Diagnostic specificity | Cohen’s kappa coefficient | |
| M. haemolytica | 18 (19.5) | 7 (7.6) | 6 (6.5) | 61 (66.3) | 0.72 | 0.91 | 0.64 | |
| P. multocida | 23 (25.0) | 4 (4.3) | 8 (8.7) | 57 (61.9) | 0.85 | 0.88 | 0.69 | |
| H. somni | 23 (25.0) | 4 (4.3) | 2 (2.1) | 63 (68.4) | 0.85 | 0.96 | 0.84 | |
C+, culture positive; C-, culture negative; PCR+, Pneumo4B positive; PCR-, Pneumo4B negative.
In the 84 sample comparison of M. bovis detection between the Pneumo4B PCR and the NVI qPCR as the reference standard (Table 2 ), the Pneumo4B test had a diagnostic sensitivity of 0.96 and a diagnostic specificity of 0.71, with moderate agreement (kappa = 0.57) between the two tests. Results for the viral targets, where the Pneumo4V PCR was compared for three viruses with the qPCRs at NVI as reference standards, are also shown in Table 2. Detailed results including Ct values, appear in Supplementary Table 3. High diagnostic sensitivities of 1.00, 1.00, and 0.92 and diagnostic specificities of 0.96, 0.95 and 0.96 for BPI3, BCoV and BRSV, respectively, were found (Table 2). Agreement between Pneumo4V and the reference method were also high for viral targets (kappa = 0.71, 0.90, 0.82 for BPI3, BCoV, BRSV, respectively). As expected, all 84 TA samples from Danish calves tested negative for BoHV-1 and BVDV.
Table 2.
Comparison of Pneumo4B multiplex qPCR for Mycoplasma bovis and Pneumo4 V multiplex RT-qPCR and reference (reference standard) PCR methods (R) for detection of bovine parainfluenza virus type 3 (BPI3), bovine corona virus (BCoV) and bovine respiratory syncytial virus (BRSV) in 84 tracheal aspirate (TA) samples.
| No. (%) of TA samples | |||||||
|---|---|---|---|---|---|---|---|
| Pathogen | R + PCR+ | R + PCR- | R-PCR+ | R- PCR- | Diagnostic sensitivity | Diagnostic specificity | Cohen’s kappa coefficient |
| M. bovis | 25 (29.7) | 1 (1.2) | 17 (20.2) | 41 (48.8) | 0.96 | 0.71 | 0.57 |
| BPI3 | 4 (4.7) | 0 (0) | 3 (3.5) | 77 (91.6) | 1.0 | 0.96 | 0.71 |
| BCoV | 19 (22.6) | 0 (0) | 3 (3.5) | 62 (73.8) | 1.0 | 0.90 | |
| BRSV | 11 (13.0) | 1 (1.1) | 3 (3.5) | 69 (82.4) | 0.92 | 0.96 | 0.82 |
R+, reference PCR positive; R-, reference PCR negative; PCR+, Pneumo4 V/4B positive; PCR-, Pneumo4 V/4B negative.
Discussion
The Pneumo4 assays were developed to provide rapid, effective detection of pathogens involved in BRD. For a total of nine pathogens the Pneumo4B and Pneumo4V multiplex qPCR assays can be run at the same time under the same PCR program, with benefits in costs and processing time for a diagnostic laboratory compared to multiple singleplex qPCR assays run in parallel. However, the efficiency and sensitivity of a multiplex PCR can be compromised due to primer and probe sets interfering, or the competition for the components in the reaction (Kalle et al., 2014, Parker et al., 2015). In this study, the commercially-available multiplex qPCR Pneumo4B and Pneumo4V assays demonstrated that their efficiencies for seven of these target pathogens (93–106% and 91–104%, respectively) were comparable to those of published singleplex qPCR assays (84.2–101.8%) (Kishimoto et al., 2017). Pneumo4B and Pneumo4V were able to detect between 10 copies of genomic DNA and 10–50 copies of target RNA per reaction, respectively, which are comparable to previously established methods which reported of 100–250 copies per reaction (Rahpaya et al., 2018). The qPCR efficiency was also similar to multiplex qPCR assays reported by others (Cornelissen et al., 2017, Wisselink et al., 2017).
The Pneumo4B assay demonstrated that a number of TA samples (n = 16) were PCR positive for M. haemolytica, P. multocida or H. somni and negative by culture methods (Table 1). These results suggest bacteria were present, but were undetected by culture possibly due to their fastidious nature or overgrowth by other organisms on culture (Bell et al., 2014, Van Driessche et al., 2017). PCR methods such as Pneumo4B are likely to overcome the effects of accompanying flora, and detect pathogens in mixed samples where traditional culture fails (Loy et al., 2018). The limitations of culture as a reference standard assay must be considered when determining the diagnostic specificity and sensitivity of Pneumo4B. If Pneumo4B correctly identified bacterial species undetected by culture, the specificity of the Pneumo4B assay would be underestimated. A similar number of samples (n = 15) were positive for M. haemolytica, P. multocida or H. somni by culture and negative in the Pneumo4B assays (Table 1), confirming that qPCR can result in false negative results, as previously reported (Bell et al., 2014). Explanations for this could be primer/probe mis-matches, degradation of nucleic acids during storage or handling, or technical errors during extraction.
In the present study, the majority of the culture positive/PCR negative samples contained low numbers of bacterial cells (Supplementary Table 2), possibly the result of low amounts of sample used for DNA extraction compared to a larger volume used for culture. However, some target bacteria could have sequence substitutions at the primer or probe recognition sites. Since the primer sequences have not been released by the company, we have no means of investigating this. According to the company, primer design included comparison of all available genome sequences of the target bacteria, and we did not observe any problems with the bacterial cultures used in kit testing. PCR inhibitors may also have caused these results, as we found some samples with culture positive/PCR negative results had unacceptably low PCR signals from the internal amplification control (IAC) (no signal or Ct >32), and became PCR positive on retesting after a 5 fold dilution of the DNA extract. However in our methods comparisons, we classified such samples as PCR negative.
The M. bovis qPCR tests identified more positives in TA samples by the Pneumo4B than by the reference method (Table 2). We cannot rule out that Pneumo4B may cross react with other species of Mycoplasma which have not been tested in the current study. However, in samples positive by both methods, Pneumo4B gave results 3–4 Ct values lower than results of standard PCR (Supplementary Table 3), suggesting this assay has a higher sensitivity for M. bovis than the reference method. Several factors influence PCR efficiency, such as sample storage and transport, differences in materials (Buzard et al., 2012) and reagents including DNA preparation method (Bowman et al., 2016, Dilhari et al., 2017). These may explain the sensitivity difference between the two methods for M. bovis detection and the moderate agreement of the two assays is also consistent with the Pneumo4B assay being more sensitive than the reference standard reference.
Various techniques including cell culture, serology (fluorescent antibody tests, ELISA) and nucleic acid testing are used in laboratory diagnosis of viral infections in BRD (Albayrak et al., 2019, Timsit et al., 2010). These differ in sensitivity and specificity, with none an optimal reference standard able to detect all viruses in all clinical situations. In this study, we used the official Danish qPCR viral detection protocols, for which the National Veterinary Laboratory is accredited, as reference standard reference singleplex assays. In comparison to these standard methods, the Pneumo4V multiplex assay was equally or slightly more sensitive. Some samples (n = 9) were positive for BPI3, BCoV or BRSV by Pneumo4V PCR and negative in standard PCRs, whereas only one was positive by standard PCR and negative by Pneumo4V. The Ct values of these positive samples were ≥30 (Supplementary Table 3). We cannot rule out that a high Ct may be generated through cross-contamination or by nonspecific amplification with other viral species not tested in this study. The nucleic acid extraction protocol included with Pneumo4 assays does not include high speed centrifugation in the initial step; however, this is also the case for the NVI protocol. It is unlikely that the Pneumo4V extraction protocol harvests free virus particles, as this requires centrifugation speeds of >30,000 g (Payne, 2018). However, since sick calves have high numbers of virus-infected neutrophils, macrophages and lymphocytes in the lung (Blodörn et al., 2015), enough target particles are likely to be obtained.
The diagnostic sensitivity and specificity of Pneumo4V were validated on samples from only Danish farms, which is a limitation since Denmark is free from BoHV-1 and BVDV. Thus the diagnostic sensitivity for BoHV-1 and BVDV could not be evaluated. The results of all 84 TA samples were indeed negative for BoHV-1 and BVDV in both Pneumo4 V and standard PCR, indicating the assays for these two viruses in Pneumo4 V do not cross-react with other bovine respiratory viruses in Denmark. For routine use in BoHV-1/BVDV free countries, it is important that the BoHV-1/BVDV assays have high specificities because false-positive detections can have regulatory and trade implications. Additional studies to evaluate the sensitivity of Pneumo4 V on clinical samples containing BoHV-1 and BVDV are needed.
Conclusions
Fast, specific and sensitive multiplex qPCR assays for the detection of M. haemolytica, P. multocida, H. somni, and M. bovis (Pneumo4B), and for BPI3, BCoV, and BRSV (Pneumo4 V) were confirmed in kits for tracheal aspirate specimens from Danish calves. These offer an alternative to standard methods in routine diagnostics.
Conflict of interest statement
J. Katholm and P. Pansri are employees of DNA Diagnostic, which produces and sells the Pneumo4B and Pneumo4V kits. None of the authors has any other financial or personal relationships that could inappropriately influence or bias the content of the paper.
Acknowledgements
This study was the research part of a GUDP project funded by Danish Ministry of Environment and Food (Research Grant 34009-16-078).
Footnotes
See: DVFA, 2017. The 2016 Annual Report on Animal Health in Denmark. https://www.foedevarestyrelsen.dk/Publikationer/Alle%20publikationer/Animal%20Health%202017.pdf (Accessed 6 January 2019).
See: USDA, 2013. NAHMS Feedlot Study 2011. https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/monitoring-and-surveillance/nahms/nahms_feedlot_studies/!ut/p/z1/lZLLcoIwFIafxQXLkhN0ALsDyggt2FalYjYM1nCZAcKQWGb69I1tN72ImE0mme87yfkTRFCMSJO-lXkqStaklVzviJ4ssbMAc4aDhXuHwfLm_oNn6AChgbafQPA4c7C9Bjm7Nljuyghd19cATxG5zn_xdOlvos3SxLY_18b5cGZYMM4fAMhw-ftLB8gEtS50whyRNhXFTdlkDMVNWtQ8ySg9VEwkXBwPJeXyMmSw3LP2DZzLe3UJOD3IL-Bv4iN6ziu2__oeVrOfmrK5jma0o5167OR2IUTLbxVQoO97NWcsr6j6ymoF_lMKxgWKf5KoraMoit-DbO2XT_XW5NZk8gHd6NGI/#feedlot11 (Accessed 6 January 2019). 3See: Fagan, J. and Fee, S., 2017. In: Barret, D. and Johnson, A., All-island Animal Disease Surveillance Report 2016. https://www.afbini.gov.uk/sites/afbini.gov.uk/files/publications/All%20Island%20Disease%20Surveillance%20Report%202016.pdf (Accessed 6 January 2019).
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.tvjl.2020.105425.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Albayrak H., Yazici Z., Ozan E., Tamer C., El Abd, Wahed A., Wehner S., Ulrich K., Weidmann M. Characterisation of the first bovine parainfluenza virus 3 isolate detected in cattle in Turkey. Vet. Sci. 2019;6:56. doi: 10.3390/vetsci6020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer H.M., Abd El, Wahed A., Shalaby M.A., Almajhdi F.N., Hufert F.T., Weidmann M. A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J. Virol. Methods. 2013;193:337–340. doi: 10.1016/j.jviromet.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angen O., Thomsen J., Larsen L.E., Larsen J., Kokotovic B., Heegaard P.M., Enemark J.M. Respiratory disease in calves: microbiological investigations on trans-tracheally aspirated bronchoalveolar fluid and acute phase protein response. Vet. Microbiol. 2009;137:165–171. doi: 10.1016/j.vetmic.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.J., Blackburn P., Elliott M., Patterson T.I., Ellison S., Lahuerta-Marin A., Ball H.J. Investigation of polymerase chain reaction assays to improve detection of bacterial involvement in bovine respiratory disease. J. Vet. Diagn. Investig. 2014;26:631–634. doi: 10.1177/1040638714540166. [DOI] [PubMed] [Google Scholar]
- Blodörn K., Hägglund S., Gavier-Widen D., Eléouët J.-F., Riffault S., Pringle J., Taylor G., Valarcher J.F. A bovine respiratory syncytial virus model with high clinical expression in calves with specific passive immunity. BMC Vet. Res. 2015;11:76. doi: 10.1186/s12917-015-0389-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S., Roffey P., McNevin D., Gahan M.E. Evaluation of commercial DNA extraction methods for biosecurity applications. Aust. J. Forensic Sci. 2016;48:407–420. [Google Scholar]
- Buzard G.S., Baker D., Wolcott M.J., Norwood D.A., Dauphin L.A. Multi-platform comparison of ten commercial master mixes for probe-based real-time polymerase chain reaction detection of bioterrorism threat agents for surge preparedness. Forensic Sci. Int. 2012;223:292–297. doi: 10.1016/j.forsciint.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Cornelissen J.B., de Bree F.M., van der Wal F.J., Kooi E.A., Koene M.G., Bossers A., Smid B., Antonis A.F., Wisselink H.J. Mycoplasma detection by triplex real-time PCR in bronchoalveolar lavage fluid from bovine respiratory disease complex cases. BMC Vet. Res. 2017;13:97. doi: 10.1186/s12917-017-1023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czuprynski C.J., Leite F., Sylte M., Kuckleburg C., Schultz R., Inzana T., Behling-Kelly E., Corbeil L. Complexities of the pathogenesis of Mannheimia haemolytica and Haemophilus somnus infections: challenges and potential opportunities for prevention? Anim. Health Res. Rev. 2004;5:277–282. doi: 10.1079/ahr200483. [DOI] [PubMed] [Google Scholar]
- De Briyne N., Atkinson J., Pokludova L., Borriello S.P. Antibiotics used most commonly to treat animals in Europe. Vet. Rec. 2014;175:325. doi: 10.1136/vr.102462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilhari A., Sampath A., Gunasekara C., Fernando N., Weerasekara D., Sissons C., McBain A., Weerasekera M. Evaluation of the impact of six different DNA extraction methods for the representation of the microbial community associated with human chronic wound infections using a gel-based DNA profiling method. AMB Express. 2017;7:179. doi: 10.1186/s13568-017-0477-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D., Credille B., Lehenbauer T.W., Berghaus R., Aly S.S., Champagne J., Blanchard P., Crossley B., Berghaus L., Cochran S., Woolums A. Agreement among four sampling methods to identify respiratory pathogens in dairy calves with acute bovine respiratory disease. J. Vet. Intern. Med. 2017;31:954–959. doi: 10.1111/jvim.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis N.F., Krogh H.V. Isolation of mycoplasmas from Danish cattle. Nord. Vet. 1983;35:74–81. [PubMed] [Google Scholar]
- Fulton R.W. Bovine respiratory disease research (1983-2009) Anim. Health Res. Rev. 2009;10:131–139. doi: 10.1017/S146625230999017X. [DOI] [PubMed] [Google Scholar]
- Grønbæk L., Westphael N., Martin H.L. Obduktioner kaster lys over årsager til kalvedødelighed. Dansk Veterinærtidsskrifts. 2016;01:30–33. [Google Scholar]
- Hay K.E., Barnes T.S., Morton J.M., Clements A.C.A., Mahony T.J. Risk factors for bovine respiratory disease in Australian feedlot cattle: use of a causal diagram-informed approach to estimate effects of animal mixing and movements before feedlot entry. Prev. Vet. Med. 2014;117:160–169. doi: 10.1016/j.prevetmed.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Howard C.J., Parsons K.R., Thomas L.H. Systemic and local immune responses of gnotobiotic calves to respiratory infection with Mycoplasma bovis. Vet. Immunol. Immunopathol. 1986;11:291–300. doi: 10.1016/0165-2427(86)90008-5. [DOI] [PubMed] [Google Scholar]
- Johnson K., Burn C., Wathes D.C. Rates and risk factors for contagious disease and mortality in young dairy heifers. Cab Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2011;6:1. [Google Scholar]
- Johnson K.F., Chancellor N., Burn C.C., Wathes D.C. Prospective cohort study to assess rates of contagious disease in pre-weaned UK dairy heifers: management practices, passive transfer of immunity and associated calf health. Vet. Rec. Open. 2017;4 doi: 10.1136/vetreco-2017-000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalle E., Kubista M., Rensing C. Multi-template polymerase chain reaction. Biomol. Detect. Quantif. 2014;2:11–29. doi: 10.1016/j.bdq.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto M., Tsuchiaka S., Rahpaya S.S., Hasebe A., Otsu K., Sugimura S., Kobayashi S., Komatsu N., Nagai M., Omatsu T. Development of a one-run real-time PCR detection system for pathogens associated with bovine respiratory disease complex. J. Vet. Med. Sci. 2017;79:517–523. doi: 10.1292/jvms.16-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klem T.B., Sjurseth S.K., Sviland S., Gjerset B., Myrmel M., Stokstad M. Bovine respiratory syncytial virus in experimentally exposed and rechallenged calves; viral shedding related to clinical signs and the potential for transmission. BMC Vet. Res. 2019;15:156. doi: 10.1186/s12917-019-1911-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugadas A., Poindexter J., Lee M.L., Bavananthasivam J., Call D.R., Brayton K.A., Srikumaran S. Growth of Mannheimia haemolytica: inhibitory agents and putative mechanism of inhibition. Vet. Microbiol. 2014;174:155–162. doi: 10.1016/j.vetmic.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Kusiluka L.J., Kokotovic B., Ojeniyi B., Friis N.F., Ahrens P. Genetic variations among Mycoplasma bovis strains isolated from Danish cattle. FEMS Microbiol. Lett. 2000;192:113–118. doi: 10.1111/j.1574-6968.2000.tb09368.x. [DOI] [PubMed] [Google Scholar]
- Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- Larsen L.E., Tjornehoj K., Viuff B., Jensen N.E., Uttenthal A. Diagnosis of enzootic pneumonia in Danish cattle: reverse transcription-polymerase chain reaction assay for detection of bovine respiratory syncytial virus in naturally and experimentally infected cattle. J. Vet. Diagn. Investig. 1999;11:416–422. doi: 10.1177/104063879901100505. [DOI] [PubMed] [Google Scholar]
- Lima S.F., Teixeira A.G.V., Higgins C.H., Lima F.S., Bicalho R.C. The upper respiratory tract microbiome and its potential role in bovine respiratory disease and otitis media. Sci. Rep. 2016;6 doi: 10.1038/srep29050. 29050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy J.D., Leger L., Workman A.M., Clawson M.L., Bulut E., Wang B. Development of a multiplex real-time PCR assay using two thermocycling platforms for detection of major bacterial pathogens associated with bovine respiratory disease complex from clinical samples. J. Vet. Diagn. Investig. 2018;30:837–847. doi: 10.1177/1040638718800170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon A. A spreadsheet for the calculation of comprehensive statistics for the assessment of diagnostic tests and inter-rater agreement. Comput. Biol. Med. 2000;30:127–134. doi: 10.1016/s0010-4825(00)00006-8. [DOI] [PubMed] [Google Scholar]
- Nickell J.S., White B.J. Metaphylactic antimicrobial therapy for bovine respiratory disease in stocker and feedlot cattle. Vet. Clin. North Am. Food Anim. Pract. 2010;26:285–301. doi: 10.1016/j.cvfa.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Pardon B., Alliet J., Boone R., Roelandt S., Valgaeren B., Deprez P. Prediction of respiratory disease and diarrhea in veal calves based on immunoglobulin levels and the serostatus for respiratory pathogens measured at arrival. Prev. Vet. Med. 2015;120:169–176. doi: 10.1016/j.prevetmed.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J., Fowler N., Walmsley M.L., Schmidt T., Scharrer J., Kowaleski J., Grimes T., Hoyos S., Chen J. Analytical sensitivity comparison between singleplex real-time PCR and a multiplex PCR platform for detecting respiratory viruses. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S. second edition. Academic Press; UK and US: 2018. Methods to Study Viruses. In: Viruses: From Understanding to Investigation; pp. 37–52. Chapter 4. [Google Scholar]
- Pedersen K.S., Johansen M., Angen O., Jorsal S.E., Nielsen J.P., Jensen T.K., Guedes R., Stahl M., Baekbo P. Herd diagnosis of low pathogen diarrhoea in growing pigs - a pilot study. Ir. Vet. J. 2014;67:24. doi: 10.1186/2046-0481-67-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K.S., Stege H., Jensen T.K., Guedes R., Stahl M., Nielsen J.P., Hjulsager C., Larsen L.E., Angen O. Diagnostic performance of fecal quantitative real-time polymerase chain reaction for detection of Lawsonia intracellularis-associated proliferative enteropathy in nursery pigs. J. Vet. Diagn. Investig. 2013;25:336–340. doi: 10.1177/1040638713480499. [DOI] [PubMed] [Google Scholar]
- Petersen M.B., Pedersen J., Holm D.L., Denwood M., Nielsen L.R. A longitudinal observational study of the dynamics of Mycoplasma bovis antibodies in naturally exposed and diseased dairy cows. J. Dairy Sci. 2018;101:7383–7396. doi: 10.3168/jds.2017-14340. [DOI] [PubMed] [Google Scholar]
- Rahpaya S.S., Tsuchiaka S., Kishimoto M., Oba M., Katayama Y., Nunomura Y., Kokawa S., Kimura T., Kobayashi A., Kirino Y. Dembo polymerase chain reaction technique for detection of bovine abortion, diarrhea, and respiratory disease complex infectious agents in potential vectors and reservoirs. J. Vet. Sci. 2018;19:350–357. doi: 10.4142/jvs.2018.19.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richeson J.T., Beck P.A., Gadberry M.S., Gunter S.A., Hess T.W., Hubbell D.S., 3rd, Jones C. Effects of on-arrival versus delayed modified live virus vaccination on health, performance, and serum infectious bovine rhinotracheitis titers of newly received beef calves. J. Anim. Sci. 2008;86:999–1005. doi: 10.2527/jas.2007-0593. [DOI] [PubMed] [Google Scholar]
- Shanthalingam S., Goldy A., Bavananthasivam J., Subramaniam R., Batra S.A., Kugadas A., Raghavan B., Dassanayake R.P., Jennings-Gaines J.E., Killion H.J. PCR assay detects Mannheimia haemolytica in culture-negative pneumonic lung tissues of bighorn sheep (Ovis canadensis) from outbreaks in the western USA, 2009-2010. J. Wildl. Dis. 2014;50:1–10. doi: 10.7589/2012-09-225. [DOI] [PubMed] [Google Scholar]
- Sudaryatma P.E., Nakamura K., Mekata H., Sekiguchi S., Kubo M., Kobayashi I., Subangkit M., Goto Y., Okabayashi T. Bovine respiratory syncytial virus infection enhances Pasteurella multocida adherence on respiratory epithelial cells. Vet. Microbiol. 2018;220:33–38. doi: 10.1016/j.vetmic.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svec D., Tichopad A., Novosadova V., Pfaffl M.W., Kubista M. How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015;3:9–16. doi: 10.1016/j.bdq.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeier C., Angen O., Ahrens P. Comparison of bacterial cultivation, PCR, in situ hybridization and immunohistochemistry as tools for diagnosis of Haemophilus somnus pneumonia in cattle. Vet. Microbiol. 2000;76:385–394. doi: 10.1016/s0378-1135(00)00259-5. [DOI] [PubMed] [Google Scholar]
- Tegtmeier C., Uttenthal A., Friis N.F., Jensen N.E., Jensen H.E. Pathological and microbiological studies on pneumonic lungs from Danish calves. Zentralblatt Veterinarmedizin Reihe B. 1999;46:693–700. doi: 10.1046/j.1439-0450.1999.00301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit E., Maingourd C., Dréan E.L., Belloc C., Seegers H., Douart A., Assié S. Evaluation of a commercial real-time reverse transcription polymerase chain reaction kit for the diagnosis of bovine respiratory syncytial virus infection. J. Vet. Diagn. Investig. 2010;22:238–241. doi: 10.1177/104063871002200211. [DOI] [PubMed] [Google Scholar]
- Van Driessche L., Valgaeren B.R., Gille L., Boyen F., Ducatelle R., Haesebrouck F., Deprez P., Pardon B. A deep nasopharyngeal swab versus nonendoscopic bronchoalveolar lavage for isolation of bacterial pathogens from preweaned calves with respiratory disease. J. Vet. Intern. Med. 2017;31:946–953. doi: 10.1111/jvim.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtala A.M., Grohn Y.T., Mechor G.D., Erb H.N. The effect of maternally derived immunoglobulin G on the risk of respiratory disease in heifers during the first 3 months of life. Prev. Vet. Med. 1999;39:25–37. doi: 10.1016/s0167-5877(98)00140-8. [DOI] [PubMed] [Google Scholar]
- Wisselink H.J., Cornelissen J., van der Wal F.J., Kooi E.A., Koene M.G., Bossers A., Smid B., de Bree F.M., Antonis A.F.G. Evaluation of a multiplex real-time PCR for detection of four bacterial agents commonly associated with bovine respiratory disease in bronchoalveolar lavage fluid. BMC Vet. Res. 2017;13:221. doi: 10.1186/s12917-017-1141-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Liu X., Liu M., Ma B., Xu L., Wang J. Development of a multiplex PCR for simultaneous detection of Pasteurella multocida, Mannheimia haemolytica and Trueperella pyogenes. Acta Vet. Hung. 2017;65:327–339. doi: 10.1556/004.2017.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.