Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein, which is not expressed by other known coronaviruses, can down-regulate the envelope (E) protein via a proteasome-dependent pathway. Here, we showed that the down-regulation of E is not dependent on the lysine residues on 8b and the reduction of polyubiquitination of E mutants is not correlated with their down-regulation by 8b, suggesting an ubiquitin-independent proteasome pathway is involved. A time-course study revealed that 8b was expressed at late-stages of SARS-CoV infection. By using Vero E6 cells stably expressing green fluorescence protein-tagged 8b, ectopic expression of 8b was shown to significantly reduce the production of progeny virus and down-regulate E expression. Taken together, these results suggest that 8b negatively modulates virus replication by down-regulating E via an ubiquitin-independent proteasome pathway.
Keywords: Severe acute respiratory syndrome coronavirus (SARS-CoV), Accessory proteins, 8b protein, Envelope (E), Ubiquitin-independent, Proteasome pathway
1. Introduction
The severe acute respiratory syndrome coronavirus (SARS-CoV) was identified as a novel coronavirus that caused a viral outbreak in 2003. It spread from Guangdong province in China to about 28 countries around the world and caused 774 deaths in 8096 infected individuals [1], [2], [3]. SARS-CoV is an enveloped, positive-sense RNA virus with a genome size of 29.7 kb, and belongs to group 2b within the Coronaviridae family [4]. The genome of SARS-CoV encodes for the replicase polyproteins (pp1a and pp1ab) and four structural proteins, spike (S), membrane (M), envelope (E) and nucleocapsid (N), that are common to all coronaviruses as well as eight unique accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b, 9b) [5], [6], [7]. Among these accessory proteins, 3a, 6 and 7a were shown to be virion-associated [8], [9], [10], [11] while 6, 7b and 8a were demonstrated to be able to modulate virus replication [12], [13], [14]. For other coronaviruses, the accessory proteins have been shown to contribute to viral virulence and pathogenesis even though they are not essential for coronavirus replication in cultured cells [15], [16].
Comparative genomic analysis shows that the genome of SARS-CoV isolated from patients in the middle phase of the epidemic has a 29-nucleotide deletion in the open-reading frame (ORF) 8 region compared to SARS-CoV isolates from animals and patients in the early phase [17], [18], [19]. It is postulated that this deletion within the ORF8 region of the SARS-CoV genome may be an evolutionary adaptation to the human host [19], [20], [21]. The 29-nucleotide deletion results in the splitting of a single ORF8ab into two overlapping ORFs, ORF8a and ORF8b. The ORF8ab region encodes for a single protein 8ab of 122 amino acids while ORF8a and ORF8b encode for proteins 8a and 8b of 39 and 84 amino acids, respectively [19].
Recent evidence shows that 8ab, which can be glycosylated, is a functional ER-resident protein, behaves like an integral membrane protein and modulates the unfolded protein response pathway by specifically activating ATF6 [22], [23], [24]. 8a was reported to promote viral replication and induce apoptosis while 8b was shown to induce DNA synthesis [13], [25]. We have also previously characterized the cellular properties of the 8a, 8b and 8ab proteins and found that 8b down-regulates the SARS-CoV E protein through a post-translational pathway [26]. Further studies have shown that proteasome inhibitors can inhibit both the degradation of 8b and 8b-mediated down-regulation of E [22]. In addition, 8b and 8ab were shown to be able to bind covalently and non-covalently to monoubiquitin and polyubiquitin. Ubiquitin is a 76 amino-acid protein that plays a role in the regulation of protein interactions and protein sorting pathways [27]. Many viruses have evolved to hijack the host ubiquitin-proteasome machinery to regulate their own viral protein levels [28]. Ubiquitin conjugation to target protein consists of three distinct enzymatic steps [28], [29], [30]. First, an activating enzyme (E1) binds ubiquitin through a thioester bond, using ATP in the process. Next, an ubiquitin-conjugating enzyme (E2) takes over the activated ubiquitin through a thioester linkage and finally, the transfer of the ubiquitin to a lysine residue on the target protein is catalyzed by an E3 ligase.
In this study, site-directed mutagenesis experiments were performed to determine the contributions of the lysine residues in 8b or E to the ubiquitination of these proteins and pathway for 8b-mediated down-regulation of E. By using Vero E6 cells stably expressing green fluorescence protein (GFP)-tagged 8b, the effects of the ectopic expression of 8b on the production of progeny SARS-CoV and expression of E in infected cells were also analyzed.
2. Materials and methods
2.1. Construction of plasmids
For the construction of pXJ40-flag-ubiquitin, the ubiquitin gene was amplified from a template, pXJMyc-ubiquitin, which was previously described in Ref. [22] by using the primers 5′-ATGGATCCATGCAGATCTT-3′ and 5′-CCGCTCGAGTTACCCACCTCTGAGAC-3′. PCR products were digested with BamHI and XhoI, and ligated into the vector pXJ40-flag. To construct pEGFP-8b, ORF8b was amplified from the template pXJ3′-8b-myc using the primers 5′-CGCTCGAGACATGTGCTTGAAGAT-3′ and 5′-CGGGATCCTTAATTTGTTCGT-3′ and the PCR products were digested with XhoI and BamHI, and ligated into the vector pEGFP-C1 (BD Biosciences Clontech). To generate mutants of 8b and E, standard PCR methods were used and the templates, pXJ3′-8b-myc and pXJ3′-E, have been previously described in Refs. [26], [31].
2.2. Culturing of stable cell lines
Vero E6 cells were transfected with 10 μg of pEGFP-8b by electroporation and plated onto a 100 mm dish. Medium containing 2 mg/ml of G418 (Invitrogen, Carlsbad, CA) was added 24 h post-transfection and cells were cultured for 3–5 days. Colonies of cells were picked and transferred into individual wells in a flat-bottomed 96-well plate and cultured for 5 days before transferring into a 24-well plate. After 3–5 days, cells were split into two 60 mm dishes for screening and freezing down. Positive clones were re-thawed and grown in a T75 flask for experiments.
2.3. Transient transfection and western blot analysis
Transient transfection was performed using Lipofectamine reagent (Invitrogen), according to the manufacturer’s protocol. Western blot analysis was performed as previously described in Ref. [26]. Some of the primary antibodies (anti-HA monoclonal, anti-GFP monoclonal (Roche Molecular Biochemicals, Indianapolis, Ind.), anti-flag polyclonal, anti-actin monoclonal (Sigma), anti-myc monoclonal, anti-myc polyclonal and anti-HA polyclonal (Santa Cruz Biotechnology, Santa Cruz, CA)) were purchased. The rabbit anti-E and anti-8b polyclonal antibodies have been described in Ref. [26].
2.4. Ubiquitination assay
For in-vivo ubiquitination assay, transfected 293T cells in 60 mm dishes were treated with 50 μM MG132 for 4 h. Cells were then scraped and lysed in ubiquitination buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Triton-X100, 1 mM EDTA, 1 mM DTT, 50 mM NaF, 0.1 mM Na3VO4). Fresh PMSF (0.1 mM), NEM (10 μM) and complete protease cocktail inhibitor (Roche) were added to the buffer for each experiment. Samples were then sonicated for 2 min at 20 s interval. Anti-myc monoclonal antibody (Santa Cruz Biotechnology) was used to precipitate the protein complexes followed by incubation with Protein-A sepharose beads (Roche). Alternatively, anti-flag monoclonal beads (Sigma) were used. The beads were washed 3× with ubiquitination buffer. Then, 20 μl of 2× Laemmli’s SDS loading buffer were added to the beads and boiled for 5 min at 100 °C and the protein complexes separated on SDS-polyacrylamide gels, which were then examined by Western blotting with an anti-flag polyclonal antibody (Sigma) or anti-HA polyclonal antibody (Santa Cruz).
2.5. Pulse-chase labeling
293T cells in 60 mm dishes were transfected with pXJ3′-E-HA, pXJ3′-E(K54R)-HA, pXJ3′-E(K64R)-HA or pXJ3′-E(K54,64R)-HA. At 24 h post-transfection, the cells were starved at 37 °C for 30 min in methionine- and cysteine-free medium (MP Biomedicals, Solon, OH) and labelled with 55 μCi of 35S-methionine and cysteine (NEN) at 37 °C for 15 min. Cells were then washed with medium containing methionine and cysteine (Invitrogen), followed by incubation with 1 ml of the medium at 37 °C and chased for 0.5–6 h. Cells were washed with 1 ml of cold PBS and lysed with 500 μl lysis buffer containing 150 mM NaCl, 20 mM Tris (pH 7.5), 1% NP40, 5 mM EDTA and 1 mM PMSF and centrifuged at 16,000× g for 10 min. Immunoprecipitation was carried out by adding anti-HA monoclonal (Roche) to the lysate and incubating at 4 °C for 1 h, followed by protein-A sepharose beads (Roche) and rotating at 4 °C overnight. The beads were washed 3× with lysis buffer. Then, 20 μl of 2× Laemmli’s SDS loading buffer were added to the beads and boiled for 5 min at 100 °C. Samples were separated on SDS-polyacrylamide gels, fixed for 30 min using fixing solution (10% acetic acid, 45% methanol), and treated with Amplify Fluorographic Reagent (Amersham Biosciences), dried and visualized by autoradiography. An imaging densitometer (Bio-Rad, Hercules, CA) was used for quantification of the intensities of specific bands on autoradiographs.
2.6. Virus infection and immunofluorescence
Vero E6 cells grown on glass slides in a 24-well plate were infected with SARS-CoV (HK39849 strain, Genbank accession number AY278491) at multiplicity of infection (M.O.I.) of 1 and harvested at 1, 8, 24 and 32 h post-infection (hpi). Cells were fixed with 4% paraformaldehyde for 1 h at room temperature and permeabilized with 0.5% Triton-X 100, followed by blocking with PBS containing 1% bovine serum albumin (BSA) for 30 min. Primary antibody incubation with rabbit anti-SΔ10 (1:200) [32] and rabbit anti-8b (1:100) [26] was carried out for 1.5 h and washed 3× using PBS containing 1% BSA before incubation with fluorescein isothiocyanate-conjugated secondary antibody (1:200) for 1 h. All incubations and washes were performed at room temperature. Slides were mounted and analyzed using a fluorescence microscope (Carl Zeiss, Hallbergmoss, Germany).
2.7. Virus infection and titration
Vero E6 cells, stably expressing GFP-8b and GFP, grown in 24-well plates were infected with SARS-CoV (Frankfurt strain 1, Genbank accession number AY291315) at M.O.I. of 0.1. While the HK39849 strain was used above, the Frankfurt strain 1 was used here. However, the sequences of these strains are 100% identical. At 1 hpi, the cells were washed twice and fresh medium was added. After 24 hpi, the virus supernatant was harvested and titration of progeny virus was carried out on a 96-well plate containing fresh Vero E6 cells as described previously in Ref. [33]. At 48 hpi, the amount of virus, determined as the 50% tissue culture infectious dose (TCID50), was calculated from the cytopathic effect induced in cell culture by serial ten-fold dilutions of the harvested virus.
2.8. Virus infection and western blot analysis
Vero E6 cells, stably expressing GFP-8b and GFP, grown in 24-well plates were infected with SARS-CoV (Frankfurt strain 1) at M.O.I. of 0.1 and 5. The cells were harvested at 18 hpi and separated on SDS-polyacrylamide gel for detection of S and E using anti-SΔ10 [32] and anti-E antibodies [26], respectively.
3. Results
3.1. Knockout of lysine residues on 8b does not prevent its polyubiquitination and ability to down-regulate E
In a previous study, 8b was shown to undergo both monoubiquitination and polyubiquitination [22]. The host ubiquitin-proteasome pathway is a commonly exploited pathway by viruses for their own protein level regulation [28] and the role of an E3 ligase is to catalyze the transfer of an activated ubiquitin from an E2 to the lysine residue of the target protein [27], [28], [29], [30]. To explore the possibility of 8b acting as an E3 ligase, a lysine knockout mutant of 8b, 8b(K4,26,81R), was generated by substituting the three lysine residues in 8b with arginine residues. As shown in Fig. 1 A, both 8b and 8b(K4,26,81R) underwent monoubiquitination and polyubiquitination (Fig. 1A, lanes 2 and 3). The monoubiquitinated product of 8b-myc was detected at ∼17 kDa while the polyubiquitinated products of 8b-myc appeared as a ladder. However, the monoubiquitinated 8b was somewhat reduced in the lysine knockout mutant (Fig. 1A, lane 7). Thus, a novel mechanism might be involved in the polyubiquitination of 8b while the lysine residues contribute partially to the monoubiquitination of 8b.
Fig. 1.
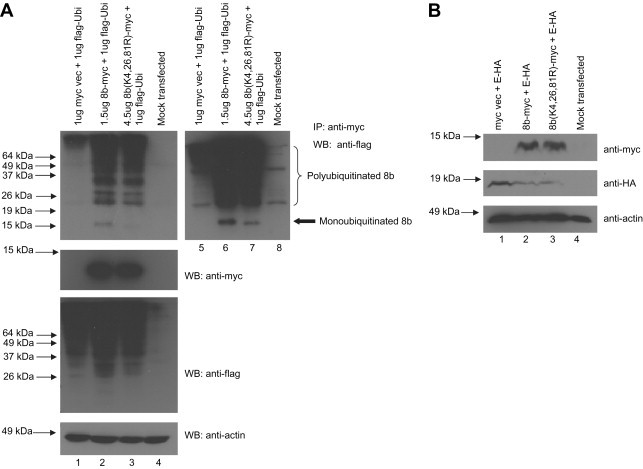
Lysine knockout mutant of 8b undergoes polyubiquitination and down-regulates E. (A) 293T cells were transfected with cDNA constructs for expressing flag-ubiquitin (flag-Ubi) and 8b-myc or 8b(K4,26,81R)-myc. The cells were harvested at 16 h post-transfection, lysed and subjected to immunoprecipitation (IP) with anti-myc mouse monoclonal antibody and protein-A beads. The amounts of polyubiquitinated 8b-myc or 8b(K4,26,81R)-myc were determined by Western blot (WB) analysis with an anti-flag rabbit polyclonal antibody (1st panel, lanes 1–4). A darker film exposure was included (1st panel, lanes 5–8). The amounts of myc-tagged and flag-tagged proteins in the lysates before IP were determined by Western blot analysis (2nd and 3rd panels). Anti-actin monoclonal antibody was used to verify that equal amounts of lysates were used in each lane (4th panel). (B) 293T cells were transfected with cDNA constructs for expressing E-HA, 8b-myc and 8b(K4,26,81R)-myc. The cells were harvested at 16 h post-transfection, lysed and subjected to Western blot analysis. The expressions of 8b and E were determined using anti-myc antibody (1st panel) and anti-HA antibody (2nd panel) respectively. Anti-actin monoclonal antibody was used to verify that equal amounts of lysates were used in each lane (3rd panel).
In addition, 8b(K4,26,81R) down-regulated the expression of E as well as 8b (Fig. 1B, lane 2 and 3). Since the polyubiquitination of 8b cannot be abolished by lysine knockout, it cannot be ascertain if the polyubiquitination of 8b is important for the 8b-mediated down-regulation of E. However, the monoubiquitination of 8b is not essential for the 8b-mediated down-regulation of E.
3.2. Substitution of K64 to R on E increases the protein stability by reducing the polyubiquitination of E
To investigate the importance of the lysine residues on E for its polyubiquitination, three substitution mutants, E(K54R), E(K64R) and E(K54,64R), were generated. As shown in Fig. 2 A, the amounts of E protein were increased in both the E(K64R) and E(K54,64R) transfected cells when compared to wild-type E transfected cells (Fig. 2A, lanes 1, 3 and 4). On the other hand, the expression of E(K54R) was the same as wild-type E (lanes 1 and 2). This result suggests that the K64R substitution can increase the protein stability of E.
Fig. 2.
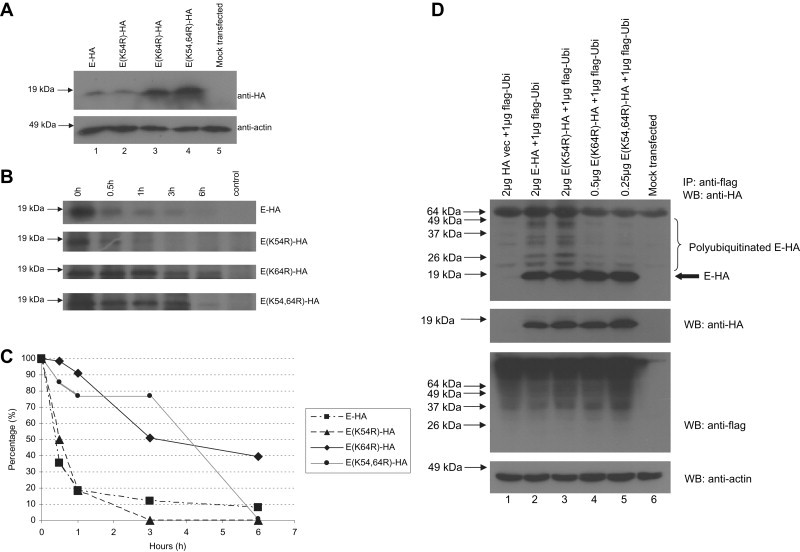
Substitution of K64 to R on E increases its stability by reducing its polyubiquitination. (A) 293T cells were transfected with 1 μg of cDNA constructs for expressing E-HA, E(K54R)-HA, E(K64R)-HA or E(K54,64R)-HA and their protein expression levels were detected using anti-HA antibody (1st panel). Anti-actin monoclonal antibody was used to verify that equal amounts of lysates were used in each lane (2nd panel). (B) Transfected cells were starved at 24 h post-transfection at 37 °C for 30 min in methionine- and cysteine-free medium. Cells were pulsed for 15 min with medium containing 35S-labelled methionine and cysteine amino acids and chased for 0–6 h before lysis, and subjected to IP with anti-HA monoclonal antibodies and protein-A beads. (C) A graph showing the amounts of 35S radio-labeled E-HA, E(K54R)-HA, E(K64R)-HA and E(K54,64R)-HA remaining at 0.5–6 h after expression. (D) 293T cells were transfected with cDNA constructs for expressing flag-ubiquitin (flag-Ubi) and E-HA, E(K54R)-HA, E(K64R)-HA or E(K54,64R)-HA. The cells were harvested at 16 h post-transfection, lysed and subjected to IP with anti-flag mouse monoclonal antibody beads. The amounts of ubiquitinated E-HA, E(K54R)-HA, E(K64R)-HA or E(K54,64R)-HA were determined by Western blot analysis with an anti-HA rabbit polyclonal antibody (1st panel). The amounts of HA-tagged and flag-tagged proteins in the lysates before IP were determined by Western blot analysis (2nd and 3rd panels). Anti-actin monoclonal antibody was used to verify that equal amounts of lysates were used in each lane (4th panel).
A pulse-chase labeling experiment was performed in a similar manner as reported by Nal et al. [34]. Consistently, the results showed that E(K64R) and E(K54,64R) decayed at slower rates than wild-type E while E(K54R) has a similar decay rate (Fig. 2B). The intensities of specific bands in the autoradiographs were quantified using an imaging densitometer and the percentages of remaining proteins after different times of decay were plotted. As shown in Fig. 2C, the half-lives of E(K64R) and E(K54,64R) are greater than 3 h while that of wild-type E and E(K54R) are less than 1 h.
Next, ubiquitination assay was performed to investigate the effects of K54R and K64R substitutions on the polyubiquitination of E. 293T cells were co-transfected with plasmids expressing flag-ubiquitin and E, E(K54R), E(K64R) or E(K54,64R), and an immunoprecipitation experiment was performed to pull-down proteins associated with the flag-tagged ubiquitin. The results showed that the K54R substitution does not affect the polyubiquitination of E(K54R) (Fig. 2D, lanes 2 and 3) but K64R substitution reduces the polyubiquitination of E(K64R) and E(K54,64R) (Fig. 2D, lanes 4 and 5). Hence, the increase in the stabilities of E(K64R) and E(K54,64R) is correlated with a decrease in their polyubiquitination.
3.3. Both lysine residues on E are important for 8b-mediated E degradation
To determine if the lysine residues on E are important for the 8b-mediated degradation pathway, the expressions of E, E(K54R), E(K64R) and E(K54,64R) were determined in the absence or presence of 8b (Fig. 3 A). The intensities of specific bands in the autoradiographs were quantified using an imaging densitometer and the percentage reduction in E expression in the presence of 8b was computed. As shown in Fig. 3B, both E(54R) and E(K64R) were less sensitive to 8b-mediated degradation than wild-type E. As the double substitution mutant, E(K54,64R), was the least sensitive to 8b-mediated degradation, it appears that the contributions of K54 and K64 are additive.
Fig. 3.

Both lysine residues on E are involved in the 8b-mediated down-regulation of E. (A) 293T cells were transfected with cDNA constructs for expressing E-HA, E(K54R)-HA, E(K64R)-HA or E(K54,64R)-HA in the absence or presence of 8b-myc and harvested at 24 h post-transfection. The amounts of HA-tagged and myc-tagged proteins expressed were determined by Western blot analysis (1st and 2nd panels). Mock-transfected cells were used as negative control (lanes 5 and 10). (B) The expression levels of E-HA, E(K54R)-HA, E(K64R)-HA and E(K54,64R)-HA were quantified with a densitometer and their expression levels in the presence of 8b relative to that in the absence of 8b are presented in the graph.
3.4. Expression of 8b increases with time in SARS-infected Vero E6 cells
In our earlier study, 8b of SARS-CoV was found to be expressed in Vero E6 cells using both Western blot and immunofluorescence assays [26]. However, Oostra et al., reported that they could not detect 8b in SARS-CoV infected cells at 8 hpi [23]. To address this discrepancy, the expressions of S and 8b in Vero E6 cells were analyzed at different time-points post infection (Fig. 4 ). The results showed that S was expressed early during infection and could be detected in almost all the cells at 8 hpi (Fig. 4, M–P). However at the 8 hpi, only a small percentage of cells have a weak expression of 8b (Fig. 4, F). At 24 and 32 hpi, the 8b expression increased and could be detected in a larger percentage of infected cells (Fig. 4, G and H, respectively). This is consistent with our previous results [26] and showed that 8b was expressed at late time-points during SARS-CoV infection.
Fig. 4.
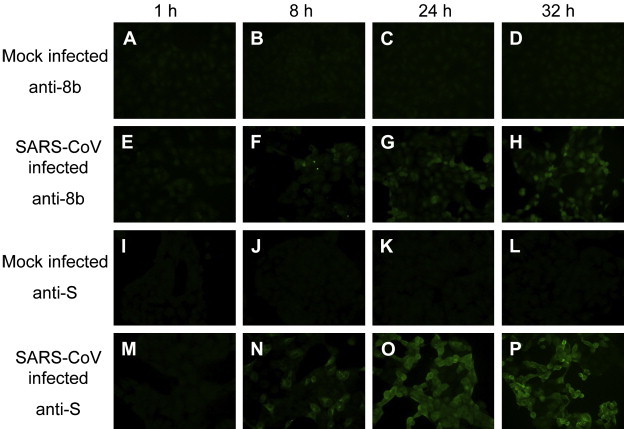
8b has a higher expression at late-stage infection of Vero E6 cells. Vero E6 cells were either infected with SARS-CoV (E–H, M–P) at M.O.I. of 1 or mock-infected as negative control (A–D, I–L). Cells were harvested at 1, 8, 24 and 32 hpi, fixed using 4% paraformaldehyde and permeabilized with 0.5% Triton-X 100. Expression of 8b and S was detected using anti-8b (A–H) and anti-S polyclonal antibodies (I–P), respectively.
3.5. 8b is a negative modulator of SARS-CoV replication
Cells stably expressing GFP (as control) and GFP-8b were generated to determine the effect of 8b on SARS-CoV replication. As shown by Western blot analysis (Fig. 5 A), GFP-8b (lane 2) is expressed at a much lower expression than GFP (lane 1). To determine if the level of GFP-8b expressed in the stable cell line is sufficient to down-regulate E, the stable cell lines were transfected with the pXJ3′-E plasmid. As shown in Fig. 5B, the expression of E was down-regulated in GFP-8b expressing cells (lane 3) but not in the control cells expressing GFP (lane 2).
Fig. 5.
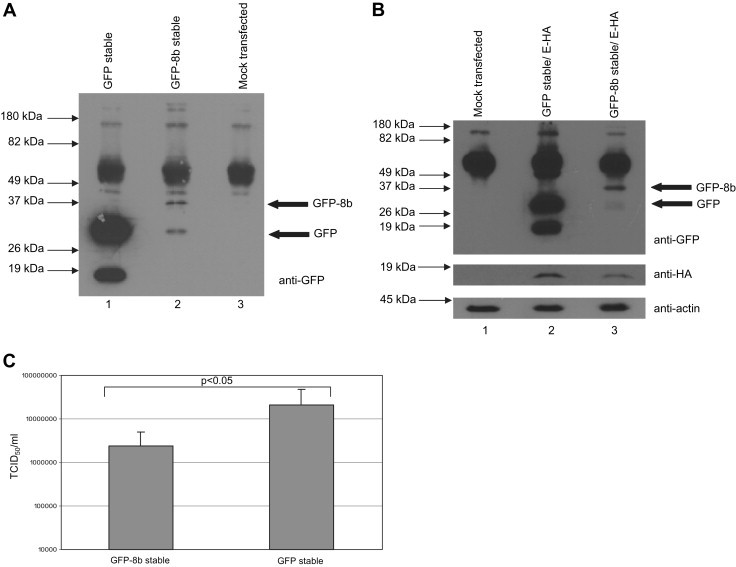
Ectopic 8b expression reduces viral replication in Vero E6 cells. (A) Cells stably expressing GFP and GFP-8b were harvested and lysed and subjected to Western blot analysis. Expressions of GFP and GFP-8b in the stable cell lines were detected using an anti-GFP antibody. (B) GFP-8b and GFP stable cells were transfected with a cDNA construct for expressing E, harvested and lysed at 24 h post-transfection. Expressions of GFP and GFP-8b were detected using an anti-GFP antibody (1st panel). The amount of E in the cell lysates was detected using anti-E antibody (2nd panel). Anti-actin monoclonal antibody was used to verify that equal amounts of lysates were used in each lane (3rd panel). (C) A graph showing the titers of the virus produced by GFP and GFP-8b cells. The average and standard deviation of readings from 12 independent experiments are plotted. Statistical analysis was performed using Student's t test assuming unequal variance and the difference between GFP and GFP-8b cells was found to be significant (p < 0.05).
Next, the replication of SARS-CoV in these two stable cell lines was compared. The stable cell lines were infected with SARS-CoV at an M.O.I. of 0.1, and at 24 hpi, the supernatant was titrated out on a 96-well plate containing Vero E6 cells. The amount of virus was deduced by the 50% tissue culture infective dose (TCID50), which was calculated from the cytopathic effect (CPE) induced in cell culture by different dilutions of the harvested virus as previously described in Ref. [33]. As shown in Fig. 5C, the amount of virus produced by the GFP-8b cells was about 10-fold lower than the GFP-expressing cells, suggesting that 8b has a negative effect on the SARS-CoV replication. The difference in viral titers is statistically significant (p < 0.05).
3.6. E is down-regulated in SARS-CoV infected GFP-8b expressing cells
To investigate the effect of 8b on the expression level of E during SARS-CoV infection, GFP-8b and GFP cells were infected with SARS-CoV at different M.O.I. and harvested at 18 hpi. At M.O.I. of 0.1, the expression of S in the GFP-8b cells was much lower than GFP cells (Fig. 6 , lanes 1 and 2) and this is probably a result of the lower viral replication in the GFP-8b cells as observed above. At M.O.I. of 5, the expressions of S in both GFP-8b and GFP-expressing cells were similar (lanes 3 and 4). However, the expression of E was significantly lower in the GFP-8b cells compared to the GFP cells (lanes 3 and 4). This result suggests that GFP-8b down-regulates the expression of E in SARS-CoV infected cells.
Fig. 6.
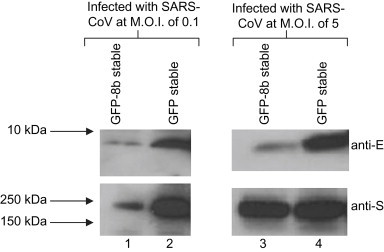
Expression of E is down-regulated in SARS-CoV infected cells stably expressing GFP-8b. Vero E6 cells stably expressing GFP and GFP-8b were infected with SARS-CoV at M.O.I. of 0.1 (lanes 1 and 2) and M.O.I. of 5 (lanes 3 and 4). Cells were harvested and lysed at 18 hpi and the expressions of S and E in the cell lysates were detected using anti-E (1st panel) and anti-S (2nd panel) antibodies, respectively.
4. Discussion
Since the outbreak of SARS-CoV in 2003, extensive studies have been carried out to understand the evolution of the virus as well as the functions of each of the viral proteins. From genomic studies, it is predicted that the reservoirs of SARS-CoV are the bats, with the palm civet acting as the amplification media before spilling the virus over to the human population [35], [36], [37]. When a RNA virus switches from host-to-host, genomic changes take place due to the high error rate of the viral RNA-dependent RNA polymerase and absence of a proof-reading mechanism, thus allowing for the evolution of new strains and selection of the fittest strains [38]. During human-to-human transmission in the middle phase of the epidemic, the ORF8 region of the SARS-CoV acquired a 29-nucleotide deletion, resulting in the removal of the 8ab protein while acquiring new proteins, 8a and 8b [19]. Whether this genomic change is a result of the instability of the genome or the acquisition of new functions for host adaptation remains to be addressed.
Previously, we have shown that 8b can down-regulate E in SARS-CoV infected cells [26] and 8b can be monoubiquitinated and polyubiquitinated [22]. It is widely known that lysine residues are important for the binding of ubiquitin to a protein for ubiquitination [27]. In this study, the results showed that the lysine residues in 8b are not required for its polyubiquitination or ability to down-regulate E (Fig. 1). Furthermore, the degradation of E by 8b is dependent on both K54 and K64 in E while the half-life and polyubiquitination of E in the absence of 8b is only dependent on K64 (Fig. 2, Fig. 3). Thus, the 8b-mediated degradation of E, which can be inhibited by proteasome inhibitors [22], seems to involve an ubiquitin-independent proteasome pathway. Several ubiquitin-independent proteolytic functions of the proteasome have been reported [39]. Although many of the ubiquitin-independent pathways have yet to be precisely defined, a recent study showed that more than 20% of cellular proteins are degraded by the proteasome via ubiquitin-independent pathways [40].
Recently, a study using reverse genetics techniques showed that deletion of group specific genes 6, 7a, 7b, 8a, 8b and 9b from the SARS-CoV genome reduced its replication titer by 5–10-fold in human CaCo2, Huh-7.5.1 and Huh-7 cells while deletion of E resulted in a 100–1000-fold reduction, compared to cells infected with wild-type SARS-CoV [41], [42]. A time-course study revealed that 8b was expressed at late time-points post infection (∼32 hpi) (Fig. 4). By comparing Vero E6 cells stably expressing GFP-8b or GFP alone, progeny virus production was found to be significantly reduced in the presence of ectopic expression of 8b (Fig. 5). Moreover, the expression of E in SARS-CoV infected cells was down-regulated in cells stably expressing GFP-8b when compared to GFP-expressing cells (Fig. 6). This is consistent with our previous observation that infected cells with high level of 8b have low level of E [26] and suggests that 8b reduces viral replication by down-regulating the expression of E protein.
Taken together, our results suggest that the overexpression of the SARS-CoV 8b protein reduces viral replication by down-regulating E via an ubiquitin-independent proteasome pathway. In virus evolution, rapid mutation is constantly taking place to achieve a strain which can have optimal replication titer for both efficient transmission and the ability to evade the host immune response [43], [44]. Based on the results obtained using Vero E6 overexpressing 8b, we speculate that the ability of 8b to reduce viral replication could be a mechanism to prevent the over-production of virus after reaching an optimal viral titer. However, future studies need to be performed to determine if 8b can modulate viral replication during natural infection. Interestingly, another SARS-CoV accessory protein, 7b, was recently shown to be an attenuating factor [12]. One unanswered question is whether the replication of a SARS-CoV without 8b in the natural host will be higher than that of the wild-type virus. Previous studies have shown the deletion of 8b and several other accessory proteins, namely 6, 7a, 7b, 8a and 9b, from the SARS-CoV genome reduced viral replication in cell cultures by 5–10-fold [41], [42]. Thus, it is possible that the weak down-regulation of viral replication by 8b can be counter-balanced by other accessory proteins. For example, Chen et al. have shown that 8a protein can increase viral replication [13]. Thus, the SARS-CoV could have acquired the 8a and 8b proteins to fine-tune its replication in the human host.
Acknowledgements
This work was supported by a grant from the Agency for Science, Technology and Research (A∗STAR), Singapore.
References
- 1.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Who . 2004. Summary of Probable SARS Cases with Onset of Illness from November 1, 2002 to July 31, 2003.http://www.who.int/csr/sars/country/table2004_04_21/en/index.html [Accessed 21.04.04] [Google Scholar]
- 3.Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan Y.J., Lim S.G., Hong W. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antivir. Res. 2006;72:78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 7.Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen S., Lin P.S., Chao Y.C., Zhang A., Yang X., Lim S.G., Hong W., Tan Y.J. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 2005;330:286–292. doi: 10.1016/j.bbrc.2005.02.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C., Ito N., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 2006;80:7287–7294. doi: 10.1128/JVI.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J. Virol. 2007;81:5423–5426. doi: 10.1128/JVI.02307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfefferle S., Krahling V., Ditt V., Grywna K., Muhlberger E., Drosten C. Reverse genetic characterization of the natural genomic deletion in SARS-Coronavirus strain Frankfurt-1 open reading frame 7b reveals an attenuating function of the 7b protein in-vitro and in-vivo. Virol. J. 2009;6:131. doi: 10.1186/1743-422X-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C.Y., Ping Y.H., Lee H.C., Chen K.H., Lee Y.M., Chan Y.J., Lien T.C., Jap T.S., Lin C.H., Kao L.S., Chen Y.M. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 2007;196:405–415. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J., Falcon A., Zhou H., Netland J., Enjuanes L., Perez Brena P., Perlman S. Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J. Virol. 2009;83:2368–2373. doi: 10.1128/JVI.02371-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Haan C.A., Masters P.S., Shen X., Weiss S., Rottier P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U S A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinese S.M.E.C. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 19.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 20.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 21.Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., Cui B., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Qi X., Chen K., Du L., Gao K., Zhao Y.T., Zou X.Z., Feng Y.J., Gao Y.F., Hai R., Yu D., Guan Y., Xu J. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le T.M., Wong H.H., Tay F.P., Fang S., Keng C.T., Tan Y.J., Liu D.X. Expression, post-translational modification and biochemical characterization of proteins encoded by subgenomic mRNA8 of the severe acute respiratory syndrome coronavirus. FEBS J. 2007;274:4211–4222. doi: 10.1111/j.1742-4658.2007.05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oostra M., de Haan C.A., Rottier P.J. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 2007;81:13876–13888. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung S.C., Chao C.Y., Jeng K.S., Yang J.Y., Lai M.M. The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology. 2009;387:402–413. doi: 10.1016/j.virol.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law P.Y., Liu Y.M., Geng H., Kwan K.H., Waye M.M., Ho Y.Y. Expression and functional characterization of the putative protein 8b of the severe acute respiratory syndrome-associated coronavirus. FEBS Lett. 2006;580:3643–3648. doi: 10.1016/j.febslet.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keng C.T., Choi Y.W., Welkers M.R., Chan D.Z., Shen S., Gee Lim S., Hong W., Tan Y.J. The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology. 2006;354:132–142. doi: 10.1016/j.virol.2006.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 28.Banks L., Pim D., Thomas M. Viruses and the 26S proteasome: hacking into destruction. Trends Biochem. Sci. 2003;28:452–459. doi: 10.1016/S0968-0004(03)00141-5. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson K.D. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 30.Ciechanover A., Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Tan Y.J., Goh P.Y., Fielding B.C., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., Lim S.G., Hong W. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keng C.T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H., Chou C.F., Loh C.B., Wang S., Fu J., Yang X., Lim S.G., Hong W., Tan Y.J. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J. Virol. 2005;79:3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akerstrom S., Mousavi-Jazi M., Klingstrom J., Leijon M., Lundkvist A., Mirazimi A. Nitric oxide inhibits the replication cycle of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:1966–1969. doi: 10.1128/JVI.79.3.1966-1969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.Y., Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- 35.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vijaykrishna D., Smith G.J., Zhang J.X., Peiris J.S., Chen H., Guan Y. Evolutionary insights into the ecology of coronaviruses. J. Virol. 2007;81:4012–4020. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U S A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Domingo E., Escarmis C., Sevilla N., Moya A., Elena S.F., Quer J., Novella I.S., Holland J.J. Basic concepts in RNA virus evolution. FASEB J. 1996;10:859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 39.Orlowski M., Wilk S. Ubiquitin-independent proteolytic functions of the proteasome. Arch. Biochem. Biophys. 2003;415:1–5. doi: 10.1016/s0003-9861(03)00197-8. [DOI] [PubMed] [Google Scholar]
- 40.Baugh J.M., Viktorova E.G., Pilipenko E.V. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeDiego M.L., Alvarez E., Almazan F., Rejas M.T., Lamirande E., Roberts A., Shieh W.J., Zaki S.R., Subbarao K., Enjuanes L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007;81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dediego M.L., Pewe L., Alvarez E., Rejas M.T., Perlman S., Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vossen M.T., Westerhout E.M., Soderberg-Naucler C., Wiertz E.J. Viral immune evasion: a masterpiece of evolution. Immunogenetics. 2002;54:527–542. doi: 10.1007/s00251-002-0493-1. [DOI] [PubMed] [Google Scholar]
- 44.Bonhoeffer S., Sniegowski P. Virus evolution: the importance of being erroneous. Nature. 2002;420:367–369. doi: 10.1038/420367a. [DOI] [PubMed] [Google Scholar]


