Abstract
The role of viruses in Acute Exacerbations of Chronic Obstructive Pulmonary Disease (AECOPD) needs further elucidation. The aim of the present study was to evaluate the molecular epidemiology of viral pathogens in AECOPD.
Patients presenting to the Emergency Room with AECOPD needing hospitalization were recruited. Oropharyngeal and sputum samples were collected in order to perform microarrays-based viral testing for the detection of respiratory viruses.
A total of 200 (100%) patients were analyzed and from them in 107 (53.5%) a virus was detected. The commonest identified viruses were the human Respiratory Syncytial Virus (subtypes A and B) (40.5%), influenza virus (subtypes A, B, C) (11%), rhinovirus (8%) and human Parainfluenza Virus (subtypes A and B) (7.5%). A bacterial pathogen was isolated in 27 (14%) patients and a dual infection due to a bacterial and a viral pathogen was recognised in 14/107 patients. Patients with AECOPD and a viral infection had a lengthier hospital stay (9.2 ± 4.6 vs 7.6 ± 4.3, p < 0.01) while the severity of the disease was no related with significant differences among the groups of the study population.
In conclusion, the isolation of a virus was strongly associated with AECOPD in the examined population. The stage of COPD appeared to have no relation with the frequency of the isolated viruses while dual infection with a viral and a bacterial pathogen was not rare.
Keywords: AECOPD, Viral infections, Characteristics, Microarrays
1. Introduction
Chronic obstructive pulmonary disease (COPD) is frequently complicated by recurrent exacerbations that are called Acute Exacerbations of Chronic Obstructive Pulmonary Disease (AECOPD). These events are associated with high morbidity, increased health care expenditures and reduced health status of the patients [1], [2], [3], [4]. The main risk factor associated with AECOPD is the occurrence of a respiratory infection although multiple other factors contribute potentially to this process such as industrial pollutants, allergens, sedatives and comorbidities that are frequently present in these patients. Up to 60% of AECOPD are reportedly caused by a virus while bacterial infections mainly due to Haemophilus influenzae, Streptococcus pneumoniae, Moraxella catarrhalis, Haemophius parainfluenzae, Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Staphylococcus aureus are responsible for approximately 40% of those events [3], [5], [6].
The importance of viral infections in AECOPD has been recognized with the evolution of newer molecular diagnostic methods since traditional diagnostic techniques (e.g. direct immunofluorescence, culture etc) are considered slow regarding the final result, present a low sensitivity threshold profile and are unable to quickly identify common or emerging viruses [7], [8], [9]. Moreover traditional techniques may fail to identify specific pathogens such as human Metapneumovirus that is not detected by serology or viral cultures. The development of nucleic acid amplification tests has enabled timely and accurate detection of respiratory viruses. Such tests have now been developed for all respiratory viruses including both traditional and emerging strains. The recent development of DNA/RNA microarrays has enabled the identification of multiple gene targets from a single pathogen or multiple pathogens in a single clinical sample [10], [11].
The aim of this study was to assess the epidemiology of viral infections and their potential contribution to AECOPD in patients requiring hospitalization using a new PCR-based arrays technique.
2. Material and methods
2.1. Demographics
All patients consecutively examined at the Emergency Department (ER) of a tertiary care hospital (“SOTIRIA” Hospital, Athens, Greece) and diagnosed with an Acute Exacerbation of Chronic Obstructive Pulmonary Disease (AECOPD) were included. The study period extended from January 2008 to December 2009. All adult patients (>18 years) with a confirmed diagnosis of COPD (classified according to GOLD criteria) and requiring hospital admission because of AECOPD were included. An AECOPD was defined as an acute in onset event in the natural course of COPD characterized by a change in the patients “baseline” dyspnea, cough and sputum (beyond the normal day-to-day variation) usually requiring a modification in regular medication of a stable COPD course [4]. Patients with a confirmed diagnosis of bronchial asthma and with no available spirometric data were excluded. The study was approved by the ethical committee of the hospital and written informed consent was obtained from all recruited patients. Immediately after the initial evaluation at the ER a detailed medical history was obtained and patients’ demographics including age, sex, severity of the disease (COPD stage), smoking habits, need for oxygen therapy, influenza vaccination, comorbidities and use of inhaled steroids were recorded. All patients included in the study underwent as part of their evaluation a complete blood count, measurement of serum C - reactive protein (CRP) levels, a Chest-X-ray, an Arterial Blood Gases (ABGs) analysis and sputum culture was sent for common bacteria. Blood cultures were obtained if the responsible physician considered this appropriate. For the classification of the patients we used the available spirometric data from the last three months. Some of the patients were able to perform a spirometry during their initial evaluation. A spirometry (pre-post bronchodilator) was performed to all of them after their discharge from the hospital. These results were compared to the available spirometric results from the last three months. This way we were certain for the correct classification of our patients. Stage I (mild COPD) patients had an FEV1/FVC < 70% and FEV1 ≥ 80% of predicted values. Stage II (moderate COPD) patients had an FEV1/FVC < 70% and 50% ≤ FEV1 < 80% of predicted values. Stage III (severe COPD) patients had an FEV1/FVC < 70% and 30% ≤ FEV1 < 50% of predicted values and Stage IV(Very severe COPD) patients had an FEV1/FVC < 70% and FEV1 < 30% of predicted values or FEV1 < 50% of values predicted plus chronic respiratory failure (GOLD criteria) [12].
2.2. Microarrays technique
After the initial evaluation and before the administration of any treatment, sputum and oropharyngeal samples were collected for microarrays technique performance. For the detection of the viruses CLART® PneumoVir kit (GENOMICA, Spain) was used. This method detects the 17 most frequent types of human viruses causing respiratory infections by identifying minimum quantities of viral genomic material using a sequence corresponding to a highly preserved region within the viral genome and binding probes specific to each respiratory virus type. Viruses analyzed include, Influenza virus type A, B and C, Parainfluenza virus type 1, 2, 3 and 4 (subtypes A and B) [hPIV1–4], human Respiratory Syncytial Virus type A and B (hRSV type A and B), Rhinovirus, human Metapneumovirus (hMPV subtypes A and B) type A and B, Enterovirus (Echovirus), Adenovirus, Coronavirus and Bocavirus. The samples were collected in Thin Prep CytoLyt® solution and centrifuged at 2000 g. The molecular procedure was followed according to the manufacturer’s instructions. Briefly, viral DNA/RNA was extracted by using 200 μl of clinical sample mixed with lysis buffer and allowed to stand for 15 min followed by addition of isopropanol and centrifugation at 13,000 rpm for 20 min. The supernatant was removed and the precipitate was resuspended with 1000 μl 70% ethanol followed by centrifugation at 13,000 rpm for 15 min. The supernatant was removed again, the sample was left to dry for 15 min (until there were no ethanol residues left) and lastly the pellet was resuspended in 20 μl of dilution solution. The viral DNA/RNA extracts were stored at –20 °C until amplification. Virus amplification was performed via two RT (reverse transcriptase) Multiplex PCR reactions of a specific 120–330 bp fragment of the viral genome. The PCR employed the following thermal cycler settings: 1 cycle of 45 min at 45 °C and 15 min at 95 °C, followed by 45 cycles of 30 s at 95 °C, 1.5 min at 50 °C and 1 min at 68 °C and 1 final cycle of 10 min at 68 °C. The visualization of the amplified product was performed on a platform based on low-density micro-arrays, the so-called ArrayTube (AT). This detection system is based on the precipitation of an insoluble product at those sites of the AT where the hybridization of the products amplified by specific molecular probes is produced. During RT-PCR, amplified products are labeled with biotin. Following amplification, they hybridize with their respective specific probes that are immobilized in concrete and known sites of the AT, after which they are incubated with a streptavidin-peroxidase conjugate. The conjugate binds via streptavidin with the biotin present in the amplified products (which are also bound to their specific probes), while in the presence of o-dianisidine, the peroxidase activity of the conjugate induces the appearance of an insoluble product which precipitates at the hybridization sites of the AT.
2.3. Statistical analysis
Normally distributed continuous data were summarized by mean ± standard deviation (SD) whereas non-normally distributed data by median and interquartile range (IQR). Categorical data were summarized by rates. Non-parametric tests such as Mann Whitney assessed differences in continuous variables between COPD groups. Categorical variables were compared with chi-square test. A p value of less than 0.05 was considered significant. All tests were two-tailed. Data were analyzed using statistical software (SPSS, version 15.0; SPSS; Chicago,IL).
3. Results
A total of 200 patients met the inclusion criteria. At ER presentation all patients had increasing dyspnea, cough and production of purulent sputum while 20 (10%) of them reported fever and 5 (2.5%) other symptoms (somnolence or excitation). The reported by patients symptoms appeared on average 5 ± 2 days before their ER evaluation while 149 patients were able to perform a spirometry in the ER in order to be classified according to GOLD criteria. The rest 51 were classified using the most recent available spirometric data since they were unable to perform spirometry due to the severity of their disease. Comorbid conditions were noted in the following order of frequency: 100 (50%) patients reported a history of chronic cardiac disease, 58 (29%) patients had known diabetes mellitus and 23 (11.5%) patients had a history of chronic renal failure. In 107 (53.5%) patients a virus was identified by arrays in at least one collected specimen (either in orophryngeal or in sputum sample) while in 93(46.5%) no virus was isolated. A mixed infection with more than one viral pathogen was identified in 41 (38.3%) patients (Table 2 ). No differences were noted between the COPD groups with regards to the isolation of a virus. The groups differed in the length of hospital stay that was longer in COPD patients where a virus was isolated (p < 0.01) (Table 1). In 27 (13.5%) patients, a bacterial pathogen was identified in the sputum culture. Twelve (6%) patients did not survive (Table 1).
Table 2.
Positive samples for viruses from sputum and oropharyngeal specimens.
| Type of virus | Oropharyngeal Lavage, N = 94 | Sputum N = 96 | p value |
|---|---|---|---|
| hRSVA | 36(18%) | 29(14.4%) | 0.4 |
| hRSVB | 21(10.5%) | 21(10.5%) | 0.8 |
| Adenovirus | 1(0.5%) | 3(1.5%) | 0.6 |
| Coronavirus | 3(1.5%) | 2(1%) | 0.3 |
| Rhinovirus | 8(4%) | 9(4.5%) | 1.0 |
| hPIV3 | 4(2%) | 11(5.5%) | 0.1 |
| hPIV4 | 0 | 2(1%) | 0.4 |
| Echovirus | 1(0.5%) | 1(0.5%) | 0.4 |
| Enterovirus B | 1(0.5%) | 0 | 1.0 |
| hMPVA | 3(1.5%) | 1(0.5%) | 0.6 |
| hMPVB | 0 | 1(0.5%) | 1.0 |
| Influenza A | 6(3%) | 6(3%) | 0.7 |
| Influenza B | 4(2%) | 7(3.5%) | 0.5 |
| Influenza C | 4(2%) | 0 | 0.1 |
hRSVA = human Respiratory Suncytial Virus A, hRSVB = human Respiratory Suncytial Virus B, hPIV3 = human Parainfluenza Virus type 3, hPIV4 = human Parainfluenza Virus type 4, hMPVA = human Metapneumovirus type A, hMPVB = human Metapneumovirus type B p < 0.05 = statistical significant.
Table 1.
All AECOPD patients with and without a virus detection.
| Parameters | All patients n = 200(100%) | Patients with virus detection n = 107(53.5%) | Patients without virus detection n = 93 (46.5%) | p valuesa |
|---|---|---|---|---|
| Demographics | ||||
| Age (years) | 69.7 ± 9.1 | 70.6 ± 9.2 | 68.8 ± 8.9 | 0.1 |
| Males | 150(75%) | 77(72%) | 73(78.5%) | 0.2 |
| Oxygen therapy | 91(45.5%) | 52(48.6%) | 39(41.9%) | 0.3 |
| Influenza vaccination | 89(44.5%) | 51(47.7%) | 40(43%) | 0.5 |
| Pack years | 65 ± 38 | 60.6 ± 38.3 | 71.4 ± 37.3 | 0.05 |
| COPD (years) | 6.41 ± 6.45 | 6.6 ± 6.8 | 5.8 ± 6.03 | 0.4 |
| Inhaled steroids | 131(65.5%) | 71(66.4%) | 57(61.3%) | 0.4 |
| Comorbidities | ||||
| 1–2 comorbidities | 109(54.5%) | 58(54.2%) | 51(54.8%) | 0.9 |
| >2 comorbidities | 10(0.05%) | 8(7.45%) | 2(2.2%) | 0.1 |
| Laboratory findings | ||||
| PO2 (mmHg) | 58 ± 12.57 | 57.3 ± 12.9 | 58.9 ± 12.2 | 0.3 |
| PCO2 (mmHg) | 46 ± 15.43 | 45.6 ± 13.3 | 47.2 ± 17.6 | 0.4 |
| pH | 7.41 ± 0.06 | 7.42 ± 0.6 | 7.42 ± 0.6 | 0.9 |
| HCO3 (mmMol) | 29 ± 6.2 | 29.3 ± 5.7 | 29.4 ± 6.7 | 0.9 |
| WBC(c/mm3 × 103) | 10.9 ± 4.5 | 10.6 ± 4.3 | 11.3 ± 4.6 | 0.2 |
| CRP (mg/dl) | 3.8 ± 5.3 | 3.9 ± 4.9 | 3.8 ± 5.6 | 0.9 |
| Microbiology | ||||
| Positive bacterial cultures | 27(13.5%) | 14(13.1%) | 13(14%) | 0.8 |
| Positive CxRb | 34(17%) | 18(16.8%) | 16(17.2%) | 0.9 |
| Length of stay (days) | 8.4 ± 4.5 | 9.2 ± 4.6 | 7.6 ± 4.3 | 0.01a |
| Outcome | ||||
| Survived | 188(94%) | 102(95.3%) | 86(92.5%) | 0.3 |
| Non-Survived | 12(6%) | 5(4.7%) | 7(7.5%) | 0.3 |
AECOPD = Acute Exacerbation of Chronic Obstructive Pulmonary Disease, COPD = Chronic Obstructive Pulmonary Disease, WBC = White Blood Cells, CRP = C-Reacting Protein.
Values are expressed as percentage or ±SD.
p < 0.05 is considered statistical significant for comparisons between patients with and without a virus detection.
Patients with findings in CxR consisted with pneumonia.
The isolated viruses in order of frequency were human Respiratory Suncytial Virus (hRSV) subtypes A and B (40.5%), influenza type A, B and C virus (12%), rhinovirus (8%), human Parainfluenza (hPIV) subtypes 3 and 4 (7.5%), coronavirus (2%), human Metapneumovirus (hMPVA) subtype A and B (2%), adenovirus (2%), enterovirus (0.5%) (Fig. 1 ). Significant discrepancies were noted in the isolation of the viruses between oropharyngeal and sputum samples. RSV was detected by arrays in 33 (55.9%) of the oropharyngeal samples where no virus was detected in the concomitantly taken sputum sample. Conversely RSV was identified in 33 (68.8%) of the collected sputum samples whereas it was not found in the simultaneously taken samples from the oropharynx. In 15 cases overall (14%) the virus was found from both samples. Rhinovirus was most successfully isolated from the oropharyngeal samples whereas hPIV and influenza viruses from the sputum (data not shown). The vast majority of positive samples were noted from January through April and in October.
Fig. 1.
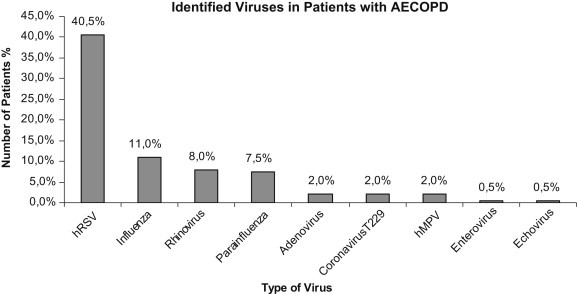
Percentage of isolated virus.
Patients’ characteristics according to COPD stage are shown in Table 3 . Specifically COPD patients of stage III/IV were more frequently O2 dependent (61.5% vs 6.66%; p < 0.001), smoked more as expressed in pack years (52 vs. 56.9 vs. 69.5; p < 0.03), had longer duration of COPD course (2.4 yrs vs. 4.35 yrs vs. 7.2 yrs; p < 0.001) and used more inhaled steroids (3 vs. 22 vs. 103; p < 0.001) than stage I and II patients. Stage III/IV COPD patients had more comorbidities in comparison with patients with a less severe disease (5% vs. 17% vs. 87%; p < 0.01) while significant differences were detected for PaCO2, pH and HCO3 values (36 vs. 38 vs. 49; p < 0.001, 7.46 vs. 7.43 vs. 7.40; p = 0.01 and 27 vs. 25 vs. 30; p < 0.001). The severity of the disease did not correlate with a statistically significant increased detection rate of viruses (p < 0.7, Table 3, Table 4 ). From the 200 patients recruited in our study only 7 died. As stated in Table 1 death was not related to the presence of a viral infection (p = 0.3) and as stated from Table 3 it was also not related to the stage of the disease (p = 0.2). The only risk factor we found to be related with non-survival was the use of oxygen. It seemed that patients who did not survive were more oxygen dependent and this result was statistically important (p = 0.001). No other comorbidity (ex cardiac disease, renal failure or diabetes mellitus) seemed to be a risk factor for negative outcome.
Table 3.
All AECOPD patients with and without virus isolated according to the severity of the disease.
| Parameters n (%) | Total Number | Stage I | Stage II | Stage III/IV | p valuea |
|---|---|---|---|---|---|
| Demographics | |||||
| Number of patients | 200(100%) | 12(6%) | 45(22.5%) | 143(71.5%) | <0.001a |
| Age (years) | 69.7 ± 9.1 | 67.1 ± 12.4 | 68.8 ± 10.7 | 70.2 ± 8.2 | 0.4 |
| Sex (Males) | 150(75%) | 10(83.3%) | 35(77.8%) | 105(73.4%) | 0.4 |
| Oxygen therapy | 91(45.5%) | 0 | 3 (6.6%) | 88(61.5%) | <0.001a |
| Influenza vaccination | 89(44.5%) | 5(41.6%) | 19(42.2%) | 76(53.1%) | 0.8 |
| Pack years | 65 ± 38 | 52 ± 30 | 56.9 ± 40 | 69.5 ± 41 | 0.03 |
| COPD (years) | 6.41 ± 6.4 | 2.4 ± 2.1 | 4.35 ± 4.1 | 7.2 ± 7 | <0.001a |
| Inhaled steroids | 131(65.5%) | 3(25%) | 22(48.9%) | 103(72%) | <0.001a |
| Comorbidities | |||||
| 1–2 Comorbidities | 109(54.5%) | 5(41.9%) | 17(37.8%) | 87(60.8%) | 0.01a |
| More than 2 | 10(0.05%) | – | 4(8.9%) | 6(4.2%) | 0.2 |
| Laboratory findings | |||||
| PO2 (mmHg) | 58 ± 12.5 | 64 ± 11 | 58 ± 8 | 57 ± 13 | 0.1 |
| PCO2 (mmHg) | 46 ± 15.4 | 36 ± 4 | 38 ± 7 | 49 ± 16 | <0.001a |
| pH | 7.41 ± 0.06 | 7.46 ± 0.03 | 7.43 ± 0.06 | 7.40 ± 0.06 | 0.01 |
| HCO3 (mmMol) | 29 ± 6.2 | 27 ± 4 | 25 ± 3 | 30 ± 6 | <0.001a |
| WBC(c/mm3 × 103) | 10.9 ± 4.5 | 8.4 ± 2.8 | 11.3 ± 4.3 | 11.0 ± 4.6 | 0.6 |
| CRP (mg/dl) | 3.8 ± 5.3 | 1.8 ± 2.5 | 4.1 ± 4.4 | 3.9 ± 5.7 | 0.9 |
| Microbiology | |||||
| Infection with a virus | 107(53.5%) | 5(41.6%) | 24(53.3%) | 78(54.5%) | 0.7 |
| Infection with a bacterial | 27(13.5%) | – | 3(6.6%) | 24(16.8%) | 0.01a |
| Dual infection (virus and bacterial) | 14(7%) | – | 1(2.2%) | 13(9.1%) | 0.1 |
| Positive CxRb | 34(17%) | 5(41.7%) | 8(17.8%) | 21(14.7%) | 0.05 |
| Length of stay (days) | 8.4 ± 4.5 | 6.3 ± 1.9 | 7.8 ± 3.4 | 8.9 ± 4.9 | 0.08 |
| Outcome | |||||
| Survived | 188(94%) | 12(100%) | 44(97.8%) | 132(92.3%) | 0.2 |
| Non – Survived | 12(6%) | – | 1(2.2%) | 11(7.9%) | 0.2 |
AECOPD = Acute Exacerbation Chronic Obstructive Pulmonary Disease, COPD = Chronic Obstructive Pulmonary Disease, WBC = White Blood Cells, CRP = C-Reacting Protein.
p < 0.05 is considered statistical significant for comparisons between COPD Stage III/IV and Stage I and II.
Number of patients with findings in CxR consisted with pneumonia, values are expressed as percentage or ±SD.
Table 4.
All detected viruses according to COPD stage.
| Type of virus | Stage of COPD |
|||
|---|---|---|---|---|
| Total number n = 200 (100%) | Stage I/II n = 57 (28.5%) | Stage III/IV n = 143(71.5%) | p values | |
| hRSVA | 56(28) | 14(24.6) | 42(29.4) | – |
| hRSVB | 38(19) | 8(14) | 30(21) | – |
| hRSV total | 81(40.5)a | 22(38.6) | 59(41.3) | 0.8 |
| Influenza A | 11(5.5) | 2(3.5) | 9(6.3) | 0.6 |
| Influenza B | 9(4.5) | 2(3.5) | 7(4.9) | 0.6 |
| Influenza C | 4(2) | 0 | 4(2.8) | 0.4 |
| Influenza total | 22(12)a | 4(7) | 18(12.6) | 0.3 |
| Rhinovirus | 16(8) | 3(5.3) | 13(9.1) | 0.4 |
| hPIV3 | 14(7) | 4(7) | 10(6) | – |
| hPIV4 | 2(1) | 1(1.8) | 1(0.7) | – |
| hPIV total | 15(7.5)a | 5(8.8) | 10(7) | 0.8 |
| Coronavirus | 4(2) | 0 | 4(2.8) | 0.3 |
| hMPVA | 3(1.5) | 2(3.5) | 1(0.7) | 0.1 |
| hMPVB | 1(0.5) | 0 | 1(0.7) | 0.8 |
| hMPV total | 4(2) | 2(3.5) | 2(1.4) | 0.5 |
| Adenovirus | 4(2) | 1(1.8) | 3(2) | 1 |
| Echovirus | 1(0.5) | 0 | 1(07) | 1 |
| Enterovirus B | 1(0.5) | 0 | 1(0.7) | 1 |
| >2 viruses | 41(20.5) | 9(15.8) | 32(22.4) | 0.3 |
hRSVA,B = human Respiratory Suncytial Virus A and B, hPIV3,4 = human Parainfluenza Virus type 3, 4 = human Parainfluenza Virus type 4, hMPVA,B = human Metapneumovirus type A, B.
p < 0.05 is considered statistical significant for comparisons between Stage III/IV and Stage I/II.
Subtypes of the virus have been detected both in the same patients.
In 14 (13.1%) AECOPD patients where a virus was isolated a simultaneous positive bacterial culture was noted. More specifically in 8 (4%) patients Pseudomonas aeruginosa was cultured, in 4 (2%) Haemophilus influenzae, in 4(2%) Klebsiella pneumoniae, in 4 (2%) Staphylococcus aureus, in 3 (1.5%) Acinetobacter baumanii, in 2 (1%) Stenotrophomomonas maltophilia and in 3 (1.5%) other bacteria (in one patient 2 bacterial pathogens were identified) (Table 5 ). Bacterial cultures were more frequently positive in patients with COPD of stage III/IV than in COPD patients of stage I and II (3vs24; p < 0.01) (Table 1, Table 3).
Table 5.
Positive sputum cultures for bacterial pathogens in patients with a positive PCR for a virus.
| COPD |
Stage I |
Stage II |
Stage III/IV |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria |
SA | HI | KP | PA | SM | Sm | AB | SA | HI | KP | PA | SM | Sm | AB | SA | HI | KP | PA | SM | Sm | AB |
| Virus | |||||||||||||||||||||
| hRSVA | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 2 | 1 | 2 | 1 | – | 1 | |
| hRSVB | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | 1 | – | – | 1 |
| Adenovirus | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – |
| Coronavirus | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 |
| Rhinovirus | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | 1 | – | – | – | – |
| hPIV3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – |
| PIV3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| PIV4 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – |
| Echovirus | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – |
| Enterovirus B | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – |
| hMPVA | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – |
| hMPVB | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – |
| Influenza A | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Influenza B | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Influenza C | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
SA = Staphylococcus aureus, HI = Haemophilus influenzae, KP = Klebsiella pneumoniae, PA = Pseudomonas aeruginosa, SM = Stenotrophomonas maltophilia, Sm = Serratia marcescens, AB = Acinetobacter baumanii, hRSVA = human Respiratory Suncytial Virus A, hRSVB = human Respiratory Suncytial Virus B, hPIV3 = human Parainfluenza Virus type 3, PIV3 = Parainfluenza Virus type 3, PIV4 = Parainfluenza Virus type 4, hMPVA = human Metapneumovirus type A, hMPVB = human Metapneumovirus type B, the numbers indicate the positive samples for a certain virus.
4. Discussion
In our study we have included a large number of patients from Greece who were admitted in the hospital with severe exacerbation of COPD. We examined samples from both upper and lower respiratory tract and we compared the results with those of the literature. This limited small study is the first to our knowledge to assess the epidemiology of respiratory viruses in this population in our country. The main findings of this study are that a) in AECOPD patients requiring hospitalization high rates of respiratory virus infections were detected, b) a higher frequency of isolation was noted for specific viruses and c) the severity of the disease was not associated with a higher detection rate of viruses.
It is well known that up to 60% of AECOPD are triggered by viral infections that are more prevalent in the winter months, and cause severe exacerbations characterized by a longer recovery period than the exacerbations triggered by other factors [13], [14]. In our study a high detection rate of viruses was noted as a result of the increased sensitivity of the applied PCR-based technique. The available diagnostic methods (serology, viral cultures, PCR) present different rates of sensitivity in detecting respiratory viruses. This sensitivity varies depending on the sample obtained and the tropism of the virus (nasopharyngeal aspirates, nasal, oropharyngeal, nose-throat swabs). This variability was observed in our study. Others have claimed that the use of PCR or real-time PCR techniques may help in overcoming such an effect [5], [15]. The role of virus tropism may be a significant factor however the highest rates of rhinoviruses were discovered in the oropharyngeal and not in the sputum samples in our studies (we did not study nasal secretions). Alternatively if a sputum sample does not have the same virus with a concomitant positive oropharyngeal sample this may denote an upper but not a lower airway infection. Clearly further work should look into the explanation of similar discrepancies. The detection rate of respiratory viruses in our study (53.5%) is comparable with other studies although some differences could be explained by the different period of sampling [16], [17]. The prolonged length of hospital stay in these patients could be explained by the more intense systemic inflammatory response characterized mainly by increased CRP and blood leucocytes [18], [19]. However, neither CRP nor the number of blood leucocytes was significantly different between patients with and without a virus.
The commonest isolated viruses in our cohort were hRSV type A and B, influenza virus and rhinovirus. The role of hRSV is well recognized as a major cause of respiratory infection in infants/young children and a significant cause of morbidity and mortality in adults and COPD patients [20], [21], [22]. In our study hRSV was the predominant virus detected in both oropharyngeal and sputum samples. This could be explained by the fact that after an acute respiratory infection with hRSV the virus frequently persists in the respiratory tract with a low viral load and can be detected later in patients with a stable COPD leading to a further decline in lung function and more frequent exacerbations. In our patients a quantitative PCR was not available in order to identify the viral load and for this reason we are not able to clarify if this virus had actually a causal relation to AECOPD or reflected a more chronic colonization in those patients. The relatively high rate of influenza virus isolation (11%) could be explained by the observed lower influenza vaccination rate in relation to other studies [5], [14]. However, no differences were noted between the immunized and non-immunized patients probably because of the varied antibody response of the vaccinated subjects. Rhinovirus is a major cause of common cold considered as a major pathogen for AECB. In our study the detection rate (8%) was smaller than for hRSV and influenza virus probably because of seasonal variability in the circulation of these viruses although we performed the study over a two-year period in order to avoid seasonal bias [14]. Most of our positive samples were collected during the period of late Autumn and Winter. Human metapneumovirus is a newly emerging pathogen initially detected in Dutch children with bronchiolitis and is considered most closely to hRSV [23]. In our study this virus was identified in 4 patients (2%) presenting a detection rate similar to the one reported by Rohde et al (2.3%). Asymptomatic carriage of this virus is unlikely while it is detectable only in AECOPD and during the winter [23], [24].
No correlation between the isolation rate of respiratory viruses and the severity of the disease in patients with AECOPD was identified. Although the available data is limited, McManus et al reported that COPD patients do not have a higher carriage rate of viruses in comparison to healthy individuals [17]. This could be explained by the delayed seeking of medical care from patients in relation to the appearance of the symptoms and the seasonality of respiratory viral infections. On the other hand the isolation rate of bacterial pathogens in our study was lower in total (13.5%) than in other studies. No differences were observed between AECOPD patients with and without a viral detection but the detection of a bacterial pathogen was higher in patients with more severe disease (COPD stages III/IV) (16.8% vs. 6.6%). The low detected rate of bacterial pathogens in relation to the other reports could be explained by the low yield from sputum cultures that could also be laboratory dependent. On the other hand the higher detected rate in patients with more severe disease (16.8% vs. 6.6%) could be explained by the fact that patients with severe COPD have more frequent hospitalizations and excessive use of antibiotics (as outpatients, although this approach is not considered appropriate) that make them more susceptible to subsequent bacterial infections with multiresistant pathogen. Thus the bacterial data should be interpreted with caution. The low rate of bacterial detection is also the reason we did not perform further analysis examining bacterial infections. The commonest bacterial pathogen was P. aeruginosa probably because the majority of the recruited patients belonged in COPD stage III and IV. Mixed infection with a bacterial and a viral pathogen was identified in a small percentage in our cohort. In these patients P. aeruginosa and H. influenzae were also the commonest detected bacterial pathogens. A dual infection with a bacterial pathogen and hRSV was more frequently observed than with other viruses. This could be explained by the fact that hRSV like other viruses alters the expression of receptor molecules on respiratory epithelial cells allowing therefore for an increased bacterial adherence and invasion. Additionally a protein called hRSV G works as a cell receptor for H. influenzae and Streptococcus pneumoniae facilitating the bacterial binding to cells infected by a virus. Alternatively, an antecedent bacterial infection may increase the susceptibility to viral infection by increasing expression of host-cell molecules that bind viruses [25], [26], [27], [28].
The PCR-based techniques seem to have higher yield in recovering viruses from sputum rather than from nasal aspirates [29], [30]. This was not seen in our study. However, one of the main drawbacks of the detection by genetic amplification is the frequently observed false negative results. These are attributed to: a) a poor quality of the extracted DNA/RNA (caused by an insufficient quantity of the initial sample, or a degradation of the virus genetic material due to the inadequate storage of the said sample or its loss during its extraction); and b) the presence of inhibitors of the enzyme mixture (RT and DNA polymerase) in those samples where virus detection is going to be performed (hemoglobin, salts, etc.). The false negative results seem to be eliminated thanks to the addition of an internal control in the clinical sample [10] confirming the correct efficiency of both the extraction and the PCR amplification reaction. In our cohort, we used a PCR-based arrays technique able to detect the 17 frequent types of human viruses which potentially can cause respiratory infections. This assay needs to be further validated in prospective studies.
The present study has a number of limitations. First, a quantitative PCR was not available in order to estimate the viral load and to clarify if the patients were carriers of a specific virus (especially hRSV) or if they had a true infection. In our study no patients with stable COPD were recruited as control subjects. Thus it was not sure whether the viruses that were identified in the sputum or oropharyngeal sample were the cause of the exacerbation or whether they represented chronic colonization/infection. In previous studies (Seemungal et al) non-RSV respiratory viruses were detected in 11% and RSV in 23% of stable COPD patients. As the rate of virus identification in patients with stable COPD was much lower than during exacerbation and most RSV infections were associated with symptomatic respiratory illness together with more intense inflammatory response (the RSV group of patients was characterized by increased CRP levels (4.03 ± 4.9-normal value<0.5) and number of leucocytes (11004.89 ± 4499.73-normal values 4000–10,000), it is highly likely that viral infections play an important role in triggering COPD exacerbations. Moreover, we could not adequately evaluate differences in positivity rates between the samples tested. The differences observed might have been even greater with nasal sampling. Larger studies evaluating sampling from several sites taking into account viral tropism for specific sites of the respiratory tract will assist in this regard. Furthermore, due to the design of the study there was no information provided concerning the specificity of the test for each of the viruses and for positive and negative controls. A significant limitation of our study is the lack of a baseline (recovery/convalescent) evaluation in stable conditions. In the absence of such a control condition, the findings at exacerbation should be carefully interpreted, since it cannot be excluded that pathogens were already present in stability. Lastly we could not account for any residual confounding with regards to risk factors for virus isolation.
5. Conclusions
In conclusion, using a molecular assays method we evaluated the viral epidemiology of AECOPD patients in a single center. Viral infections were strongly related to AECOPD and were associated with lengthier hospital stay in our patients. However, the stage of COPD was not related to the frequency of virus isolation. Further studies regarding the role of viral infection in COPD course are necessary in order to develop new algorithms that will be more pathogen-directed for the treatment of COPD exacerbations. This will lead ease the health burden of AECOPD and change the quality of life of these patients.
References
- 1.Rabe K.F., Hurd S., Anzueto A., Barnes P.J., Buist S.A., Calverley P. Global strategy for the diagnosis, management and prevention of COPD-2006 update. Am J Respir Crit Care Med. 2007;176(6):532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 2.MacNee W. Acute exacerbations of COPD. Swiss Med Weekly. 2003;133:247–257. doi: 10.4414/smw.2003.10025. [DOI] [PubMed] [Google Scholar]
- 3.Sapey E., Stockley R.A. COPD exacerbations 2: aetiology. Thorax. 2006;61(3):250–258. doi: 10.1136/thx.2005.041822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burge S., Wedzicha J.A. COPD exacerbations: definitions and classifications. Eur Respir J Suppl. 2003;41:46s–53s. doi: 10.1183/09031936.03.00078002. [DOI] [PubMed] [Google Scholar]
- 5.Papi A., Bellettato C.M., Braccioni F., Romagnoli M., Casolari P., Caramori G. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am J Respir Crit Care Med. 2006;173(10):1114–1121. doi: 10.1164/rccm.200506-859OC. [DOI] [PubMed] [Google Scholar]
- 6.Sykes A., Mallia P., Johnston S.L. Diagnosis of pathogens in exacerbations of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2007;4(8):642–646. doi: 10.1513/pats.200707-101TH. [DOI] [PubMed] [Google Scholar]
- 7.Smith C.B., Kanner R.E., Golden C.A., Klauber M.R., Renzetti A.D., Jr. Effect of viral infections on pulmonary function in patients with chronic obstructive pulmonary disease. J Infect Dis. 1980;141(3):271–280. doi: 10.1093/infdis/141.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethi S. Infectious etiology of AECB. Chest. 2000;117(5 Suppl. 2):s380–s385. doi: 10.1378/chest.117.5_suppl_2.380s. [DOI] [PubMed] [Google Scholar]
- 9.Henrickson K.J. Advances in laboratory diagnosis of viral respiratory disease. Pediatr Infect Dis J. 2004;23(1 Suppl.):S6–S10. doi: 10.1097/01.inf.0000108187.63151.ea. [DOI] [PubMed] [Google Scholar]
- 10.Chin C.Y., Urisman A., Greenhow T.L., Rouskin S., Yagi S., Schnurr D. Utility of DNA microarrays for detection of viruses in acute respiratory tract infections in children. J Pediatr. 2008;153(1):76–83. doi: 10.1016/j.jpeds.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernet G. Use of molecular assays for the diagnosis of influenza. Expert Rev Anti-infective Ther. 2007;15(1):89–104. doi: 10.1586/14787210.5.1.89. [DOI] [PubMed] [Google Scholar]
- 12.Global Initiative for Chronic Obstructive Lung Disease. Pocket guide to COPD diagnosis, management and prevention. A guide for healthcare professionals. A 2008 Update. Available from: http/www.goldcopd.org/Guidelineitem.asp?l1=2&;12=1&intId=996 (accessed 24.04.09).
- 13.Wedzicha J.A., Semungal T.A.R. COPD exacerbations defining their cause and prevention. Lancet. 2007;370:786–796. doi: 10.1016/S0140-6736(07)61382-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seemungal T., Harper-Owen R., Bhowmik A., Moric I., Sanderson G., Message S. Respiratory viruses, symptoms and inflammatory markers in acute exacerbations and stable chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;164(9):1618–1623. doi: 10.1164/ajrccm.164.9.2105011. [DOI] [PubMed] [Google Scholar]
- 15.Meerhoff I.J., Houben M.L., Coenjnerts F.E.J., Kimpen J.I., Hofland R.W., Schellevis F. Detection of multiple respiratory pathogens during primary respiratory infection: nasal swab versus nasopharyngeal aspirate using real-time polymerase chain reaction. Eur J Clin Microb Infect Dis. 2010;29:365–371. doi: 10.1007/s10096-009-0865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohde G., Wiethege A., Borg I., Kauth M., Bauer T.T., Gillissen A. Respiratory viruses in exacerbations of chronic obstructive pulmonary disease requiring hospitalization: a case control study. Thorax. 2003;58(1):37–42. doi: 10.1136/thorax.58.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McManus T.E., Mariey A.M., Baxter N., Christie S.N., O’ Neil H.J., Elborn J.S. Respiratory viral infection in exacerbation of COPD. Respir Med. 2008;102:1575–1580. doi: 10.1016/j.rmed.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohde G., Borg I., Wiethege A., Kauth M., Jerzinowski S., Duong Dihn Tan. Inflammatory response in acute viral exacerbations of COPD. Infection. 2008;36:427–433. doi: 10.1007/s15010-008-7327-5. [DOI] [PubMed] [Google Scholar]
- 19.Sethi S., Murphy T.F. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med. 2008;359(22):2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- 20.Falsey A.R., Hennessey P.A., Formica M.A., Cox C., Walsh E.E. Respiratory syncytial virus infection in elderly and high risk patients. N Engl J Med. 2005;352(17):1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson T.M., Donaldson G.C., Johnston S.L., Opens haw P.J., Wedzicha J.A. Respiratory syncytial virus, airway inflammation and FEV1 decline in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(8):871–876. doi: 10.1164/rccm.200509-1489OC. [DOI] [PubMed] [Google Scholar]
- 22.Ramaswamy M., Groskreutz D.J., Look D.C. Recognizing the importance of respiratory syncytial virus in COPD. COPD. 2009;6(1):64–75. doi: 10.1080/15412550902724024. [DOI] [PubMed] [Google Scholar]
- 23.Rohde G., Borg I., Arinir U., Kronsbein J., Rausse R., Bauer T.T. Relevance of human metapneumovirus in exacerbations of chronic obstructive pulmonary disease. Respir Res. 2005;21(6):150. doi: 10.1186/1465-9921-6-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinello R.A., Esper F., Weibel C., Ferguson D., Landry M.L., Kahn J.S. Human meta-pneumovirus and exacerbations of COPD. J Infect. 2006;53(4):248–254. doi: 10.1016/j.jinf.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy T.F., Brauer A.L., Schiffmacher A.T., Sethi S. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;170(3):266–272. doi: 10.1164/rccm.200403-354OC. [DOI] [PubMed] [Google Scholar]
- 26.Avadhanula V., Wang Y., Portner A., Adderson E. Haemophilus Influenzae and Streptococcus pneumoniae bind respiratory syncytial virus glycoprotein. J Med Microbiol. 2007;56:1133–1137. doi: 10.1099/jmm.0.47086-0. [DOI] [PubMed] [Google Scholar]
- 27.Avadhanula V., Rodriguez C.A., Devincenzo J.P. Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species and cell type-dependent manner. J Virol. 2006;80:1629–1636. doi: 10.1128/JVI.80.4.1629-1636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sajjan U.S., Jia Y., Newcomb D.C., Bentley J.K., Lukacs N.W., LiPuma J.J. H.influenzae potentiates airway epithelial cell responses to rhinovirus by increasing ICAM-1 and TLR3 expression. FASEB J. 2006;20:2121–2123. doi: 10.1096/fj.06-5806fje. [DOI] [PubMed] [Google Scholar]
- 29.Falsey A.R., Formica M.A., Treanor J.J., Walsh E.E. Comparison of a quantitative reverse transcription-PCR to viral culture for assessment of respiratory syncytial virus shedding. J Clin Microbiol. 2003;41(9):4160–4165. doi: 10.1128/JCM.41.9.4160-4165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borg I., Rohde G., Löseke S., Bittscheidt J., Schultze-Werninghaus G. Evaluation of a quantitative real-time PCR for the detection of respiratory syncytial virus. Eur. Respir. J. 2003 doi: 10.1183/09031936.03.00088102. [DOI] [PubMed] [Google Scholar]


