Highlights
-
•
Middle East respiratory syndrome coronavirus (MERS-CoV) sub-genomic mRNAs indicates that the virus is replicative.
-
•
Sub-genomic mRNA was detected in lower respiratory tract specimens for up to 4 weeks after symptoms developed.
-
•
In upper respiratory tract specimens, the detection of sub-genomic mRNA and genomic RNA did not correlate.
Keywords: Coronavirus infections, RNA, Messenger, Respiratory system
Abstract
Background
Information on the duration of replicative Middle East respiratory syndrome coronavirus (MERS-CoV) shedding is important for infection control. The detection of MERS-CoV sub-genomic mRNAs indicates that the virus is replicative. This study examined the duration for detecting MERS-CoV sub-genomic mRNA compared with genomic RNA in diverse respiratory specimens.
Methods
Upper and lower respiratory samples were obtained from 17 MERS-CoV-infected patients. MERS-CoV sub-genomic mRNA was detected by reverse transcription PCR (RT-PCR) and MERS-CoV genomic RNA by real-time RT-PCR.
Results
In sputum and transtracheal aspirate, sub-genomic mRNA was detected for up to 4 weeks after symptoms developed, which correlated with the detection of genomic RNA. In oropharyngeal and nasopharyngeal swab specimens, the detection of sub-genomic mRNA and genomic RNA did not correlate.
Conclusions
These findings suggest that MERS-CoV does not replicate well in the upper respiratory tract.
Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) genomic RNA can persist for more than 1 month in respiratory specimens (Memish et al., 2014). However, the detection of MERS-CoV RNA may overestimate the duration of shedding of replicative virus. Coronaviruses have a unique mechanism of discontinuous transcription with the synthesis of sub-genomic mRNAs (Sawicki et al., 2007). The MERS-CoV has at least seven distinct sub-genomic mRNA species and the detection of these indicates that the virus is replicative (Woo et al., 2016). The objectives of this study were to examine the duration for detecting MERS-CoV sub-genomic mRNA vs. genomic RNA in different respiratory specimens.
Methods
Respiratory samples were collected from 17 patients admitted to three Seoul National University (SNU) affiliated hospitals during the 2015 MERS outbreak in Korea. The patients were categorized into severe (A–I) or mild (J–Q) groups depending on their oxygen supplementation requirements. The severe group required oxygen supplementation to maintain arterial saturation above 90%. Patients A–E received ventilator therapy, while patients F–I did not (Park et al., 2015). The institutional review board at SNU Hospital provided study approval and waived the requirement for written consent.
Oropharyngeal and nasopharyngeal swabs were collected using a UTM kit containing viral transport medium (Copan Diagnostics Inc., Murrieta, CA, USA). The RNA was extracted from respiratory samples using a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, USA). To detect the MERS-CoV genomic RNA, multiplex quantitative real-time reverse transcription (rRT)-PCR was performed using the PowerChek MERS (upE & ORF1a) Real-time PCR Kit (Kogenebiotech, Seoul, South Korea) and all assays were performed using a ViiA7 Real-time PCR system (Applied Biosystems, Grand Island, NY, USA). The results of genomic RNA titers have been presented in part in a previous publication (Oh et al., 2016).
The MERS-CoV sub-genomic mRNA was detected using AccuPower RT-PCR PreMix (Binder Inc., Alameda, CA, USA). PCR primers were designed to detect sub-genomic mRNA that codes for the spike (S) (433 bp) and nucleocapsid (N) (662 bp) proteins (Table 1 ). Forward primer was elaborated from the leader sequence and backward primers of 5′ untranslated region (UTR)-S and 5′UTR-N were from gene sequences coding for proteins S and N, respectively. The PCR reactions were performed as follows: initial denaturation at 94 °C for 5 min and 40 cycles of denaturation at 94 °C for 20 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min. Sub-genomic mRNA was sequenced using a DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad) and the ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Grand Island, NY, USA). If the sequences included the leader sequence and were consistent with the MERS-CoV genome by ≥98% using Basic Local Alignment Search Tool (BLAST) software, they were confirmed as sub-genomic mRNA.
Table 1.
PCR primer pairs used in this study.
| Forward | Sequence (5′ → 3′) | Reverse | Sequence (5′ → 3′) | |
|---|---|---|---|---|
| 5′UTR-S | 1For | GATTTAAGTGAATAGCTTGGCTA | S21751Rev | TGAGAATAGTTAGCTACAAACAACT |
| 5′UTR-N | 1For | GATTTAAGTGAATAGCTTGGCTA | N29121Rev | TCCTGATCTTGAACCTTGTGAACT |
Results and discussion
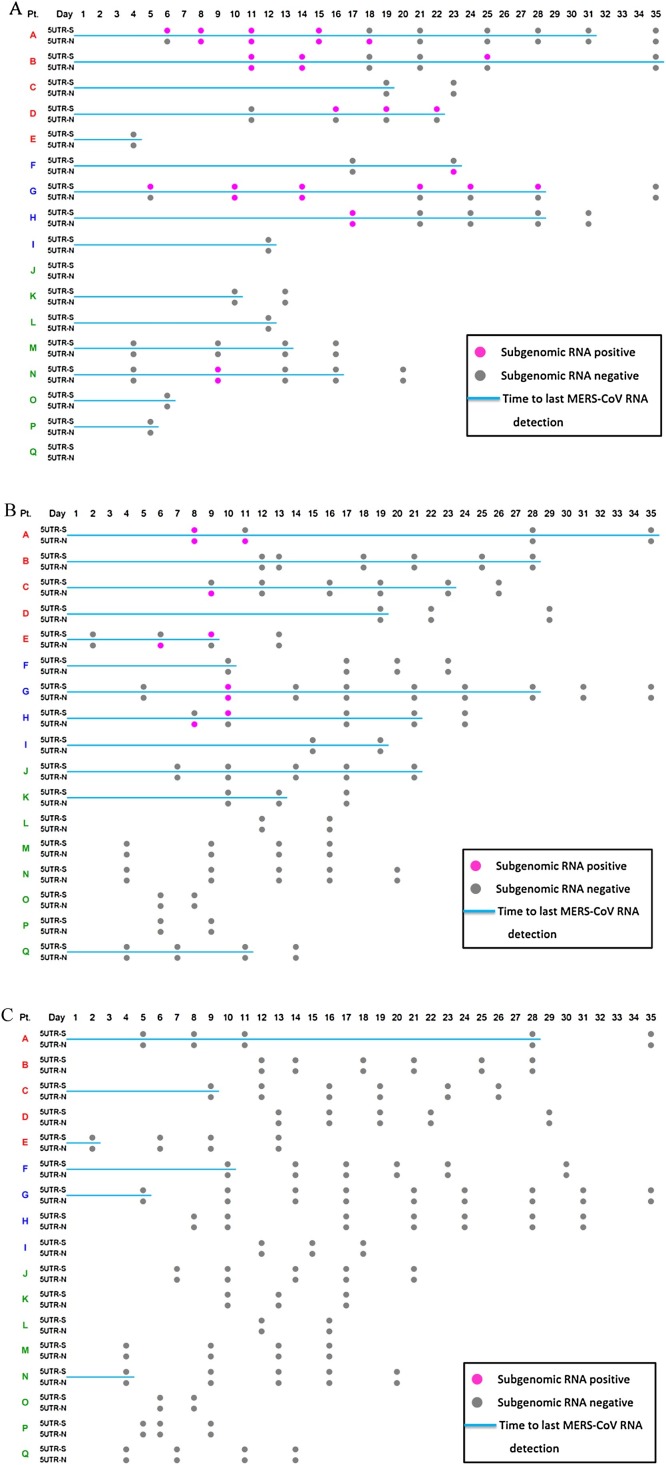
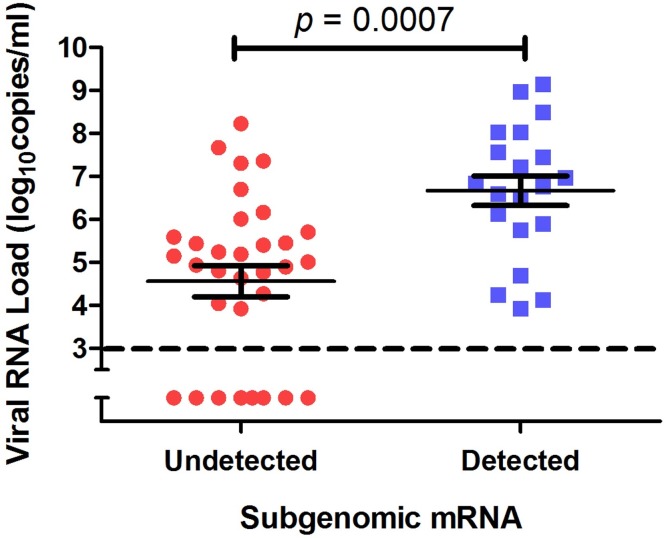
In sputum and transtracheal aspirates, the detection of MERS-CoV sub-genomic mRNA was more frequent in the severe group than in the mild group (Figure 1 A). Sub-genomic mRNA was detected ≤28 days after the illness onset. In the severe group, the period for detecting sub-genomic mRNA strongly correlated with the duration for detecting MERS-CoV genomic RNA (Pearson correlation coefficient = 0.803, p = 0.009). The MERS-CoV genomic RNA titer was significantly higher in the specimens with sub-genomic mRNA detection than in those where sub-genomic mRNA was not detected (P = 0.0007) (Figure 2 ).
Figure 1.
Detection of Middle East respiratory syndrome coronavirus (MERS-CoV) sub-genomic mRNA in (A) sputum and transtracheal aspirates; (B) throat swabs; (C) nasopharyngeal swabs. A pink dot denotes a positive RT-PCR result for MERS-CoV sub-genomic mRNA, while a gray dot indicates a negative result. The blue lines indicate the duration from symptom onset to the last positive result for MERS-CoV genomic RNA. Patients A–I were in the severe group and patients J–Q were in the mild group. Among the severe group subjects, patients A–E received ventilator therapy, while patients F–I did not. 5UTR-S and 5UTR-N denote the sub-genomic mRNA of spike protein and nucleocapsid protein, respectively.
Figure 2.
Middle East respiratory syndrome coronavirus (MERS-CoV) genomic RNA (upE) titers in sputum and transtracheal aspirates with vs. without sub-genomic mRNA detection. Solid lines indicate the mean and standard error of the mean. The dashed line indicates the detection limit.
In oropharyngeal swab specimens, sub-genomic mRNA was detected only in the severe group for up to 11 days after the illness onset (Figure 1B), and the period for detecting sub-genomic mRNA was not significantly correlated with that for MERS-CoV genomic RNA (Pearson correlation coefficient = 0.335, p = 0.378). No sub-genomic mRNA was detected in nasopharyngeal swab specimens (Figure 1C).
In the present study, replicative MERS-CoV was detected in sputum or transtracheal aspirate for up to 4 weeks after symptom development in MERS-CoV-infected patients with severe pneumonia. This result is consistent with the findings of previous studies that have tested MERS-CoV genomic RNA (Memish et al., 2014, Corman et al., 2016, Min et al., 2016). On the basis of these results, infection prevention and control precautions should be thoroughly applied for at least 1 month after symptom onset if the patient with MERS-CoV infection has severe pneumonia.
The differences in the detection of replicative viruses between upper and lower respiratory tract specimens may have originated from differences in viral titers. Several studies have demonstrated that the viral titer of MERS-CoV RNA in upper respiratory tract specimens is lower than that in lower respiratory tract specimens (Oh et al., 2016, Corman et al., 2016). In the present study, sub-genomic mRNA was not detected in any of the nasopharyngeal specimens. The current guidelines recommend that isolation should continue until two consecutive upper respiratory tract specimens taken at least 24 h apart test negative by RT-PCR (WHO, 2018). However, the present study suggests that, if possible, lower respiratory tract specimens should be used to determine the duration of isolation and that nasopharyngeal swab specimens should be avoided.
This study has a few limitations. First, differences in sensitivity between the real-time RT-PCR used to detect genomic RNA and the conventional RT-PCR for sub-genomic mRNA may have affected the results. Second, RT-PCR methods for sub-genomic mRNA have not been validated elsewhere. Other methods, such as detecting live virus, should be performed to validate the methods used in this study.
In conclusion, replicative MERS-CoV was detected in lower respiratory tract specimens for up to 4 weeks after symptom development, which was well correlated with the detection of genomic RNA. In upper respiratory tract specimens, the detection of sub-genomic mRNA and genomic RNA did not correlate. These findings suggest that MERS-CoV does not replicate well in the upper respiratory tract.
Acknowledgements
This work was supported by a grant from the Korean Healthcare Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number HI15C3227).
Acknowledgments
Conflict of interest
None.
Corresponding Editor: Eskild Petersen, Aarhus, Denmark
Contributor Information
Malik Peiris, Email: malik@hku.hk.
Myoung-don Oh, Email: mdohmd@snu.ac.kr.
References
- Corman V.M., Albarrak A.M., Omrani A.S., Albarrak M.M., Farah M.E., Almasri M. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin Infect Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Assiri A.M., Al-Tawfiq J.A. Middle East respiratory syndrome coronavirus (MERS-CoV) viral shedding in the respiratory tract: an observational analysis with infection control implications. Int J Infect Dis. 2014;29:307–308. doi: 10.1016/j.ijid.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min C.K., Cheon S., Ha N.Y., Sohn K.M., Kim Y., Aigerim A. Comparative and kinetic analysis of viral shedding and immunological responses in mers patients representing a broad spectrum of disease severity. Sci Rep. 2016;6 doi: 10.1038/srep25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.D., Park W.B., Choe P.G., Choi S.J., Kim J.I., Chae J. Viral load kinetics of MERS coronavirus infection. N Engl J Med. 2016;375:1303–1305. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- Park W.B., Perera R.A., Choe P.G., Lau E.H., Choi S.J., Chun J.Y. Kinetics of serologic responses to mers coronavirus infection in humans, South Korea. Emerg Infect Dis. 2015;21:2186–2189. doi: 10.3201/eid2112.151421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2018. Management of asymptomatic persons who are rt-PCR positive for Middle East respiratory syndrome coronavirus (MERS-CoV). Interim guidance. Updated 3 January 2018. Available at: http://apps.who.int/iris/bitstream/10665/180973/1/WHO_MERS_IPC_15.2_eng.pdf?ua=1&ua=1. [Aaccessed 21 March 2018] [Google Scholar]
- Woo P.C., Lau S.K., Fan R.Y., Lau C.C., Wong E.Y., Joseph S. Isolation and characterization of dromedary camel coronavirus uae-hku 23 from dromedaries of the Middle East: minimal serological cross-reactivity between MERS coronavirus and dromedary camel coronavirus uae-hku 23. Int J Mol Sci. 2016 doi: 10.3390/ijms17050691. [DOI] [PMC free article] [PubMed] [Google Scholar]