Highlights
-
•
The data and strategies used for the surveillance of infectious diseases are listed.
-
•
The role of clinical microbiology laboratories in this surveillance is highlighted.
-
•
A review of all of the surveillance systems described from 2009 to June 13, 2014 is provided.
Keywords: Surveillance, Infectious diseases, Epidemiology, Clinical microbiology laboratories
Summary
Background
Infectious diseases remain a major public health problem worldwide. Hence, their surveillance is critical. Currently, many surveillance strategies and systems are in use around the world. An inventory of the data, surveillance strategies, and surveillance systems developed worldwide for the surveillance of infectious diseases is presented herein, with emphasis on the role of the microbiology laboratory in surveillance.
Methods
The data, strategies, and systems used around the world for the surveillance of infectious diseases and pathogens, along with current issues and trends, were reviewed.
Results
Twelve major classes of data were identified on the basis of their timing relative to infection, resources available, and type of surveillance. Two primary strategies were compared: disease-specific surveillance and syndromic surveillance. Finally, 262 systems implemented worldwide for the surveillance of infections were registered and briefly described, with a focus on those based on microbiological data from laboratories.
Conclusions
There is currently a wealth of available data on infections, which has been growing with the recent emergence of new technologies. Concurrently with the expansion of computer resources and networks, these data will allow the optimization of real-time detection and notification of infections.
1. Introduction
Classified as the second leading cause of death in humans by the World Health Organization, with approximately 15 million deaths worldwide every year,1 infectious diseases remain a serious public health problem in the 21st century. Among them, HIV/AIDS, tuberculosis, and malaria have been nicknamed the ‘big three’ because of their important impact on global human health. In 2011, tuberculosis infected two billion people and killed 1.3 million, malaria infected 207 million people and killed 62 700, and HIV infected 35.3 million people and killed 1.6 million.2
For adequate measures to be taken to detect and fight infectious diseases, their surveillance is essential. Surveillance consists of “the ongoing systematic collection, analysis, and interpretation of health data essential to the planning, implementation, and evaluation of public health practice, closely integrated with the timely dissemination of these data to those who need to know”.3 Attempts to survey infectious diseases are not recent. One of the best known examples is the use of the London Bills of Mortality by clerks, starting in 1603, for weekly monitoring of the number of deaths in London.4 Later, in 1854, John Snow performed a topographic study in London by systematically recording the addresses of people infected with cholera, to identify the source of the pathogen.5 Currently, many surveillance strategies and systems are available around the world. Computer resources have expanded considerably, but infectious disease surveillance remains challenging. The 2009 H1N1 influenza pandemic6 and the current Ebola outbreak in West Africa7 are recent examples showing that infectious diseases cannot be predicted and modelled reliably.
Nevertheless, the detection and investigation of abnormal health-related events effectively allow the identification of true epidemic events. The abnormal increase in the number of young homosexual men infected by Pneumocystis carinii in Los Angeles between 1980 and 19818 prompted the discovery of the HIV virus in 1983.9 Similarly, the outbreak of a severe respiratory illness of unknown origin that affected 180 people who had attended a state American Legion convention in Philadelphia in July 1976 allowed the identification of Legionella pneumophila.10 In late 2002, an unknown respiratory disease with no identifiable cause was diagnosed and reported in several people living in Guangdong Province in China. The syndrome, termed ‘severe acute respiratory syndrome’ (SARS), rapidly crossed borders and became a worldwide threat.11 A coronavirus (SARS-CoV) was finally identified as the causative agent of the syndrome.12
With consideration of these aspects, an inventory of the data, surveillance strategies, and surveillance systems developed worldwide for the surveillance of infectious diseases is presented herein, with emphasis on the role of the microbiology laboratory in surveillance.
2. Infectious disease surveillance — Data used for surveillance
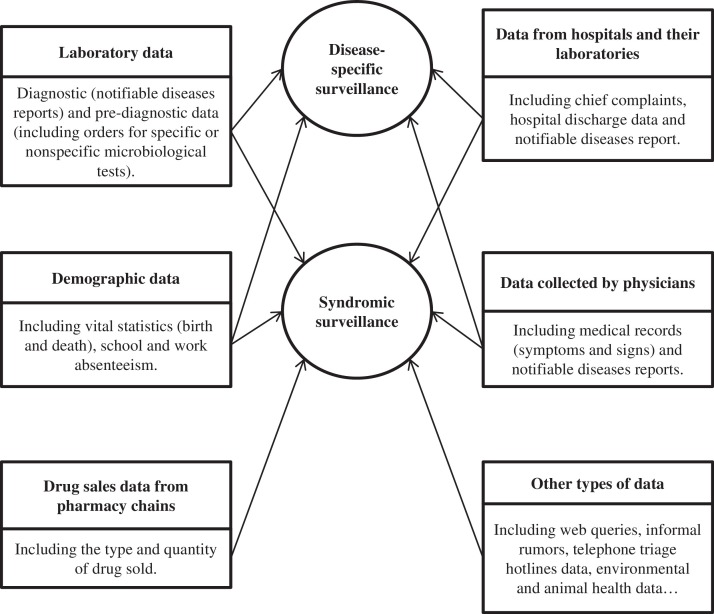
Fig. 1 summarizes the main types of data available for surveillance. These were classified according to the outbreak detection continuum published by Texier and Buisson.13
Fig. 1.
Main sources of data used by syndromic and disease-specific surveillance systems.
2.1. Human environment
2.1.1. Environmental data
Environmental data include water pollution, weather, and air pollution. For example, water quality testing from samples collected at water treatment facilities could be used to explain an increase in the number of patients presenting to emergency departments because of gastrointestinal disorders, as was done in the case of ice made during the massive outbreak of Cryptosporidium in Milwaukee.14
2.1.2. Animal health
Animal health data come directly from wild and domestic animals and are particularly valuable for the surveillance of zoonotic diseases such as plague, rabies, and monkeypox. For example, Chaintoutis et al. successfully used serum collected from sentinel juvenile domestic pigeons for the early detection of West Nile virus in Central Macedonia, Greece.15
2.2. Human behaviour
2.2.1. Internet use
The Internet can be used for infectious disease surveillance.16 Studies on influenza have proven the efficacy of the use of Web queries to complement existing surveillance methods. For example, the use of Web data to predict flu in Canada using FluWatch was done successfully and at a low cost, which inspired Google to develop Google Flu Trends, a free forecasting tool allowing the real-time surveillance of influenza activity in the USA .17 However, such an approach should not be used to replace traditional epidemiological surveillance networks as flu-tracking techniques based on Web data are more likely to be affected by changes in people's search behaviour.18 Another type of internet-based data usable for surveillance is health-related dispatches. These data include, among others, press dispatches from various origins like Reuters (http://fr.reuters.com/) and Humanitarian News (http://humanitariannews.org/), electronic mail-based discussion groups like ProMED-mail (http://www.promedmail.org/), press articles, and institutional and non-institutional warning networks like the World Health Organization.19, 20
2.2.2. Telephone triage hotlines
Telephone triage hotlines receive numerous phone calls from people requiring immediate health care assistance. Electronic data extracted from these hotlines can be a valuable source of data for surveillance. Although hotlines are inherently non-specific, data can be produced regardless of the day of the week, weather conditions, or holidays if the triage hotline is operated around the clock. These data normally include the precise time of the call, basic information about the caller, their area of residence, and some description of the symptoms.21
2.2.3. Drug sales
When people fall sick, they either go to see the doctor, treat themselves with home remedies, or practice self-medication by purchasing non-prescription remedies from a drugstore. In the latter case, sales data are entered electronically in store databases. These data are interesting for infectious disease surveillance because they reflect customer behaviour. Indeed, the class of drug sold, the quantity, and the date of purchase can provide significant information on the age distribution, size, and level of access to health care of a given population.21
2.2.4. Absenteeism
Absenteeism data include work and school absenteeism declarations. These can be used for the early detection of outbreaks, as has been demonstrated with influenza in France.16 Absenteeism data can also provide critical information about an outbreak, such as the place where people have been infected.16 Therefore, if several children or students are unable to go to school within a short span of time because of stomach pain, and if they all ate in the same place the day before, it is reasonable to suspect that they are all affected by the same food-borne pathogen.
2.3. Health care
2.3.1. Sentinel surveillance
Sentinel physicians are physicians who agree to notify the public health authorities at regular intervals of patients presenting certain specific symptoms (e.g., influenza-like illness). Public health authorities analyse the data obtained and assess the activity and strains of diseases circulating in the population of interest.16
2.3.2. Chief complaints
Chief complaints consist of short sentences or codes summarizing the reason for the emergency department admission (for example ‘headache’ or ‘abdominal pain’).16, 21 Multiple chief complaints can be registered for the same patient, the first one being the most important for infectious disease surveillance.16 Once registered, chief complaints can be classified into syndromic surveillance categories manually or using algorithms.21
2.3.3. Medical records
When patients attend for medical examinations, physicians ask them questions to collect useful information on their health status, including the date of appearance of the symptoms and their progression over time. These data help physicians define the symptoms affecting the patient. Next, the physicians physically examine the patients to collect data on the signs of the disease. Together, signs and symptoms help the physicians to assign a differential diagnosis and create a list of probable diseases that may be affecting the patient. These records are translated into codes using the International Classification of Diseases, ninth revision (ICD-9). These codes can then be grouped into syndromic categories.16, 21, 22
2.3.4. Hospital discharge data
Hospital discharge data include ICD-9 codes, hospital zip code, home zip code, the patient's age, and the patient's dates of admission and discharge.16 These data can be useful for the surveillance of infectious diseases.
2.3.5. Microbiology orders
Physicians and hospitals may ask laboratories to perform microbiological tests to confirm or exclude the presence of pathogens in patients. These tests include, among others, culture tests, nucleic acid amplification tests, serological tests, and agglutination tests. These tests can be used for the pre-diagnostic surveillance of pathogens. The number of a particular specific or non-specific test performed per unit of time can be a good indicator of the presence of a particular pathogen in a given population.21 However, as the results of these tests are either positive or negative, they allow only low specificity surveillance, which can lead to the investigation of the wrong epidemiological events.
2.3.6. Notifiable disease reports
Notifiable disease reports consist of mandatory reporting by mail, phone, fax, or using a computer, of diseases defined by health departments as being a threat to the community of interest.16 The list of diseases may be defined at the national level or at the state level, and varies according to fluctuations in the incidence and prevalence of pathogens over time.16, 21
2.4. Demographics
Vital statistics include data on birth, death, and marital status.16 Birth data can provide information on the cause of premature delivery or birth anomalies, but can equally be used for the surveillance of infant mortality.16 Death certificates are also valuable in surveillance because they include the cause, age, and the place of death.16
3. Infectious disease surveillance — Surveillance strategies
Disease surveillance can be divided broadly into disease-specific surveillance, syndromic surveillance, and event-based surveillance (Table 1 ). The latter surveillance class falls outside the scope of this review and is not considered comprehensively herein.
Table 1.
Advantages and limitations of the main types of surveillance system developed to follow infectious diseases around the world
| Type of surveillance system | Principle | Advantage(s) | Limitation(s) |
|---|---|---|---|
| Disease-specific surveillance system | Surveillance of specific pathogens, diseases, or syndromes in a target population | • Surveillance of a wide range of pathogens • Useful to follow global trends of surveyed pathogens • Can be used to monitor public health measures taken to fight precise pathogens |
• Standardization of data used is necessary • Limited capacities can lead to underestimated prevalence of the surveyed event • Targets (pathogens, diseases, syndromes, and populations) must be clearly identified before starting the surveillance |
| Syndromic surveillance system | Real-time or near real-time collection, analysis, interpretation, and dissemination of health-related data for the early identification of potential health threats | • Can be used in emergency cases • High sensitivity because laboratory confirmation is not needed • Possible deployment in low-incomes countries • Rapid to implement |
• Efficiency depends on pathogens and patient characteristics • Lack of human and technological resources can affect data collection, management, timeliness, and sharing • Low specificity |
| Event-based surveillance system | Real-time or near-real-time manual or automatic collection and analysis of unstructured information from numerous text sources and in various languages to detect potential or confirmed health hazards | • Use of potentially unverified information from medical and non-medical informal sources of data • Can be used in low-income countries with no public health surveillance • Rapid detection and report of possible health hazards Gathering and analysis of data published in various different languages |
• Detected events need to be confirmed using reliable sources of data |
3.1. Disease-specific surveillance
Disease-specific surveillance consists of the surveillance of a selection of diseases, syndromes, and risk exposures considered as public health threats for the population of interest.16, 23 This is the traditional form of surveillance, based on notifiable disease reporting using clinical case reports sent by sentinel structures or general practitioners and positive results reported from clinical laboratories.16, 23 These systems can be deployed at the national level.
A good example is the National Tuberculosis Surveillance System (NTSS),24 which was first implemented in 1953 in the USA for the collection of data on tuberculosis cases. Briefly, if a patient is positive for Mycobacterium tuberculosis, the state health department sends an anonymous report to the NTSS. Reports summarizing the data are then published on the Centers for Disease Control and Prevention website (http://www.cdc.gov/tb/topic/default.htm). In France, various disease-specific surveillance networks have been developed under the leadership of the Institut National de Veille Sanitaire (INVS), the French National Institute for Public Health Surveillance. Among these, special mention can be made of LaboVIH, a laboratory-based surveillance system implemented in 2001 for the national specific surveillance of HIV activity.25 Twice a year, the INVS contacts all of the French biomedical laboratories (approximately 4300 laboratories in 2014) to collect data on the number of people tested for HIV and the number of people found to be positive for the first time in each laboratory in the network.25 The analysis is then shared through the weekly INVS epidemiology report (http://www.invs.sante.fr). Such surveillance can be implemented internationally, as has been done for the European Gonococcal Antimicrobial Surveillance Programme (Euro-GASP).26 This surveillance system was implemented in 2004 by the European Surveillance of Sexually Transmitted Infections Project (ESSTI) to provide data on the susceptibility of gonococci to various antibiotics by studying the evolution of gonococcal antibiotic resistance in the European Union and the European Economic Area.26 All of the states included in the ESSTI were asked to participate in the Euro-GASP to contribute to the collection and antibiotic susceptibility testing of gonococcal strains in their laboratories.26
3.2. Syndromic surveillance
According to Sala Soler et al., syndromic surveillance is based on data that are “non-specific health indicators including clinical signs, symptoms as well as proxy measures”, which “are usually collected for purposes other than surveillance and, where possible, are automatically generated” for allowing “a real-time (or near real-time) collection, analysis, interpretation, and dissemination of health-related data to enable the early identification of the impact (or absence of impact) of potential human or veterinary public health threats”.27 Syndromic surveillance systems collect and analyse health indicators such as nurse calls and school or work absenteeism rates.21, 22, 27 These systems are known to be non-specific but are sensitive and timely because data can be collected automatically without extra work.22, 27, 28 Moreover, as data sources can be varied, these systems allow interconnectivity among participants, increasing the capacity of public health authorities to manage possible epidemic situations.22 Finally, such surveillance systems can assist public health leaders in their decision-making on the guidance, implementation, and evaluation of programs and policies for the prevention and control of infectious diseases.29
A good example of a syndromic surveillance system is ESSENCE (the Electronic Surveillance System for the Early Notification of Community-Based Epidemics).30 The implementation of ESSENCE started as a collaboration between the US Department of Defense and the Johns Hopkins University Applied Physics Laboratory more than a decade ago.30 The first version of ESSENCE, ESSENCE I, is currently used to perform worldwide monitoring of the army personnel in all US military treatment facilities.31 The latest version of ESSENCE, ESSENCE II, performs integrated surveillance by analysing de-identified data from the National Capital Region military and civilian health department data.31 The data collected by ESSENCE II include information on military ambulatory visits and prescription medications, and various data from civilian databases including chief complaint data from civilian emergency departments.31 Once received, the data are archived and analysed. ESSENCE II transfers information to its users via secure websites.31 To summarize, users can see data and results in different formats, including a map of the geographic distribution of data sent by users and clusters obtained by scan statistics or lists of alerts emitted after the detection processes.31 ESSENCE II normally analyses data every 4 h but can also alter the processing period if real-time data are available.31 The French armed forces developed a real-time syndromic surveillance system, “le système d’Alerte et Surveillance en Temps réel” or ASTER.32, 33, 34 Briefly, every 10 minutes, the system collects medical data routinely transmitted via secure Internet connections by doctors, paramedics, and nurses who live with the French armed forces deployed outside of the country. The data include the numbers of military personnel suffering from various symptoms, including cardiovascular, gastrointestinal, and respiratory symptoms. Data are routinely analysed using the Current Past Graph method and the mean ± 2 or 3 standard deviations method . At the end of the process, a dashboard summarizing the epidemiological situation is presented to the health service of the armed forces based in Marseille, France, and doctors, paramedics, and nurses deployed with the armed forces obtain real-time feedback on the health status of the military personnel with whom they live.
3.3. Event-based surveillance
Event-based surveillance consists of the real-time or near-real-time scanning, collection, and analysis of unstructured information from diverse Internet sources (such as news or online discussion platforms) for detecting potential or confirmed health hazards occurring worldwide from reports and rumours.35, 36 Among the most well known examples of event-based surveillance systems are the Program for Monitoring Emerging Diseases (ProMED-mail, http://www.promedmail.org/),37 the Global Public Health Intelligence Network (GPHIN),38 MedISys (http://medisys.newsbrief.eu/medisys/homeedition/fr/home.html),36 BioCaster, EpiSPIDER, and HealthMap (http://www.healthmap.org/).19 These surveillance systems are crucial sources of epidemic intelligence and contribute greatly to the global detection of true outbreaks.35
4. Role of laboratories in the surveillance of infectious diseases
Laboratories produce some data currently usable for infectious disease surveillance.
4.1. Different types of laboratory
According to Wagner et al., laboratories can be classified as follows:16
-
(1)
Clinical laboratories, which provide a wide range of services from rapid screening tests to confirmatory analyses usable for the diagnosis and treatment of patients. This category of laboratory includes laboratories of clinics, departments of health, veterinary hospitals, and those in health care facilities (hospitals, physician offices, etc.).
-
(2)
Environmental laboratories, which perform analyses on environmental samples to determine their physical, chemical, and microbiological characteristics.
-
(3)
Commercial laboratories, which are large, independent laboratories that can perform tests on clinical and/or environmental samples. As these laboratories produce a large quantity of data, they can be central structures for surveillance systems.
-
(4)
Governmental laboratories, which include federal laboratories that perform reference laboratory testing and participate in the development of new laboratory technologies, state laboratories that are involved in the surveillance of various diseases, including communicable diseases but also in public health programs, local public health laboratories that play a role in the screening of diseases including tuberculosis or sexually transmitted infections, and other state or local laboratories that can provide valuable test results.
Their functions generate a wide variety of information, making them significant sources of data on infectious diseases. Indeed, they can confirm the presence of target pathogens, diseases, or syndromes in a population. Moreover, their activity can be used for a syndromic monitoring system (e.g., the weekly number of tests performed, etc. ). Finally, laboratory data can be used to investigate epidemiological events (Fig. 1).
4.2. Current impact of laboratories on infectious disease surveillance
To evaluate the importance of laboratories in the worldwide surveillance of infectious diseases, a PubMed search of papers in English published between 2009 and June 13, 2014 was conducted. The following keywords, always followed by “infectious diseases”, were used: “surveillance system”, “laboratory-based surveillance”, “syndromic surveillance”, “sentinel surveillance”, “integrated surveillance”, and “population-based surveillance”. Only surveillance systems using laboratory data were registered. If more than one article described the same surveillance system, the surveillance system was recorded only once. The systems were then tagged according to what they monitor (bacteria, viruses, fungi, parasites, or others), whether they are recognized nationally or internationally, and whether they perform syndromic or disease-specific surveillance (Supplementary Material Table S1; Fig. 2 ).
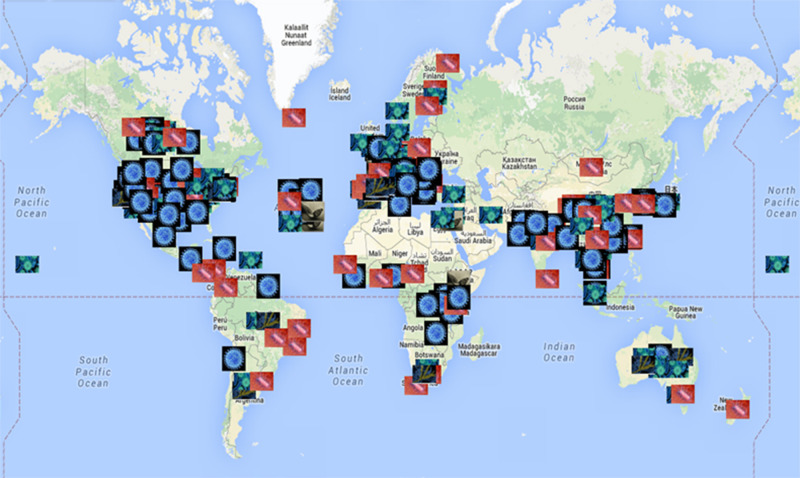
Fig. 2.
Infectious disease surveillance systems described around the world, January 2009 to June 13, 2014. The map is available at https://www.google.com/maps/d/edit?mid=z4TNutoSpTfw.k7NzPhL00pmc. The virus image represents surveillance systems focusing on viruses, the bacterium image represents surveillance systems focusing on bacteria, the fungus image represents surveillance systems focusing on fungi, and the polymicrobial image represents surveillance systems monitoring various different pathogens .
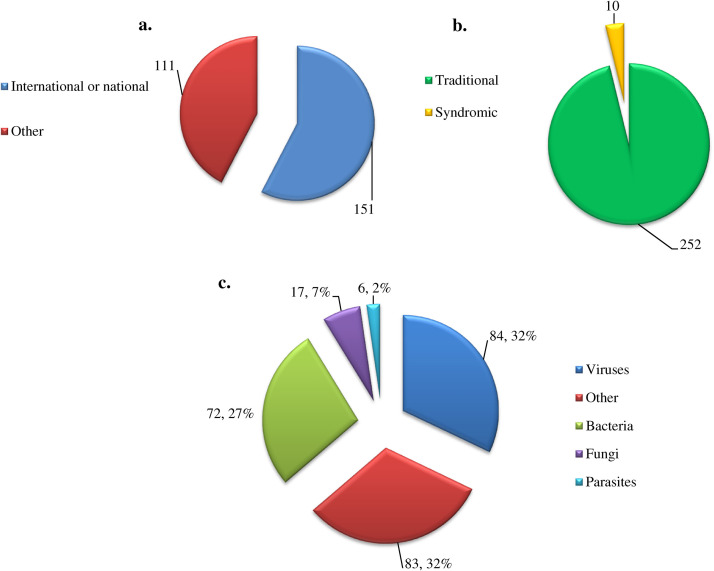
The analysis of the characteristics of the 262 surveillance systems is summarized in Fig. 3 . Briefly, most of the surveillance systems are recognized internationally or nationally, and perform disease-specific surveillance. Most were implemented for the surveillance of viruses (84 surveillance systems) and bacteria (72 surveillance systems). Amongst the 76 pathogens monitored by the surveillance systems, influenza was found to be the most surveyed virus (monitored in 31 countries/groups of countries) and Listeria spp and Salmonella spp the most surveyed bacterial species (14 for each).
Fig. 3.
Summary of the main characteristics of the 262 surveillance systems registered from January 2009 to June 13, 2014. (a) Number of international and national surveillance systems, and those that are neither. (b) Number of surveillance systems that are disease-specific (traditional surveillance) and those that are syndromic. (c) Classification of the surveillance systems according to what they monitor.
5. The future of infectious disease surveillance
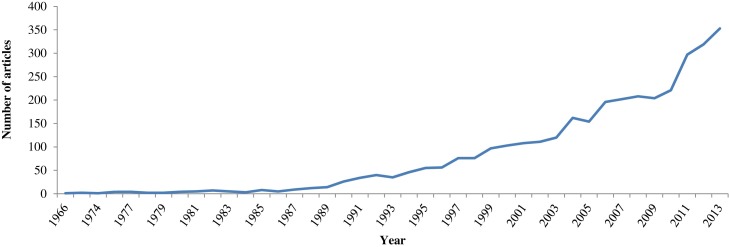
Developing surveillance systems to fight infectious diseases is a fast-growing and evolving field (Fig. 4 ), engaging more and more countries and resources. Disease-specific surveillance has allowed the effective management of numerous epidemics and infectious diseases. Smallpox was successfully eradicated in 1978 after the World Health Organization initiated a smallpox-specific mass vaccination program that included laboratory investigations.39 Similar results were seen in the case of rinderpest, a disease directly infecting Artiodactyla species (cattle, eland, buffalo, etc.) causing famines. This disease was declared eradicated on May 25, 2011 after long-term efforts to fight it through the development of the disease-specific Global Rinderpest Eradication Programme.40 Nevertheless, thanks to the global spread of Internet use and the unprecedented interconnectivity of people that this allows, it can be speculated that, in the future, improved syndromic surveillance systems will be coupled with disease-specific surveillance systems, as has already been done for influenza surveillance.17
Fig. 4.
Global change in the number of publications dealing with “surveillance system” AND infect* from 1966 to 2013.
Many surveillance systems survey few pathogens and not necessarily throughout the year. The surveillance of infections needs to be more global and instantaneous. This is a prerequisite for the detection and notification of abnormal events related to infections, appropriate prioritization of public health threats, and the implementation of optimal strategies and policies. Such approaches appear increasingly feasible with the tremendous expansion of computer resources, networks, and the real-time acquisition and sharing of data worldwide.
Funding
This work was supported by the Centre National de la Recherche Scientifique and the IHU Méditerranée Infection.
Ethical approval
Approval was not required.
Conflict of interest
None to declare.
Acknowledgements
We thank American Journal Experts for English corrections.
Corresponding Editor: Eskild Petersen, Aarhus, Denmark.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2016.04.021.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Morens D.M., Folkers G.K., Fauci A.S. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourzac K. Infectious disease: beating the big three. Nature. 2014;507:S4–S7. doi: 10.1038/507s4a. [DOI] [PubMed] [Google Scholar]
- 3.Thacker S.B., Berkelman R.L. Public health surveillance in the United States. Epidemiol. Rev. 1988;10:164–190. doi: 10.1093/oxfordjournals.epirev.a036021. [DOI] [PubMed] [Google Scholar]
- 4.Graunt J. Roycroft and Dicas; London: 1652. Natural and political observations made upon the Bills of Mortality. [Google Scholar]
- 5.Newsom S.W. Pioneers in infection control: John Snow, Henry Whitehead, the Broad Street pump, and the beginnings of geographical epidemiology. J. Hosp. Infect. 2006;64:210–216. doi: 10.1016/j.jhin.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Viboud C., Simonsen L. Global mortality of 2009 pandemic influenza A H1N1. Lancet Infect. Dis. 2012;12:651–653. doi: 10.1016/S1473-3099(12)70152-4. [DOI] [PubMed] [Google Scholar]
- 7.Piot P., Muyembe J.J., Edmunds W.J. Ebola in West Africa: from disease outbreak to humanitarian crisis. Lancet Infect. Dis. 2014;14:1034–1035. doi: 10.1016/S1473-3099(14)70956-9. [DOI] [PubMed] [Google Scholar]
- 8.Curran J.W., Jaffe H.W. AIDS: the early years and CDC's response. MMWR Surveill. Summ. 2011;60(Suppl 4):64–69. [PubMed] [Google Scholar]
- 9.Barre-Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention Follow-up on respiratory illness—Philadelphia. 1977. MMWR Morb. Mortal Wkly Rep. 1997;46:50–56. [PubMed] [Google Scholar]
- 11.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 12.Gerberding J.L. Faster. but fast enough? Responding to the epidemic of severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:2030–2031. doi: 10.1056/NEJMe030067. [DOI] [PubMed] [Google Scholar]
- 13.Texier G., Buisson Y. From outbreak detection to anticipation. Rev. Epidemiol. Sante Publique. 2010;58:425–433. doi: 10.1016/j.respe.2010.06.169. [DOI] [PubMed] [Google Scholar]
- 14.MacKenzie W.R., Hoxie N.J., Proctor M.E., Gradus M.S., Blair K.A., Peterson D.E. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 15.Chaintoutis S.C., Dovas C.I., Papanastassopoulou M., Gewehr S., Danis K., Beck C. Evaluation of a West Nile virus surveillance and early warning system in Greece, based on domestic pigeons. Comp. Immunol. Microbiol. Infect. Dis. 2014;37:131–141. doi: 10.1016/j.cimid.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Wagner M.M., Moore A.W., Aryel R.M. 1st ed. Academic Press; USA: 2006. Handbook of biosurveillance. [Google Scholar]
- 17.Butler D. Web data predict flu. Nature. 2008;456:287–288. doi: 10.1038/456287a. [DOI] [PubMed] [Google Scholar]
- 18.Butler D. When Google got flu wrong. Nature. 2013;494:155–156. doi: 10.1038/494155a. [DOI] [PubMed] [Google Scholar]
- 19.Lyon A., Nunn M., Grossel G., Burgman M. Comparison of web-based biosecurity intelligence systems: BioCaster, EpiSPIDER and HealthMap. Transbound. Emerg. Dis. 2012;59:223–232. doi: 10.1111/j.1865-1682.2011.01258.x. [DOI] [PubMed] [Google Scholar]
- 20.Grein T.W., Kamara K.B., Rodier G., Plant A.J., Bovier P., Ryan M.J. Rumors of disease in the global village: outbreak verification. Emerg. Infect. Dis. 2000;6:97–102. doi: 10.3201/eid0602.000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babin S., Magruder S., Hakre S., Coberly J., Lombardo J.S. Part I: System design and implementation. In: Lombardo J.S., Buckeridge D.L., editors. Disease surveillance: a public health informatics approach. John Wiley & Sons; Hoboken: 2006. pp. 43–91. [Google Scholar]
- 22.Chen H., Zeng D., Yan P. Springer; New York: 2010. Infectious disease informatics: syndromic surveillance for public health and bio-defense. [Google Scholar]
- 23.Koski E. Clinical laboratory data for biosurveillance. In: Zeng D., Chen H., Castillo-Chavez C., Thurmond M., editors. Infectious disease informatics and biosurveillance. Springer; New York: 2011. pp. 67–87. [Google Scholar]
- 24.Vinnard C., Winston C.A., Wileyto E.P., Macgregor R.R., Bisson G.P. Isoniazid resistance and death in patients with tuberculous meningitis: retrospective cohort study. BMJ. 2010;341:c4451. doi: 10.1136/bmj.c4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cazein F., Le Strat Y., Sarr A., Ramus C., Bouche N., Le Vu S. Dépistage de l’infection par le VIH en France, 2003-2013. Bull Epidémiol. Hebd. 2014;32-33:534–540. [Google Scholar]
- 26.Cole M.J., Unemo M., Hoffmann S., Chisholm S.A., Ison C.A., van de Laar M.J. The European Gonococcal Antimicrobial Surveillance Programme, 2009. Euro. Surveill. 2011;16(42) Article 4 ( http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19995) [PubMed] [Google Scholar]
- 27.Sala Soler M., Fouillet A., Viso A.C., Josseran L., Smith G.E., Elliot A.J. Assessment of syndromic surveillance in Europe. Lancet. 2011;378:1833–1834. doi: 10.1016/S0140-6736(11)60834-9. [DOI] [PubMed] [Google Scholar]
- 28.M’ikanatha N.O., Lynfield R., Van Beneden C.A., de Valk H. 2nd ed. Wiley-Blackwell; Chichester, United Kingdom: 2013. Infectious disease surveillance. [Google Scholar]
- 29.Mostashari F., Hartman J. Syndromic surveillance: a local perspective. J. Urban Health. 2003;80:i1–i7. doi: 10.1093/jurban/jtg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis S.L., Feighner B.H., Loschen W.A., Wojcik R.A., Skora J.F., Coberly J.S. SAGES: a suite of freely-available software tools for electronic disease surveillance in resource-limited settings. PLoS ONE. 2011;6:e19750. doi: 10.1371/journal.pone.0019750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lombardo J.S., Burkom H., Pavlin J. ESSENCE II and the framework for evaluating syndromic surveillance systems. MMWR Morb. Mortal Wkly. Rep. 2004;53(Suppl):159–165. [PubMed] [Google Scholar]
- 32.Meynard J.B., Chaudet H., Texier G., Queyriaux B., Deparis X., Boutin J.P. [Real time epidemiological surveillance within the armed forces: concepts, realities and prospects in France] Rev. Epidemiol. Sante Publique. 2008;56:11–20. doi: 10.1016/j.respe.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Chaudet H., Pellegrin L., Meynard J.B., Texier G., Tournebize O., Queyriaux B. Web services based syndromic surveillance for early warning within French Forces. Stud. Health Technol. Inform. 2006;124:666–671. [PubMed] [Google Scholar]
- 34.Jefferson H., Dupuy B., Chaudet H., Texier G., Green A., Barnish G. Evaluation of a syndromic surveillance for the early detection of outbreaks among military personnel in a tropical country. J. Public Health (Oxf). 2008;30:375–383. doi: 10.1093/pubmed/fdn026. [DOI] [PubMed] [Google Scholar]
- 35.Keller M., Blench M., Tolentino H., Freifeld C.C., Mandl K.D., Mawudeku A. Use of unstructured event-based reports for global infectious disease surveillance. Emerg. Infect. Dis. 2009;15:689–695. doi: 10.3201/eid1505.081114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohigas P.A., Santos-O’Connor F., Coulombier D. Epidemic intelligence and travel-related diseases: ECDC experience and further developments. Clin. Microbiol. Infect. 2009;15:734–739. doi: 10.1111/j.1469-0691.2009.02875.x. [DOI] [PubMed] [Google Scholar]
- 37.Morse S.S., Rosenberg B.H., Woodall J. ProMED global monitoring of emerging diseases: design for a demonstration program. Health Policy. 1996;38:135–153. doi: 10.1016/0168-8510(96)00863-9. [DOI] [PubMed] [Google Scholar]
- 38.Mykhalovskiy E., Weir L. The Global Public Health Intelligence Network and early warning outbreak detection: a Canadian contribution to global public health. Can J. Public Health. 2006;97:42–44. doi: 10.1007/BF03405213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arita I. Virological evidence for the success of the smallpox eradication programme. Nature. 1979;279:293–298. doi: 10.1038/279293a0. [DOI] [PubMed] [Google Scholar]
- 40.Morens D.M., Holmes E.C., Davis A.S., Taubenberger J.K. Global rinderpest eradication: lessons learned and why humans should celebrate too. J. Infect. Dis. 2011;204:502–505. doi: 10.1093/infdis/jir327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.