Abstract
Objective
We examined the feasibility of combining communication by e-mail and self-collection of nasal swabs for the prospective detection of acute respiratory infections in a non-medical setting.
Methods
The study was conducted among a convenience sample of employees (n = 53) at a research institution (December 2009–April 2010). Real-time data on the occurrence of acute respiratory symptoms and a nasal self-swab were collected prospectively, with automated weekly e-mails as a reminder mechanism. Reverse transcription polymerase chain reaction (RT-PCR) was used to detect respiratory viral pathogens in the swabs.
Results
Fifty-one out of 53 participants completed the study. The study design was well accepted. Thirty (∼57%) participants reported at least one episode of acute respiratory infection and returned the nasal swab during the study period (eight participants reported two episodes). The majority had no difficulties taking the self-swab and preferred this to swabbing by study personnel. Most participants obtained and returned the swabs within the recommended time. Viral respiratory pathogens were detected in 19 of 38 swabs (50%), with coronaviruses 229E/NL63 and OC43 and rhinoviruses A and B constituting 17 positive swabs (89%).
Conclusions
Combining e-mail-based symptomatic surveillance with nasal self-swabbing promises to be a powerful tool for the real-time identification of incident cases of acute respiratory infections and the associated pathogens in population-based studies.
Keywords: Acute respiratory infections, Self-collected nasal swabs, Modern communication tools
1. Introduction
Research on acute respiratory infections in human populations is limited by certain methodological difficulties. First, their acute nature makes a timely diagnosis difficult. Second, symptoms are usually not unique for specific pathogens. These difficulties impede collecting epidemiologic (e.g., risk factors for acute respiratory infections) and clinical (e.g., the course and severity of infections) data, as well as biosamples for pathogen identification. In particular, the real-time collection of diagnostic specimens such as nasal or nasopharyngeal swabs during an acute respiratory infection season is necessary to link symptomatic data with specific pathogens.1 Therefore, there is an urgent need to develop epidemiologic research tools that ensure the timely detection of incident acute respiratory infections and the collection of diagnostic biosamples during the episode.
Most epidemiologic studies on acute respiratory infections have been based in medical settings or have been conducted in specific target populations such as trained medical personnel. Data on the occurrence and severity of symptoms have been collected in a few studies only, either retrospectively,2 usually at the end of an acute respiratory infection season, or prospectively, for instance by using diary-based questionnaires.3, 4 Recently, modern communication tools such as weekly e-mails5 and internet-based questionnaires6, 7 have been introduced in population-based studies to collect real-time data on respiratory infections. In a recent study of influenza infection, Short Message Service (SMS) was used in addition to e-mail.8 The main limitation of these studies is the lack of pathogen identification during specific episodes of acute respiratory infections. Nasal swabs for pathogen detection are usually collected by study personnel at the study site or in hospital. However, due to logistic problems and higher costs, this is difficult to organize in population-based studies with their inherently larger sample sizes. In several recent studies subjects were asked to obtain swabs from their own nares (‘self-swabbing’) to detect viral respiratory pathogens, but most of these studies were performed in healthcare environments. For instance, parents collected swabs from their symptomatic children when presenting for pediatric medical evaluation,9 or nurses self-swabbed during symptomatic episodes in a study comparing two measures for the prevention of influenza transmission.10 In the UK, swabs for the collection of nasal specimens were sent to individuals who contacted the health advice and information service (‘NHS Direct’) because of influenza-like symptoms.11 The only population-based study was conducted among parents who collected nasal swabs from their children at home.12 Thus, the feasibility of nasal self-swabbing for the detection of respiratory pathogens in population-based studies of adults remains to be demonstrated. Moreover, little is known about whether any added benefit results when active symptomatic surveillance is conducted to ensure the timely self-collection of swabs during the time window in which causative pathogens are detectable. We therefore examined the feasibility of combining e-mail-based active symptomatic surveillance with nasal self-swabbing for the detection of viral respiratory pathogens in a prospective study spanning one acute respiratory infection season.
2. Methods
2.1. Sample and study design
We conducted a prospective study among employees of the Helmholtz Centre for Infection Research (HZI) in Braunschweig, Germany, from December 2009 to April/May 2010. In December 2009, invitations to participate in the study were sent to all employees (age 18–69 years) through the internal e-mail system. This invitation contained a link to the institutional intranet where information about the study was made available. Subjects not eligible for study participation were those vaccinated against seasonal influenza in the season 2009/2010, staff of the Department of Infection Genetics (due to ethical considerations), and those who planned to leave Braunschweig during the study period. The study was approved by the Ethics Committee of the State Board of Physicians of the German Federal State of Lower Saxony.
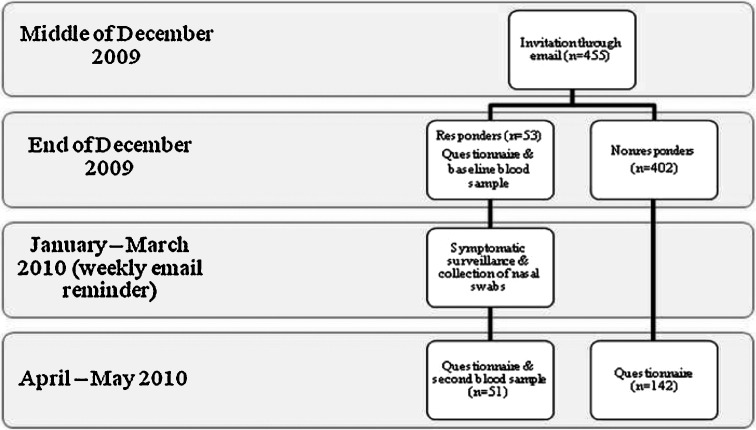
Fifty-three participants responded to the invitation e-mail, corresponding to a response rate of approximately 12% (Figure 1 ). All subjects gave written informed consent before entering the study. At baseline (December 2009), information on socio-demographics (sex, age, education, profession, country of birth, number of individuals living in the household, etc.), contacts with animals, history of vaccination against influenza, and general health status was collected through a self-administered questionnaire. The pre-season blood sample was also obtained at this time. In April/May 2010 the study participants were reinvited to give the post-season blood sample and fill in a short questionnaire. Every participant who completed the post-study questionnaire and blood sampling received a remuneration of 20 €. Serum samples were stored at −70 °C. However, at the conclusion of the study it was decided to forgo the originally planned influenza hemagglutination inhibition assays because of the unexpectedly low incidence of influenza infection in Germany during the study period.
Figure 1.
Presentation of the study design.
2.2. Surveillance of acute respiratory symptoms and collection of nasal specimens
During January–March 2010 the participants were asked to take a swab from one of the anterior nares and return it to the study site as soon as possible if they had at least one of the following acute respiratory symptoms: sudden onset of stuffy or running nose, cough, sore throat, or fever >38 °C. They received instruction by a physician (S.K.) on how to perform the nasal swab. Briefly, the swab was to be inserted into one nostril to a depth of 1–2 cm, rotated three times, and then placed into transport medium. Two kits for nasal swabbing containing a regular flocked swab with molded breakpoint (Copan, Brescia, Italy; product number 359C) and viral universal transport medium were given to the participants. During symptomatic surveillance, weekly automated e-mail messages were sent to the participants containing (1) a reminder to take a swab at the onset of at least one of the above-mentioned symptoms and (2) instructions on how to collect, store, and return the swab. Visual instructions on how to collect the swab were also available on the package. Participants were instructed to store the self-collected swab in the refrigerator (+4 °C) until returning it, as soon as possible, to the study team. Upon receipt at the study site, swabs were held at −70 °C until analysis.
2.3. Non-responder survey
Because of the low response rate, we conducted a non-responder survey in April 2010 (Figure 1). The survey was done through a self-administered questionnaire, which was sent via the internal e-mail system. To maintain anonymity, responding individuals (n = 142) were asked to return the completed questionnaire by in-house mail. Among other items, information was collected regarding reasons for not participating in the study and the occurrence of respiratory infections during the study period.
2.4. Laboratory analysis: detection of viral respiratory pathogens
Nucleic acids were extracted from 200-μl aliquots of transport medium (UTM Kit, Copan, Brescia, Italy) with the QIAamp MinElute Virus Spin Kit (Qiagen GmbH, Hilden, Germany). cDNA was synthesized with the Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics GmbH, Mannheim, Germany) and tested by multiplex PCR (Seeplex RV12 ACE Detection kit, Seegene Germany, Eschborn, Germany) for the presence of any of 12 human viral respiratory pathogens: adenovirus, metapneumovirus, coronavirus 229E/NL63 and OC43, parainfluenza virus 1–3, influenza virus A and B, respiratory syncytial virus A and B, and rhinovirus A/B.
2.5. Statistical analysis
Data were described as percentages for categorical variables and medians with range for continuous variables. Differences between groups were tested with the Chi-square test (for categorical variables) and the Mann–Whitney U-test (for continuous variables).
3. Results
3.1. Description of the samples
Overall, there were only minor differences between the participants and the non-responders (Table 1 ). Participants were slightly younger than non-responders (p = 0.03); approximately 17% of the participants and 9% of the non-responders were born outside Germany (p = 0.09). About half of the subjects in both groups had a university degree (including universities of applied sciences). There was a slightly higher proportion of smokers among the responders (p = 0.02). Reasons for not participating were: ‘did not meet inclusion criteria’ (∼31%), ‘no time’ (∼27%), ‘absent because of illness or vacation’ (∼16%), ‘did not read the invitation e-mail’ (∼12%), ‘fear of blood draw’ (∼7%), ‘concern of inadequate data protection’, ‘no interest’ (∼6% each), and ‘information about the study was unclear’ (∼5%) (subjects were allowed to give more than one reason).
Table 1.
Characteristics of the study subjects
| Characteristic | Responders (n = 53), % | Non-responders (n = 142), % | p-Value |
|---|---|---|---|
| Sex | 0.59 | ||
| Female | 75.5 | 71.6 | |
| Male | 24.5 | 28.4 | |
| Age (years), median | 29.0 | 37.0 | 0.03b |
| University degreea | 48.2 | 49.6 | 0.63 |
| Country of birth | 0.09 | ||
| Germany | 83.0 | 91.5 | |
| Other | 17.0 | 8.5 | |
| Number of household members | 0.32 | ||
| Living alone | 28.3 | 20.4 | |
| + 1 member | 41.5 | 38.7 | |
| + ≥2 members | 30.2 | 40.9 | |
| Means of getting to the work place | 0.09 | ||
| On foot | 32.1 | 19.7 | |
| By public transport | 13.2 | 9.2 | |
| By car | 54.7 | 71.1 | |
| Smoking status | 0.07 | ||
| Non-smoker | 54.7 | 69.3 | |
| Ex-smoker | 24.5 | 21.2 | |
| Current smoker | 20.8 | 9.5 |
Including universities of applied sciences.
Mann–Whitney U-test.
3.2. Weekly surveillance of acute respiratory symptoms
Out of 53 participants, 30 provided 38 nasal swabs during the symptomatic follow-up period (eight individuals had two symptomatic episodes). Thus, the clinical attack rate of acute respiratory infection, based on at least one reported symptom, was 53.6% (Table 2 ). About 38% of the participants reported at least two, 18% at least three, and 9% at least four symptoms. Similar proportions of non-responders reported three or more symptoms. However, the proportion of individuals who reported mild infections (one or two symptoms) was significantly lower among the non-responders than the study participants.
Table 2.
Clinical attack rates of acute respiratory symptoms during January to March, 2010
| Acute respiratory symptoms | Proportion of respondersa (n = 53) | Proportion of non-respondersb (n = 142) | p-Value |
|---|---|---|---|
| At least one (any) symptom | 53.6 | 31.7 | 0.02 |
| ≥2 symptoms | 37.5 | 25.4 | 0.09 |
| ≥3 symptoms | 17.9 | 18.3 | 0.94 |
| ≥4 symptoms | 8.9 | 7.0 | 0.65 |
Symptoms of acute respiratory infections based on prospective active symptomatic surveillance.
Symptoms of acute respiratory infections based on retrospective self-reported data at the end of the season.
3.3. Acceptance and compliance
Fifty-one out of 53 participants completed all aspects of the study. One participant had to leave the study because he moved away and one participant was lost to follow-up. Nearly all study participants found the study design acceptable (Table 3 ), and the vast majority of participants would participate again in such a study. Only six percent found a weekly e-mail reminder to take the swab unacceptable. The majority of those who collected a nasal swab reported no difficulties in self-swabbing. Only one participant reported difficulties opening the swab tube. About 15% felt discomfort while performing the swab.
Table 3.
Acceptance of the study design combining active symptomatic surveillance and self-collection of nasal swabs
| Characteristics | Agree/strongly agree, % | Neither nor, % | Disagree/strongly disagree, % |
|---|---|---|---|
| The overall study design was acceptable (n = 51) | 98.0 | 2.0 | 0 |
| I would participate in this kind of study again (n = 51) | 96.1 | 3.9 | 0 |
| Weekly e-mail reminder was acceptable (n = 51) | 90.2 | 3.9 | 5.9 |
| The instructions how to take the swab were understandable (n = 42) | 78.6 | 2.4 | 19.0 |
| Taking a nasal swab by myself is acceptable (n = 40) | 97.5 | 2.5 | 0 |
| I prefer taking a swab by myself rather than having it taken by medical personnel (n = 40) | 90.0 | 10.0 | 0 |
| I felt comfortable when taking a swab (n = 28)a | 66.6 | 18.5 | 14.8 |
| Nasal swabs were easy to perform (n = 28)a | 96.4 | 3.6 | 0 |
Only those who obtained the nasal swab.
3.4. Pre-analytical phase
All participants who reported acute respiratory symptoms during the period of symptomatic surveillance (n = 30) self-collected the nasal swab. Eight subjects reported two episodes of respiratory infections, and all eight returned two swabs. At the end of the study period one person reported that he/she took the swab but did not bring it to the study center. The reason stated was “I forgot to bring the swab to the study center”. Of these participants with symptoms of an acute respiratory infection, 85.7% collected the swab within the first 3 days of the onset of symptoms. One person collected the swab on the sixth day. Half of the participants brought the swab to the study center on the day of taking it. The maximum time between swab collection and delivery was 2 weeks.
3.5. Laboratory results
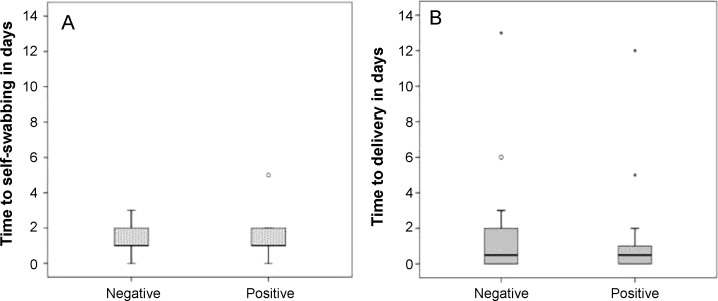
Respiratory pathogenic viruses were detected by reverse transcription polymerase chain reaction (RT-PCR) in about 50% of the swabs (Table 4 ), and the most frequently recorded ones were human coronaviruses 229E/NL63 (8/38 swabs, ∼21%) and OC43 (5/38 swabs, ∼13%). Co-infections were detected in two specimens. Influenza viruses were not detected. There were no differences in the proportion of positive (for any viruses) and negative swabs in terms of participant sex, age, and level of education. Also, there were no differences in the time elapsed between the onset of symptoms and self-swabbing (Figure 2A) or the time between self-swabbing and arrival of the swab at the study center (Figure 2B). Likewise, we did not detect any effects on viral detection when we controlled for potential effects of each variable (time (change per one day) between symptoms and swabbing, adjusted odds ratio (AOR) 0.98, 95% confidence interval (CI) 0.50–1.93; time between swabbing and delivery, AOR 0.96, 95% CI 0.76–1.21).
Table 4.
Laboratory-confirmed respiratory infections
| n (%) | |
|---|---|
| RT-PCR in nasal swabs | n = 38 |
| No pathogen detected | 19 (50) |
| Human coronavirus 229E/NL63 | 8 (21.1) |
| Human coronavirus OC43 | 5 (13.2) |
| Human rhinovirus A/B | 3 (7.9) |
| Human metapneumovirus | 1 (2.6) |
| Co-infection (metapneumovirus and parainfluenza virus 1) | 1 (2.6) |
| Co-infection (coronavirus 229E/NL63 and rhinovirus A/B) | 1 (2.6) |
| Influenza A or B | 0 (0) |
| Adenovirus | 0 (0) |
| Parainfluenza virus 1, 2, or 3 | 0 (0) |
| Respiratory syncytial virus A or B | 0 (0) |
RT-PCR, reverse transcription polymerase chain reaction.
Figure 2.
Lack of association between the detection of a pathogenic virus in self-collected nasal swabs (x-axis) and (A) the time (in days) elapsed between the onset of first respiratory symptoms and self-swabbing (y-axis) (Mann–Whitney U-test, p = 0.82) and (B) the time (in days) elapsed between self-swabbing and arrival of the swab at the study center (y-axis) (Mann–Whitney U-test, p = 0.64).
4. Discussion
4.1. Feasibility of the approach
We tested the feasibility of combining real-time symptomatic surveillance with nasal self-swabbing for the prospective collection of epidemiologic and virological data on acute respiratory infections. In the pre-analytical phase, this novel approach turned out to be highly feasible in that acceptance, satisfaction, compliance, and timeliness of logistics were high. Notably, more than 90% of the participants who self-swabbed reported that the swab was easy to obtain and that they preferred self-collection to collection by study personnel. The reason for this high degree of satisfaction may be that self-swabbing reduces duration and frequency of contact with study personnel as well as travel to a study site. The resulting greater convenience would very likely impact positively on compliance in any large-scale prospective study. Another important finding of the presented study is that neither the time between onset of symptoms and self-swabbing nor the time between self-swabbing and specimen arrival at the laboratory influenced the viral detection rate. This agrees well with results from a study in the UK,11 and is noteworthy, since in population-based studies employing self-swabbing, shipping time needs to be included in the time between swab collection and expected arrival of the specimen at the study center.
Factors other than time that could not be addressed in this study may influence the viral detection rate. One obvious candidate is the swabbing technique. The study physician instructed the participants in a standardized manner in the proper application of the technique. We do not believe that an inadequate technique impacted negatively on the rate of pathogen detection, since the detection rate of 50% recorded by us corresponds to what has been reported in other studies employing staff-collected swabs and similar detection technology.13 Also, two recent studies showed that there were no differences in pathogen detection between self- and staff-collected swabs.14, 15 Another factor that may influence the detection rate is the type of nasal swab used for specimen collection. Recently, Smieja et al. developed a flocked nasal mid-turbinate swab and compared it with the gold standard (e.g., rayon nasal and nasopharyngeal swabs). The mid-turbinate swab turned out to provide better results (based on epithelial cell counts) than the gold standard.16 We used a regular flocked swab. Thus, using the mid-turbinate flocked swab might have resulted in a higher detection rate in our study.
4.2. Specific applications of this approach in population-based studies
The non-responder survey allowed us to compare prospectively (participants) and retrospectively (non-responders) collected data on acute respiratory infection symptoms, and it revealed that the prospective approach resulted in a higher rate of detection of mild infections. Thus, one immediate strength of using active symptomatic surveillance in population-based studies on acute respiratory infections would be the more efficient identification of individuals with reduced susceptibility to infection, which would constitute an invaluable asset for large-scale studies on genetic determinants of infection susceptibility and resistance in humans. In such a study, using self-swabbing instead of swabbing by study personnel to detect specific pathogens would be immensely attractive due to its anticipated lower cost. Indeed, expenses for personnel and logistics were estimated to be 80% less if self-swabbing was used instead of swabbing by study personnel.17 Previous methods of active symptomatic surveillance have been, for example, weekly telephone calls18 or daily symptom diaries.19 However, these methods might be costly and have lower compliance rates.
4.3. Limitations
Limitations not addressed above include the representativeness of the study population. For instance, we had higher-than-expected proportions of female (∼75%) and highly educated subjects (∼50%). Moreover, due to working in a research institution, the participants could be expected to be more receptive to the study design than the general population. Since e-mail has become the primary tool of communication in professional work environments, including our institution, the population sampled for the present study may have a higher acceptance of modern communication tools than the general population. Further studies are needed regarding the use of electronic communication methods in population-based studies, particularly those targeting the less educated and the elderly. Lastly, due to the unexpected near absence of influenza infection during the study period, we could not evaluate the usefulness of the study design for the detection of influenza infection. Indeed, considering that upper respiratory symptoms occur only in about 60% and fever in about 35% of episodes of influenza infection,20 inclusion of surveillance questions about other influenza-associated symptoms (e.g., myalgia or headache) would likely increase the efficiency of screening for influenza infection with e-mail-based surveillance.
5. Conclusions
Combining e-mail-based active symptomatic surveillance with self-collection of nasal swabs ensured prospective, accurate collection of data on incident episodes of acute respiratory infections and timely sample collection for the detection of respiratory pathogens. It promises to be an efficient and cost-effective approach in population-based studies on the epidemiology of respiratory infections.
Acknowledgements
We would like to thank the participants for their kind participation in the study. We thank Prof. Udo Buchholz (Robert Koch Institute, Berlin, Germany) for helpful comments on the study design and a critical reading of the manuscript. This work was supported with intramural funds from the Helmholtz Association (Program Infection and Immunity).
Conflict of interest: The authors declare that they have no competing interests.
Corresponding Editor: Jane Zuckerman, London, UK
References
- 1.Casalegno J., Frobert E., Escuret V., Bouscambert-Duchamp M., Billaud G., Mekki Y. Beyond the influenza-like illness surveillance: the need for real-time virological data. Euro Surveill. 2011:16. pii 19756. [PubMed] [Google Scholar]
- 2.Elder A.G., O’Donnell B., McCruden E.A., Symington I.S., Carman W.F. Incidence and recall of influenza in a cohort of Glasgow healthcare workers during the 1993-4 epidemic: results of serum testing and questionnaire. BMJ. 1996;313:1241–1242. doi: 10.1136/bmj.313.7067.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepperd S., Perera R., Bates S., Jenkinson C., Hood K., Harnden A. A children's acute respiratory illness scale (CARIFS) predicted functional severity and family burden. J Clin Epidemiol. 2004;57:809–814. doi: 10.1016/j.jclinepi.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Michiels B, Philips H, Coenen S, Yane F, Steinhauser T, Stuyck S, et al., The effect of giving influenza vaccination to general practitioners: a controlled trial [NCT00221676]. BMC Med 2006; 4:17. [DOI] [PMC free article] [PubMed]
- 5.Marquet R.L., Bartelds A.I., van Noort S.P., Koppeschaar C.E., Paget J., Schellevis F.G. Internet-based monitoring of influenza-like illness (ILI) in the general population of the Netherlands during the 2003–2004 influenza season. BMC Public Health. 2006;6:242. doi: 10.1186/1471-2458-6-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friesema I.H., Koppeschaar C.E., Donker G.A., Dijkstra F., van Noort S.P., Smallenburg R. Internet-based monitoring of influenza-like illness in the general population: experience of five influenza seasons in The Netherlands. Vaccine. 2009;27:6353–6357. doi: 10.1016/j.vaccine.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 7.Tilston N.L., Eames K.T., Paolotti D., Ealden T., Edmunds W.J. Internet-based surveillance of influenza-like-illness in the UK during the 2009 H1N1 influenza pandemic. BMC Public Health. 2010;10:650. doi: 10.1186/1471-2458-10-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams C.J., Schweiger B., Diner G., Gerlach F., Haaman F., Krause G. Seasonal influenza risk in hospital healthcare workers is more strongly associated with household than occupational exposures: results from a prospective cohort study in Berlin, Germany, 2006/07. BMC Infect Dis. 2010;10:8. doi: 10.1186/1471-2334-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esposito S., Molteni C.G., Daleno C., Valzano A., Tagliabue C., Galeone C. Collection by trained pediatricians or parents of mid-turbinate nasal flocked swabs for the detection of influenza viruses in childhood. Virol J. 2010;7:85. doi: 10.1186/1743-422X-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeb M., Dafoe N., Mahony J., John M., Sarabia A., Glavin V. Surgical mask vs N95 respirator for preventing influenza among health care workers: a randomized trial. JAMA. 2009;302:1865–1871. doi: 10.1001/jama.2009.1466. [DOI] [PubMed] [Google Scholar]
- 11.Elliot A.J., Powers C., Thornton A., Obi C., Hill C., Simms I. Monitoring the emergence of community transmission of influenza A/H1N1 2009 in England: a cross sectional opportunistic survey of self sampled telephone callers to NHS Direct. BMJ. 2009;339:b3403. doi: 10.1136/bmj.b3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert S.B., Allen K.M., Nolan T.M. Parent-collected respiratory specimens—a novel method for respiratory virus and vaccine efficacy research. Vaccine. 2008;26:1826–1831. doi: 10.1016/j.vaccine.2008.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brittain-Long R., Nord S., Olofsson S., Westin J., Anderson L.M., Lindh M. Multiplex real-time PCR for detection of respiratory tract infections. J Clin Virol. 2008;41:53–56. doi: 10.1016/j.jcv.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luinstra K., Petrich A., Castriciano S., Ackerman M., Chong S., Carruthers S. Evaluation and clinical validation of an alcohol-based transport medium for preservation and inactivation of respiratory viruses. J Clin Microbiol. 2011;49:2138–2142. doi: 10.1128/JCM.00327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larios O.E., Coleman B.L., Drews S.J., Mazzulli T., Borgundvaag B., Green K. Self-collected mid-turbinate swabs for the detection of respiratory viruses in adults with acute respiratory illnesses. PLoS One. 2011;6:e21335. doi: 10.1371/journal.pone.0021335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smieja M., Castriciano S., Carruthers S., So G., Chong S., Luinstra K. Development and evaluation of a flocked nasal mid-turbinate swab for self-collected respiratory virus diagnostic testing. J Clin Microbiol. 2010;48:3340–3342. doi: 10.1128/JCM.02235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akmatov M.K., Pessler F. Self-collected nasal swabs to detect infection and colonization: a useful tool for population-based epidemiological studies? Int J Infect Dis. 2011 Jun 3 doi: 10.1016/j.ijid.2011.04.009. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Wilde J.A., McMillan J.A., Serwint J., Butta J., O’Riordan M.A., Steinhoff M.C. Effectiveness of influenza vaccine in health care professionals: a randomized trial. JAMA. 1999;281:908–913. doi: 10.1001/jama.281.10.908. [DOI] [PubMed] [Google Scholar]
- 19.Lambert S.B., Allen K.M., Druce J.D., Birch C.J., Mackay I.M., Carlin J.B. Community epidemiology of human metapneumovirus, human coronavirus NL63, and other respiratory viruses in healthy preschool-aged children using parent-collected specimens. Pediatrics. 2007;120:e929–e937. doi: 10.1542/peds.2006-3703. [DOI] [PubMed] [Google Scholar]
- 20.Carrat F., Vergu E., Ferguson N.M., Lemaitre M., Cauchemez S., Leach S. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008;167:775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]