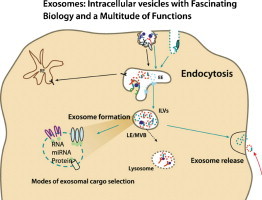
Graphical abstract
Exosomes are nanometer sized cellular vesicles released into surrounding bio-fluids. Containing the parent cell's RNA, DNA, protein and lipid component, they reflect the cell's condition and hold promise as biomarkers of exquisite specificity.
Abbreviations: mRNA, messenger RNA; miRNA or miR, micro RNA; MVB, multivesicular bodies; ATPase, adenosine triphosphatase; DNA, deoxyribonucleic acid; TSG101, tumor susceptibility gene 101; ILV, intraluminal vesicles; Rab GTPases, “Ras-related in brain” GTPase; GTPase, guanosine triphosphatase; SNAREs, SNAP (soluble NSF attachment protein) receptor; AP2, clathrin adaptor protein; Hsc70, heat shock cognate 71 kDa protein; PI3K, phosphatidyl inositol-3-kinase; CD, cluster of differentiation; iDex, immature D cell derived exosomes; ICAM1, inter-cellular adhesion molecule 1; APCs, antigen presenting cells; TGF, tumor growth factor; TNF, tumor necrosis factor; IL, interleukin; MHC, major histocompatibility complex; DC, dendritic cell; LFA1, lymphocyte function-associated antigen 1; CXCL5, C-X-C motif chemokine 5; MIF, macrophage migration inhibitory factor; CCR1, chemokine (C–C motif) receptor 1; EMMPRIN, extracellular matrix metalloproteinase inducer; HIV, human immunodeficiency virus; EBV, Epstein Barr virus; VLP, virion-like particles; HLA-DR, MHC class II cell surface receptor; SARS, severe acute respiratory syndrome; VEGF, vascular endothelial growth factor; ESCRT, endosomal sorting complex required for transport; HLA-DR, MHC class II receptor encoded by human leukocyte antigen complex; TSPAN, tetraspanin; KRAS, protein involved in cell cycle regulation; EGFR, epidermal growth factor receptor; RT-PCR, reverse transcriptase PCR; PSA, prostate-specific antigen; PCA3, prostate CAncer gene 3; I/R, ischemia reperfusion; AQP1, aquaporin 1; NKCC2, sodium–potassium–chloride co-transporter; AKI, acute kidney injury; CKD, chronic kidney disease; ATF3, cyclic AMP-dependent transcription factor; WT1, Wilms tumor protein; ELISA, enzyme linked immunosorbent assay; FACS, fluorescence activated cell sorting
Keywords: Exosomes, Biomarkers, Fluidome, Microenvironment, Secretome, Microvesicles
Abstract
Exosomes are bioactive vesicles derived from the cell's endosomal membrane system and secreted into surrounding body fluids. Exosomes contain cell and cell-state specific cargos of protein, mRNA and miRNA. Exosome formation, cargo content, and delivery to surrounding cells is of immense biological interest considering the role that exosomes are believed to play in various pathological conditions. They aid antigen presentation by immune cells and can exhibit either anti-inflammatory or pro-inflammatory properties depending on the parent antigen-presenting cell's conditioning. Viruses can hijack a host cell's exosomal machinery to evade host defense systems aiding in the trans-infection of viruses.
Tumor derived exosomes may help establish an oncogenic niche systemically via delivery of protein, mRNA, and miRNA that can aid angiogenesis, cell proliferation, and cell survival. Exosomes have also been implicated in the spread of neurodegenerative diseases.
Studies have shown that exosomes are selectively taken up by cells distal from their release. They can reprogram the recipient cells due to their active molecular cargo. Cell-lineage and state-specific exosomes imply that they may therefore harbor body fluid-based biomarkers of unparalleled accuracy, particularly for tissues that are difficult or impossible to access.
Exosome-specific membrane proteins provide markers enabling exosome identity and selection, while cell type and cell condition-specific protein, mRNA and miRNA cargo provide a rich potential source of biomarkers. This review serves to provide an overview of the current state of the science in the burgeoning field of exosome biology.
1. Cells communicate with their environment via multiple mechanisms
Organisms, tissues and cells must all continuously correspond with their environment in order to best adapt to their surrounding microcosm. Classically, cells were known to secrete protein cues into extracellular space by shedding the plasma membrane with the associated protein and lipid components or by expelling a variety of vesicular particles containing a subset of the cell's protein and membrane content. To date the long distance transmission of signals between cells and tissues has commonly been described by protein-based signaling systems exemplified by enzymes, hormones, cytokines, and chemokines.
The mode by which proteins aid cellular communication is varied and classified primarily on the basis of the “range of action” of the signaling molecules. For example, juxtacrine signaling (also known as contact-dependent signaling), involves actual cell-to-cell contact with the exchange of cytoplasmic materials through nanotubes or gap junctions (exemplified by the Notch signaling pathway). Paracrine signaling refers to the signaling effect of released molecules in close proximity to the source/producing cell.
Finally in endocrine signaling, protein molecules produced by a cell are transported systemically through the blood stream to act distally, and may act either generically on all or many cells or specifically on select recipient cells containing the appropriate molecular characteristics (i.e., correct receptor, etc.). The function of such protein-based signaling cues is generally tri-phasic involving, (i) binding to a cell surface receptor, (ii) internalization into the cell, and (iii) downstream signaling that translates into a measurable response in the cell.
In contrast to well-studied protein based cell-to-cell signaling discussed above, the possibility of RNA (ribonucleic acid) based cell-to-cell signaling has come to the forefront only recently. This resulted from the discovery that cell-extruded membranous bioactive vesicles called ‘exosomes’ transfer both functional protein and RNA species from one cell to another resulting in a change in the recipient cell [1], [2], [3]. Because of these properties, exosomes may be a rich source of stable and reproducible biomarkers in a variety of conditions.
The present review provides a brief description of the biogenesis and molecular properties of exosomes as established to date. It reviews studies that have implicated functional activities of exosomes in a few model systems and finally, reviews the biomarker potential of exosomes with respect to soluble/accessible biomarkers.
2. Exosomes
Cells secrete a variety of small, defined vesicles variously termed “exosomes”, “microparticles”, “microvesicles”, and “apoptotic blebs”. In general, there remains a state of ambiguity regarding classification of particles into these categories and the terms are often used interchangeably. The classification of a particle strictly as an exosome is based primarily on size, density, and membrane composition. The presence of late endosome components like Alix, TSG101 (tumor susceptibility gene 101) and tetraspanins reveals the multivesicular body (MVB) origin of exosomes [4].
2.1. Discovery and biogenesis of exosomes from the endosomal system
Exosomes were first described in 1981 as exfoliation from monolayer cultures containing ecto-ATPase (adenosine triphosphatase) and ecto-5′ exonuclease activity. This suggests that they are derived from specific sub-domains of the plasma membrane [5]. Early research by Johnstone et al. established that such microvesicles help maturing erythrocytes get rid of transferrin receptors and other obsolete proteins [6]. Visualization by electron microscopy and correlation of the vesicle release kinetics with the well established endocytosis kinetics of markers such as the transferrin receptor (TfR), revealed that a subset of endosomal vesicles are released intact to the cell exterior in a mechanism distinct from endocytic recycling [6].
In a process that involves invagination of the limiting membrane to form a structure budding into the lumen of the late endosome, these exosomes are known to encapsulate material from the cell cytoplasm including RNA, protein and DNA (deoxyribonucleic acid) (Fig. 1 ). Two membrane inversions occur during the formation of an exosome from a membrane patch on the plasma membrane. These two membrane inversions allow exosomes to present trans membrane cargo in the same orientation relative to the cytoplasm as at the plasma membrane [7], [8]. One inversion occurs at the plasma membrane during endocytic internalization as the plasma membrane invaginates prior to budding off. The second inversion occurs as the intraluminal vesicles (ILVs) bud off into the lumen of the late endosome (Fig. 2a).
Fig. 1.
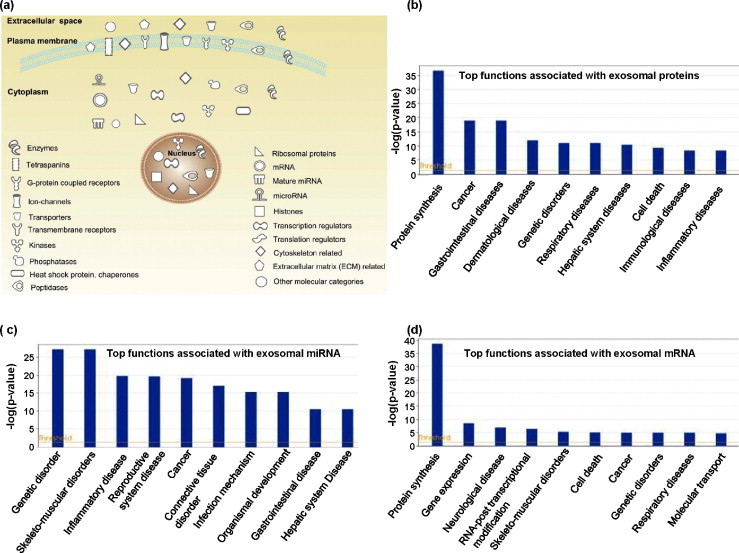
Exosomes contain diverse types of proteins, mRNA and miRNA. This figure shows the exosomal content of exosomes derived from MC9 mouse cells [2]. The exocarta compendium was queried for this exosome study with protein, mRNA and miRNA content recorded from the same samples. Considering the very large number of entities, we considered only those proteins and RNA that are part of the top physiological functional groups. Ingenuity Pathway Analysis was used to cluster the proteins by molecular category, subcellular localization and functional group. In the diagram, the top right panel (a) indicates the subcellular localization of exosomal proteins and the mRNAs (as representatives of proteins they may encode if all the mRNAs are functional and get translated). The symbol and legend are indicated on the figure. The top 10 functional clusters of proteins (b), miRNA (c) and mRNA (c) are graphically depicted. These data represent the top 10 hits from Ingenuity generated canonical pathway analysis for functional protein clusters. According to recent reports exosomes also contain mitochondrial DNA and oncogenic/amplified DNA.
Fig. 2.
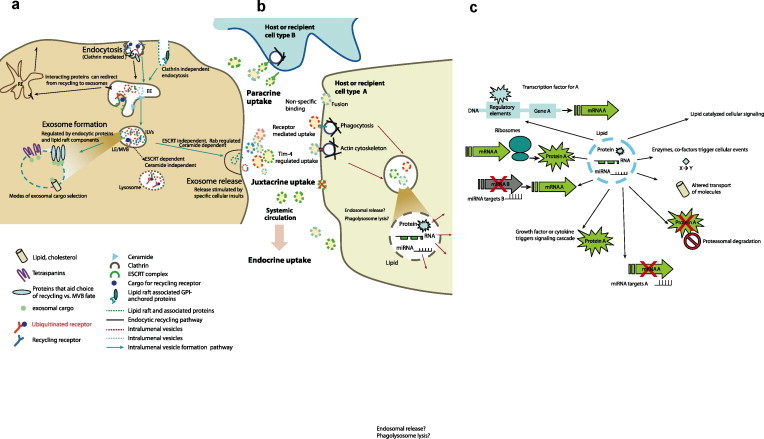
The biogenesis and molecular relevance of exosomes. (a) Exosomes arise from the cell′s endosomal system. The plasma membrane invaginates and buds off during endocytic internalization. Cargo destined for lysosomal degradation is ubiquitinated. At the early endosome the decision to recycle or be directed to the late endosome relies on specific protein interactions and domain based sorting. The early endosome matures into a late endosomal compartment where inward budding of intralumenal vesicles (ILVs) allows the capture of cytoplasmic cargo. Distinct subsets of ILVs may target cargo for lysosomal degradation (ESCRT dependent, ceramide independent) or release cargo to the cell′s exterior via exosomes. The formation of exosomes is ceramide biosynthesis dependent and relatively ESCRT independent. Exosomes retain similar membrane topology as the plasma membrane due to the two membrane inversion events. Sorting of cargo into exosomes relies on specific interactions with lipids and proteins on membrane sub-domains as well as interactions with the tetraspanin web. (b) Exosomes are internalized by cells close to the donor or originating cell (juxtacrine uptake), some distance from the donor cell (paracrine uptake) or they may spread through systemic circulation and be taken up by distant tissues (endocrine uptake). There is specificity in exosome uptake as a function of recipient cell lineage. Exosome surface markers and host cell surface markers contribute to such selectivity in uptake. Exosomes may begin protein based signaling functions even as they adhere to the host cell. Uptake of exosomes occurs primarily through phago-lysosome intermediates or utilizing specific receptors for exosomal surface proteins or the Tim-4 receptor. Once internalized, exosomal content is released via the phagolysosome or localized in the late endosome. (c) The exosomal lipid, protein, RNA, miRNA and DNA content can alter a recipient cell′s molecular profile by various mechanisms which alter gene expression. Some possibilities for exosomal cargo mediated recipient cell reprogramming may include altered transcriptional or translational profile, transcript or protein modification, regulation of signaling cascades, protein localization and turnaround, key enzymatic reactions and cellular homeostasis. The source and condition of both the donor and recipient cell may determine which of these molecular mechanisms predominate. Intriguingly, exosomes may also have recipient cell type specific functions for the same molecule. EE, early endosome, MVB, multivesicular body, LE, late endosome, ILV, intraluminal vesicle, RE, recycling endosome, Rab, Ras superfamily GTPases, ESCRT, endosomal sorting complex required for transport.
Rather than being directed into a lysosomal compartment as is the fate for a well characterized branch of the late endosome MVB population, fusion of the multivesicular bodies with the plasma membrane releases exosomes into the extracellular space. This process of directed transport relies on several components of the endocytic machinery such as the Rab GTPases (Ras-related in brain) Rab 11, Rab 27 a and b, cytoskeleton regulatory proteins, molecular motors such as myosins, and SNAREs (SNAP (soluble NSF attachment protein) receptor) for targeted fusion (Fig. 2a).
While cells of the hematopoietic lineage such as B-cells, dendritic cells, mast cells, T-cells and platelets were initially characterized for exosome production, in later years non-hematopoietic cells such as various epithelial cell types, neurons, and a very wide variety of tumor cells have also been shown to produce exosomes [9]. Of particular interest are tumor cells that exhibit enhanced production of exosomes. The mechanism or need for such enhanced production of exosomes is not understood but may indicate a role for exosomes in tumor development [10]. Since exosome production is a continuous cell function, certain cell types like platelets can be treated with agonists to induce degranulation and enhanced release of microvesicles and exosomes [11]. Exosome secretion is also increased on free radical stress or UV irradiation, decreased membrane cholesterol, and increased intracellular calcium. In vivo, exosomes have been characterized in most circulating body fluids such as plasma, urine, saliva, broncheoalveolar lavage (BAL), cerebrospinal fluid (CSF), and others [12], [13].
Exosomes from different sources have a common core set of proteins that help to characterize them as endosome-derived cellular vesicles. The extent of common mRNA and miRNA content is not well established due to the limited number of global studies which have profiled both species. Online resources exist, such as the Exocarta compendium of all exosome based studies, which compile protein, mRNA, and miRNA data from different sources into a searchable format [14]. Additionally, the urinary exosome resource from the Knepper Lab at NIH also provides a detailed dataset of urine exosomal protein content (Urinary Exosome Protein Database). Augmenting this core set of components, the biology of exosomes originates from the cell type and cell condition specific cargo of protein, lipid, RNA, miRNA and DNA (Fig. 1). This dependence of biomolecular content on cell context, and variations between normal and diseased cells, has garnered much interest in exosomes as harbors of disease biomarkers which could be thought of as a “fluidome” or “liquid biopsies” where extraction of exosomes from body fluids can provide a snapshot into the donor cell condition [15]. The results obtained in such biomarker studies to date has prompted some authors to suggest that an exosome is a miniature cell in terms of healthy vs. normal status [16].
2.2. Routing and sorting of cargo into exosomes
Exosomes arise within the cellular endosomal compartments so it is reasonable to presume that cytoplasmic cargo must be represented in exosomes. This is exemplified by the presence of membrane markers on the exosome like Tsg101 and Alix which are involved in MVB biogenesis. The mono ubiquitination of cargo and its sorting for lysosomal degradation in MVBs is a well-studied process. MVB topology serves as a sorting station where ubiquitinated cargo is directed to ILVs fated for degradation while membrane sequestration allows other proteins to be directed to the recycling compartment.
Endosomal cargo can be rerouted from a recycling fate and delivered to exosomes by lateral segregation on the membrane with GPI-anchored or lipid-raft enriched micro-domains, antibody mediated higher order oligomerization, and via aggregation (Fig. 2a). Other proteins undergo a recycling or exosomal containment fate depending on the binding partners. For example, during reticulocyte maturation, decreased clathrin adaptor AP2 levels cause the transferrin receptor to associate with Hsc70 (heat shock cognate protein) and Alix rather than binding to AP2. Highlighting the intricacy of such a molecular balance, the same binding motif is utilized in both interactions.
There remains a significant gap in understanding the mechanisms that allow the rerouting of proteins (or possibly entire ILVs) marked for degradation to exosomes for release. Exosome biology suggests the existence of separate subsets of MVBs, where some rely on ESCRT-dependent (ESCRT, endosomal sorting complex required for transport) sorting of signaling receptors. Others appear formed primarily by ceramide synthesis as a driving factor for exosome formation [7], [17] (Fig. 2a). The molecular mechanisms that direct the two processes are likely to be distinct and require further elucidation.
One of the most intriguing aspects of exosome biology remains the packaging of diverse proteins and RNA into exosomes (Fig. 1, Fig. 2a). Initial reports suggest that it is unlikely that exosomal content simply reflects the non-specific stoichiometric abundance of proteins and RNA throughout the cytoplasm. For example, the study from Skog et al. [3] demonstrated that glioblastoma derived microvesicles were enriched (relative to unfractionated glioblastoma cells) for several proteins, mRNAs and miRNAs that would aid tumor growth through angiogenesis, cell cycle disruption, control of translation, and immune suppression.
Similarly, “stemness-imparting characteristics” are enriched in stem-cell derived exosomes, demonstrating a cell context specific selection of exosomal cargo [1]. It is noteworthy that several molecules and adapters from the clathrin-coated pit are also found in exosomes [14], suggesting that membrane deformation and cargo selection may utilize the same molecular machinery as the plasma membrane.
Recently tetraspanin proteins have been implicated in exosome cargo selection along with providing the mechanistic framework for the formulation of exosomes [18]. It's interesting to note that the ESCRT complex which has been implicated in exosome formation is also involved in selective protein sorting, e.g. binding of bicoid RNA to the ESCRT II complex allows sorting into the endosomal system [19]. Furthermore, a recent report that the MVB system is linked with a maturing RISC complex [20] provides an alternatively novel mechanism for the loading of RNA and miRNA species into exosomes.
Recent studies indicate that a subset of plasma miRs are bound to Ago2 or LDL and this may be yet another mechanism that allows loading into exosome like vesicles exiting the cell [21], [22]. Given the variety of potential mechanisms of exosome formation and exosomal loading, there remains a clear need to establish the adaptors or sorting motifs and the lipid–protein, lipid–RNA/miRNA, RNA–protein, miRNA–protein, protein–mRNA–miRNA interactions that allow specific cargo packaging into exosomes.
3. Functions of exosomes
Originally, exosomes-derived from cancer cell lines were credited with nucleotidase and ATPase activity [5]. The paper also suggested that exosomes were taken up by recipient cells. Exosomes were later described as sinks for disposal of obsolete proteins such as the transferrin receptor, acetylcholinesterase, cytochalasin B binding (glucose transporter) nucleoside transporter and excess plasma membrane as reticulocytes matured into erythrocytes.
In recent times, the dominant mode of exosome uptake into recipient cells appears to be phagocytic processes dependent on dynamin2 and PI3K (phosphatidyl inositol-3-kinase). The uptake process also relies on a variety of specific cell surface molecules on the receptor cell (T-cell membrane protein 4 Tim4, but not Tim1, has been shown to be involved in exosomes uptake) as well as proteins like integrins, annexins, galectin, and ICAM1 (inter-cellular adhesion molecule 1) on the exosomal surface. Uptake of exosomes is possibly a non-random event and recent papers [18] suggest a mechanistic basis to target cell selection by invoking tetraspanins as entry selection markers. This is supported by a defined set of exosome-associated tetraspanins that appear to direct targeting to endothelial cells to promote angiogenesis and vasculogenesis. It has also been shown by Valadi et al. [2]. that DC-derived exosomes are efficiently taken up only by DC lineage cells. In recent papers there is well documented evidence that exosomes can act systemically and behave as endocrine moieties in that respect [23]. Long range targeting, tissue specificity in uptake and stability of exosomes in systemic circulation is yet another interesting and potentially therapeutically exploitable aspect of exosome biology.
Post internalization, exosomes have been found in the phagolysosome along with phagocytosis-inducing biospheres [24], or found co-localized with the late endosomal compartment. This indicates that there may be cell type specific trajectories in the recipient cell post-uptake (Fig. 2b). The mode of release of exosomal content into the cytoplasm is not known although it has been suggested that exosomal surface proteins may perform some signaling functions at the host cell surface as they adhere to the cell for internalization. The release of exosomal cargo of protein, DNA, lipids, mRNA and miRNA can cause alterations in the recipient cell that are similar to a cellular signaling based response system; with exosomes being the global signaling moiety that signals a cascade of responses within the cell (Fig. 2c). The range of molecular effects is quite diverse and is summarized in Fig. 2c to reflect the role that the various cargo identified in exosomes to date can have upon host cell gene expression profiles.
3.1. Role of exosomes in immune function
Interest in exosomes in immune response burgeoned in 1996 with the discovery that antigen presenting cells (APCs) enhance the immune cell response by utilizing exosomes for antigen presentation [25]. Endosomes of APCs are enriched for MHC (major histocompatibility complex) class II molecules, MHC class II-associated chaperone Ii, and proteases that are required for Ii degradation and antigen processing [26]. Exosomes from APCs are similarly enriched in MHC I, MHC II, and immuno-modulatory proteins. A strong immunogenic response by dendritic cell (DC) exosomes (also known as dexosomes), generally relies on the presence of co-stimulatory molecules on the APC. However in some cases dexosomes have been shown to stimulate a sufficient immune response dependent solely on the MHC I concentration and in the absence of APCs and corresponding co-stimulatory molecules [27]. B-cell derived exosomes elicit an immune response by direct antigen presentation to T-cells [28].
However, in the most common mechanism of exosome-stimulated immune response exosomes do not interact directly with T-cells [25]. Immature dendritic cell-derived exosomes (immature Dex or “iDex”) induce an indirect T-cell activation via presentation of MHC/antigen complex to receptor DCs. CD4+ (CD cluster of differentiation) T-cells are activated by MHC-II/antigen complex transfer to recipient DCs. Similarly, MHC-I/antigen complex transfer to DCs aids polarization of Tc1 CD8+ T-cells [29], [30]. The iDex response in vivo relies on CD8α+ DCs in a process dependent on LFA-1 (lymphocyte function-associated antigen 1) for exosome capture, and continues without exosome internalization and processing [31].
Tc1 response requires the administration of DC maturing agents like Toll like receptors TLR-3 and TLR-9 indicating the requirement for APCs in inducing the Dex response. Cytotoxic T-lymphocyte target cell recognition induces rapid polarization of both Rab-11+ recycling endosomes as well as Rab-27+ exosomes in a process mediated by hm-munc13. This vesicle mobilization is necessary for the release of cytotoxic granules [32]. Additionally, immature DC derived exosomes may transfer processed antigen-MHC complexes to other DCs and thus increase the effective number of APCs.
While DC exosome to DC MHC-antigen presentation is common, it has also been shown that mast cell derived exosome peptide cargo may be presented on the surface of recipient B-cells and DCs [33]. In terms of mechanisms for inducing an immune response, recent reports suggest that exosomes carry bioactive cytokines exemplified by the presence of IL-1β and other inflammasome components [34]. The concentration of tumor necrosis factor TNF-α in dexosomes indicates that in addition to a role in adaptive immunity, exosomes could also trigger an innate immune response and in fact mediate the amplification of such a response via the cargo of protein, RNA and miRNA that it can transfer to responsive tract epithelium [35], [36].
There has continued to be an intensive amount of research derived on such findings and many studies have established that exosome content and functions depend on the precise maturation stage, cell lineage, and stimulation state of the parent immune cell. For example, immature DCs produce a larger number of MVBs and exosomes then do immature DCs; however, at the level of inducing an antigen specific T-cell response, mature DC derived exosomes show twice the potency [36]. Immune functions of DC-derived exosomes can be tailored to be immunogenic or tolerizing, depending on the presence of specific molecular cargo. This has increasingly found uses in creating highly specialized therapeutics termed ‘designer dexosomes’ that are utilized in cancer therapy, vaccine development, and transplant tolerance induction as recently reviewed in Viaud et al. [36].
3.2. Role of exosomes in the virus life cycle
Exosomes and viruses seem to have developed in close association prompting some virologists to suggest that they co-opted molecular machineries as they evolved. Immature DCs are tasked with delivering viral antigens from the periphery to lymphoid organs and an additional mechanism significantly increases the number of virus challenged DCs. This amplification involves mature DCs phagocytosing antigen-bearing DC's, or antigen-loaded exosomes produced by infected DCs. The exosome antigen dissemination pathway intrinsic to DCs involves an enrichment of functional epitope–MHC complexes on the exosome, and their release from the cell followed by uptake by other immune effector cells.
Such an exosomal packaging system allows the virus to survive within the endocytic system without lysosomal degradation, and allows subsequent trans-infection of CD4+ T-cells (Fig. 3b). HIV (human immunodeficiency virus) and other retroviruses utilize a dexosome based and predominantly MHC-II antigen complex presentation schema to trans-infect CD4+ T-cells [37]. Immature DCs exhibit a budding of HIV into a structure that becomes an exosome at the stage of reverse budding in the late endosome.
Fig. 3.
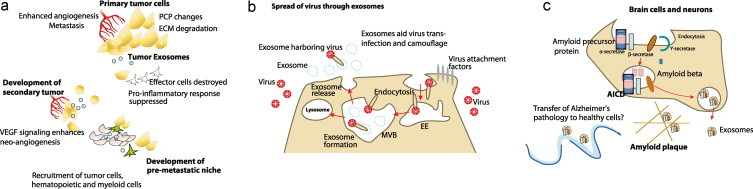
Exosomes can alter the molecular profile of their microenvironment and recipient cells and can affect tissue health. (a) Exosomes help to establish a metastatic niche to aid tumor growth and tumor metastasis. Utilizing VEGF and cytokine cargo, exosomes enhance recruitment of hematopoietic and endothelial precursor cells to enhance neo-angiogenesis in the tumor. Exosomes have also been suggested to influence planar cell polarity and the extracellular matrix to allow tumor cell mobilization. To aid tumor growth, exosomes are additionally known to trigger apoptotic reduction in anti-tumor immune cells through FasL and TNF-pathways. The growth factor and cytokine cargo helps exosomes to attract disseminated tumor cells from the primary tumor site as well as cells of the myeloid and erythropoietic lineages to the pre-metastatic niche to aid tumor spread. (b) Exosomes share many characteristics with viruses in the molecular machinery of budding from the plasma membrane. Retroviruses like HIV utilize the cell-encoded intercellular vesicle traffic as a means to protect themselves from the host cell's proteolytic system. Segregating into the exosomes during the exosome formation event, the virus is camouflaged and avoids the host surveillance system and proteolysis. The exosome encapsulated virus is furthermore able to disseminate and move between cells utilizing the exosome pathway for entering a host cell without utilizing its own fusion machinery as it searches for optimal target cells. Exosomes thus aid the trans-infection of viruses. (c) Amyloid plaques of the Alzheimer's disease afflicted brain have aggregates of amyloid beta peptide processed from amyloid precursor protein (APP). APP is processed by at least three proteases termed α-, β-, and γ-secretases, which lead to the generation of a number of proteolytic fragments, including Aβ when APP is cleaved by β- and γ-secretases. Exosomes have been found to contain APP, Aβ and other proteolytic fragments of APP, α-, β-, and some components of γ-secretase. It has also been suggested that exosomes from an unhealthy brain cell may cause spread of cellular insults in the form of RNA and protein to surrounding healthy cells and thus aid the spread of neurological disorders. EE, early endosome, AICD, amyloid precursor protein intracellular cytoplasmic/C-terminal domain.
On a per particle basis, exosome associated particles are 10-fold more infectious than cell-free virus particles [37]. Mature DC-internalized viral particles need to compete with virion-like particles (VLP) and exosomes at the vesicle internalization stage for segregation into lipid-raft like, cholesterol enriched micro domains on the DC. Glycosphingolipids from the exosome-producing cells are known to be critical for the process of VLP and exosome internalization at the mature host DC plasma membrane. This probably occurs through facilitating interaction of the exosome or viral membrane with the host [37], [38].
Conversely, as an indicator of how viruses hijack the host system to survive and propagate, the “Trojan exosome hypothesis” proposes that enveloped retroviruses like HIV utilize the host cell's endosomal system, including the exosome pathway, to move between APCs while searching for the best target cells (Fig. 3b) [39]. It is well known that HIV viral budding occurs in an MHC II-enriched compartment similar to the MVB.
The generation of ILVs/exosomes result from the binding of the hepatocyte growth factor tyrosine kinase substrate (Hrs) to ubiquitinated cargo, followed by recruitment of Tsg101. HIV-1 Gag mimics Hrs and thereby usurps Tsg101 and other components of the MVB machinery in order to facilitate viral budding. The similarity of the exosome and viral budding pathways prompted comparative studies which indicated that viruses disguised as ILVs may be co-opted in the exosome pathway and released with the typical exosomal membrane and lipid composition. This allows propagation without engaging the host immune response. In this mechanism of disguised propagation the virus is also able to conserve resources on fusion with the cell and utilizes the exosome internalization machinery to enter the host cell [40].
3.3. Role in tumorigenesis via altering microenvironment (tumor growth, angiogenesis)
It has been shown that part of the physiological role of exosomes is their ability to alter their micro-environment through their protein and RNA based cargo. For instance, studies on tumor derived exosomes reveal that they may perform several functions aiding tumor survival and metastasis (Fig. 3a). The presence of HLA-G in exosomes secreted by melanoma cells might indicate a role in creating a tumor tolerance associated response [41]. Tolerogenic activity of exosomes has been associated with Fas- and TRAIL-expression in exosome membranes inducing FasL- and TRAIL-mediated apoptosis of anti-tumor T-cells [42]. Incubation with tumor derived exosomes can disrupt the maturation of differentiating CD14+ and HLA-DRlow/neg monocyte precursors into DCs [43]. The resulting lack of T-cell stimulatory effects on exosome co-culture is primarily due to the altered monocyte cytokine profile exemplified by inflammatory cytokines such as TNF-α, interleukin IL-6, and pro-angiogenic and immuno-suppressive factors, e.g. TGF-β, IL-8 and IL-10.
In support of this result, peripheral blood of melanoma patients revealed the predicted CD14+ and HLA-DRlow/neg (MHC class II cell surface receptor) tumor growth factor TGF-β secreting myeloid cell population which was absent in healthy individuals [44]. Tumor derived exosomes contain several cytokines and growth factors such as tumor necrosis factor TNF-α, IL-1β, and TGF-β as well as functional receptors like TNFR1 and TfR2. Consequently, tumor derived exosomes by their very nature support neo-angiogenesis since they are enriched in tetraspanins which enhance the secretion of VEGF (vascular endothelial growth factor) and metalloproteinases from target cells [45]. A striking result from Skog's study on glioblastoma derived microvesicles revealed that tumor derived exosomes could support cell culture based angiogenesis of HUVEC (human umbilical vein endothelial cells) to almost the same extent as extraneously added growth factors [3].
Melanoma exosomes were shown to induce endothelial cell spheroid formation and moved between endothelial tubule cells using tunneling nanotube structures and acting in a primarily paracrine manner [46]. In an adenocarcinoma model, exosomes induced several angiogenesis related genes including von Willebrand factor, Tspan8, chemokines CXCL5 (C-X-C motif chemokine 5), MIF (macrophage migration inhibitory factor), and chemokine receptor CCR1. This indicated involvement of VEGF independent processes in exosome-mediated angiogenesis [47], [48].
A recent study showed that exosomes from a mutant KRAS allele bearing cell line have greater invasive potential than exosomes from an autologous cell line with the mutant KRAS removed by homologous recombination [10]. Such properties could clearly aid the formation of pre-metastatic niches. Exosomes also aide the spread of the human tumor Epstein Barr Virus, EBV oncogene protein called “latent membrane protein 1” to uninfected cells. EBV can enrich exosomes with proteins that are cancer associated and thus induce the enhanced cell growth, angiogenesis, and transformation of other cells. They do this while evading the immune response by being encapsulated in the circulating exosomal delivery system [49].
3.4. Potential role in propagating neurodegenerative diseases
Exosomes have additional functions where they can mediate the spread of cell damage or alter the microenvironment in various metabolic and nervous system disorders [4]. As an example, the microenvironment altering properties of exosomes have been implicated in Alzheimer's disease (AD) (Fig. 3c) and have been associated with processed Alzheimer's proteins and enzymes [50], [51], [52]. The “Trojan Exosome” model of Alzheimer's (similar in concept to the Trojan Exosome model in viral infection), proposes that the initial insult of Alzheimer's may spread to surrounding cells via exosomes laden with amyloid beta (Aβ) peptide and other proteins and RNA associated with precipitating Alzheimer's pathogenesis. This is supported by the finding of depositions of exosome proteins in amyloid plaques in AD brains [53]. Finally, exosomes have also been associated with the spread of prion proteins indicating multiple roles for exosomes in neurodegenerative diseases.
4. Applications of exosomes
Having discussed the various physiological and pathophysiological roles exosomes may play in different cell and tissue types, it is clear that many opportunities exist to harness exosome biology as either a therapeutic or treatment strategy. As an example, the capacity to differentially challenge DCs to produce dexosomes with either pro- or anti-inflammatory properties is finding exciting uses in the field of cancer vaccines, humoral immunity, SARS (severe acute respiratory syndrome) vaccine and transplant tolerance induction, delayed type hypersensitive response, rheumatoid arthritis treatment, etc. [36]. Additional studies are ongoing to establish exosomes as nanocarriers for delivery of a variety of therapeutic modalities [23].
It is formally possible, although currently unlikely, that targeting exosome release could represent a viable therapeutic strategy. However, should future research demonstrate that exosomes are critical for certain pathological disease processes, a strategy of targeting the formation and/or release of exosomes in specific cell types (e.g. primary tumor cells, etc.) would represent a novel route of therapeutic intervention. Designing a strategy that is specific to the formation of exosomes in a single cell type rather than creating a general “exosomal poison” is likely to be both tantamount, and a tremendous challenge, to the development of such a drug. The specificity of exosomal disruption would be critical since it is unlikely that a general shutdown of exosome formation would be tolerated without adverse consequences. Achieving such specificity of destruction or modulation might be a challenge due to the non-redundant nature of most exosome formation regulators and the difficulty of identifying a molecule with the specificity required to target a cell-specific exosomal formation process in the body. However, we believe this approach can’t be ruled out as a potential therapeutic strategy in the years to come. With innovative therapeutic modalities emerging today (bi-, tri-specific antibodies, etc.) and others on the horizon, such a pharmacological approach may become feasible should exosomes be conclusively implicated in disease in the future.
In this review, we will discuss in detail strides that have been made to align exosome biology with biomarker development to aid better health care standards.
4.1. Biomarkers
In terms of exosome-based applications the major facet of exosome biology that is finding utilization is the usage of cell- and condition-specific cargo as biomarkers. Given that an exosomal profile reflects the cell's content, exosomes provide a means for a “liquid biopsy” which is minimally invasive and highly enlightening. Table 1 summarizes the main exosome derived biomarkers associated with different disease conditions and from cell culture based studies. This emerging field is still relatively young considering the vast wealth of information that emerges regularly about new exosomal content. For example, it has been discovered fairly recently that exosomes contain DNA. This is represented by mitochondrial DNA as well as retro-transposon and amplified DNA of oncogenes in tumor cell derived exosomes [10].
Table 1.
Exosome-associated biomarkers.
| Disease | Biosample/methodology | Exosome/microvesicle BM | Reference |
|---|---|---|---|
| Bladder cancer | Urine/LC–MS/MS | Up regulated proteins epidermal growth factor receptor pathway, the alpha subunit of GsGTP binding protein, resistin, and retinoic acid-induced protein 3 | [62] |
| Urine and bladder cancer cell lines/LC–MS/MS | Basigin, galectin-3, trophoblast glycoprotein (5T4) | [61] | |
| Human colon cancer cell line | Cell media/A33 affinity capture + MS | Cadherin-17, carcinoembryonic antigen, epithelial cell surface antigen (EpCAM), proliferating cell nuclear antigen, epidermal growth factor receptor, mucin 13 | [55] |
| Human ovarian cancer | Plasma/RT-PCR | miR-21, -141, -200a, -200b, -200c, -203, -205, -214 | [72] |
| Malignant ascites fluid, ovarian cancer plasma/MS | Tumor progression-related proteins (L1CAM, CD24, ADAM10 and EMMPRIN) | [57] | |
| Plasma/MS | Claudin | [74] | |
| Breast cancer | Plasma/RT-PCR | miR-21 | [75] |
| Prostate cancer | Urine/Western blot | PSA, PCA3 | [63], [64] |
| Nasopharyngeal carcinoma (NPC) (EBV infected) | Cell media and plasma/RT-PCR | BART viral miRNA | [76] |
| NSCLC diagnosis and prognosis | Plasma/RT-PCR | let-7f, miR-20b, miR-30e-3p, miR-223 and miR-301 | [77] |
| Apoptotic Jurkat cells | Cell media/nucleic acid visualization, RT-PCR | Laddered DNA | [78] |
| Acute kidney injury | Urine/MS | Fetuin-A | [67] |
| Renal I/R injury | Urine/Western blot | Reduced aquaporin-1 (AQP1) | [79] |
| Nephrotic-range proteinuria | Urine/MS | Neprilysin, aquaporin-2, and podocalyxin | [80] |
| Bartter syndrome type I patients | Urine/Western blot | Absence of the NKCC2 sodium–potassium–chloride cotransporter | [60] |
| Sjogren's syndrome | Saliva/RT-PCR | miRs | [70] |
|
List of biofluid based nucleic acid based biomarkers, probably contain exosomal origin nucleic acids | ||
|---|---|---|
| Disease/pathology | Differentially expressed or modified circulating nucleic acids | Reference |
| Diffuse large B-cell lymphoma (DLBCL) | miR-21, miR-155, miR-210 | [81] |
| Diffuse large B-cell lymphoma (DLBCL) relapse free survival | miR-21 | [81] |
| Acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL) | miR-92a | [82] |
| Gastric cancer (GC) | miR-17-5p, miR-21, miR-106a, miR-106b, let-7a | [82] |
| Pancreatic ductal adenocarcinoma | miR-21, miR-155, miR-196a, miR-210 | [82] |
| Pancreatic cancer | miR-210 | [83] |
| Hepatocellular carcinoma | miR-500 | [82] |
| Colorectal cancer | miR-17-3p, miR-92, miR-29a, miR-92a | [82] |
| Melanoma | Tyrosinase mRNA | [84] |
| Lung cancer | miR-25, miR-223, miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212, miR-214, miR-1, miR-30d, miR-486, miR-499 | [82] |
| Lung cancer survival | miR-7i, miR-146b, miR-206, miR-21 | [85] |
| NSCLC survival prediction | miR-486, miR-30d, miR-1 and miR-499 | [85] |
| NSCLC | Hypermethylation pattern of DAP kinase in serum and plasma DNA | [84] |
| Squamous vs. non squamous NSCLC | miR-205 | [86] |
| Oral and squamous cell cancer | miR-184, miR-31, miR-24 | [82] |
| Squamous head and neck cancer | Human papilloma virus (HPV) DNA | [84] |
| Medulloblastoma | miR-124, miR-125a, miR-9 | [87] |
| Medulloblastoma stratification | miR-21, mir-17/92 cluster | [87] |
| Glioma | miR-21, miR-124, miR-128 | [3], [87] |
| Breast cancer | miR-155, miR-195, miR-10b, miR-34a | [82] |
| Ovarian cancer | miR-21, miR-29a, miR-92, miR-93, miR-99b, miR-126, miR-127 and miR-155 | [88] |
| Cervical cancer | Human papilloma virus (HPV) DNA | [84] |
| Prostate cancer | miR-16, miR-34b, miR-92a, miR-92b, miR-103, miR-107, miR-197, miR-328, miR-485-3p, miR-486-5p, miR-574-3p, miR-636, miR-640, miR-766, miR-885-5p | [82] |
| Acute liver injury | mir-122 | [89] |
| Alzheimer's disease CSF | miR-15b, miR-142-5p, miR-146, mir-181a, miR-191, miR-204, miR-221 | [90] |
| Myocardial infarction | miR-499, miR-208b | [91] |
| Heart failure | miR-423-5p | [92] |
| Myocardial infarction | Elevated plasma DNA | [84] |
| Stroke | Elevated plasma DNA | [84] |
| Acute tissue injury | Elevated plasma DNA | [84] |
| Systemic lupus erythematosus | Elevated extrachromosomal plasma DNA | [84] |
| Systemic sclerosis | Elevated extrachromosomal plasma DNA | [84] |
| Sex-linked diseases | Maternal circulation plasma DNA | [84] |
| Paternally inherited diseases | Y-chromosome DNA | [84] |
The power of exosomes as biomarkers relies on the enrichment of markers within the exosome which otherwise constitute only a very small proportion of the secretome, e.g. exosomal proteins represent less than 0.01% of the total plasma proteome. Healthy human circulating exosomes contain about 66 proteins [54] with an enrichment of cell trafficking and protein sorting related proteins reflecting the endosomal origin of exosomes (Fig. 1). The very highly enriched protein entities in plasma and urine will normally dampen out the minor presence of relevant biomarkers. The exosome biomarker strategy therefore truly exemplifies “less being more” since the small number of proteins and RNA nevertheless give very insightful information into a disease or pathology.
An enrichment at source, exemplified by cargo sorting into the exosome, helps the discovery of relatively lowly expressed biomarkers that normally would go undetected. Several studies have shown that purification of fluid level protein and RNA can also provide biomarkers (Table 1). The authors suggest a practical and case-by-case based scenario where the sensitivity provided by exosome isolation may be crucial for certain diagnostics; while the biomarker differential may be high enough in other disease conditions that the isolation of exosomes can be bypassed and fluid based biomarker analysis may be sufficient.
The multitude of secretomes converging in body fluids, and the variety of cell types contributing to the secretome can confound a fluid-based analysis system. For example, plasma exosomes in normal individuals originate from a variety of cell sources including red blood cells, leukocyte lineages and platelets. Tissues that directly or indirectly contact the blood stream such as endothelial cells and large fenestrated organs like the placenta and liver also allow passage of exosome vesicles into the body fluids. In cases where the biomarker change can be indicative of more than one condition, exosomes would be the method of choice to trace the cell lineage producing the change.
Another advantage of using exosomes as biomarkers is the ability to immuno-isolate exosomes from particular tissue types. The exosomal membrane reflects a cell's plasma membrane and can therefore be immuno-isolated utilizing a significantly enriched protein on the membrane surface. For example, plasma exosomes of hepatic origin can be enriched using an antibody specific to the relatively liver-specific enzyme carbamoyl phosphate synthetase 1. In cell culture, A33 affinity-isolation of colorectal cancer cell line exosomes [55], and CD45 isolation of Jurkat T-cell line exosomes [56] has yielded very specific cell based signatures and holds the promise of being applied similarly to plasma exosome isolation. In a study that compared ovarian carcinoma cell line derived exosomes vs. exosomes from malignant ascites and sera from ovarian carcinoma patients, tumor progression-related proteins like L1CAM (L1 cell adhesion molecule), CD24, and EMMPRIN (extracellular matrix metalloproteinase inducer) were found enriched on exosomes. This study also indicated that the exosomes become systemic via the blood stream [57].
Urinary exosome profiling studies have probably been established in greatest detail owing to easy accessibility of urine samples. Seminal work by the Knepper and Star Labs established conditions for collecting and storing samples, and optimization of design for mass spectrometry or gel chromatography based techniques for urinary exosome analysis [58], [59]. It has been established that urinary exosomes contain the majority of renal proteins that indicate renal cell injury including, but not limited to, aquaporins, polycystins and podocyn [60]. Successful studies have been performed for various disease conditions where a cellular contact with urine or glomerular filtrate would be possible, e.g. acute kidney injury (AKI) models (cisplatin or ischemia-reperfusion), prostate cancer, bladder cancer, etc.
Bladder cancer cell line derived exosomes contain about 350 proteins and 18 of these were also found in the urine derived exosomes from a small number of bladder cancer patients [61]. Urine micro particles from bladder cancer patients revealed that eight proteins were elevated relative to controls, and many of these proteins were linked to the epidermal growth factor receptor (EGFR) signaling pathway [62]. Such examples suggest that the exosomal biomarker discovery approach can be ‘topside up’ or ‘topside down’. An initial screen in conditioned media from cultured cell lines can be used to build a predictive model for pathways over or under represented in exosomes. These models can then be translated to in vivo models and biological fluid sampling from a large patient cohort.
Prostasomes are exosomes released from the prostate epithelium into semen or urine. Analysis of urine exosome content revealed that prostate cancer patients have tumor signatures, e.g. PSA (prostate specific antigen) and PCA3 (Prostate CAncer gene 3) [63], [64]. PC-3 cell culture media derived exosomes and urine prostasomes both exhibit enhanced δ-catenin [65]. Rotavirus related gastroentitis patients display exosome associated RV antigen in stool samples [66].
In animal models of kidney injury, exosomal Fetuin-A was identified as a marker reflecting nephrotoxic damage with a 50-fold increase over healthy animals within hours of injury and prior to the detection of the standard biomarker urine creatinine. In renal ischemia-reperfusion (I/R), decreased exosomal aquaporin-1 (AQP1) in rats correlated with decreased AQP1 levels in a person undergoing renal transplant [67].
In a pilot study on Bartter syndrome type I patients, the sodium–potassium–chloride co-transporter, NKCC2 was absent in urinary exosomes. Additionally, it has been shown by Zhou et al. that transcription factors that are undetectable in whole urine could be detected in urine derived exosomes, e.g. urinary ATF3 (cyclic AMP-dependent transcription factor) might be a novel AKI renal tubular cell injury biomarker; while WT-1 (Wilms tumor protein) might be a critical biomarker for chronic kidney disease (CKD) associated with podocyte injury [68]. It is noteworthy that currently the biomarker perspective for CKD is a clearly unmet need with no consensus on predictive or prognostic biomarkers, particularly in the context of early detection and sensitivity.
This discussion reflects the strength of exosomes as fluid-based, minimally invasive biomarkers but also identifies the need for larger cohort studies with well established protocols for a clinical study. Such studies must have built in positive and negative controls to validate the study design at various steps of sample collection, timing, storage and processing as well as the protocols for protein biomarker identification based on Western blot analysis, ELISA (enzyme linked immunosorbent assay) or FACS (fluorescence activated cell sorting) analysis.
In the context of RNA-based biomarkers, there have been only a few studies to date. RNA composition of the human urinary exosomes represents all regions of the nephron and the collecting duct as determined by RT-PCR [69]. In a significant study on the specificity of exosomal RNA as a biomarker, urinary exosomes of the V-ATPase B1 subunit, knockout mice were found to faithfully reflect the loss of this specific subunit mRNA while normal amounts of the other V-ATPase B2 subunit and aquaporin 2 were detected [69]. Preliminary studies have shown that salivary exosome protein and miRNA content can be potentially useful to detect disease conditions such as Sjorgen's syndrome [70].
Increased breast tumor miR-195 levels were reflected in systemic circulation exosomes [71]. Glioblastoma derived exosomes contain a diagnostic set of miRs in addition to mutant EGFR vIII [3]. Prostate cancer plasma exosomes revealed up-regulated miR-145 [64]. Circulating levels of miR-195 and let-7a decreased in cancer patients post-operatively suggesting feasibility as a prognosis biomarker as well. In this study, the level of certain miRs also correlated well with clinico-pathological variables like nodal status and estrogen receptor status. Significant changes to both proteins and miRNAs was also reported for circulating exosomes from patients suffering from ovarian cancers [72]. The study found that a set of 8 miRs could distinguish benign vs. malignant ovarian tumors. Peripheral blood exosomal miRs showed the same profile as the lung adenocarcinoma in stages I–IV patients [73]. The predictive capacity of exosomal miRNA was quite outstanding as exemplified by two exosomal miRNAs accurately discriminating acute lymphoid from acute myeloid leukemia.
5. Future perspectives
The authors propose the term “fluidome” to capture the complexity of secreted proteins, lipids, RNA, and DNA components that allow insight into cellular processes at a biofluid or cell culture medium level. In terms of RNA content, the earliest reports of circulating RNA hypothesized that body fluid derived RNA must somehow be protected from the degradative RNAse-rich content of the body fluid. Many interesting theories have arisen over time to answer this conundrum such as the presence of a novel conformation that imparts resistance to nucleases, confinement in exosome-like vesicles and, recently, the presence of Argonaute bound and LDL bound RNA species in plasma. Other fascinating questions in the field of exosome biology include cargo sorting mechanisms, exosome uptake specificity, and the correlation between the mRNA, miRNA and protein content of exosomes in terms of selection for expulsion as well as in the landscape of effects on the recipient cell.
So far, studies have focused on these individual biologics in the exosome and there has been no dedicated effort to correlate the effect of the molecules as integrated results at the cell function level. Thus, it is obvious that basic research is needed to answer some of these outstanding questions on exosome biology and further strengthen the knowledge base in the field.
In conclusion, with continued advancements in technology for profiling of exosomal content in a high throughput manner, combined with the elucidation of more of the basic biology of the system, exosomes provide a novel biomarker discovery and therapeutic platform with great promise. Continued basic research will set the foundation for transition of exosome-based diagnostic and prognostic biomarkers to the clinic and a better understanding of the molecular mechanisms of disease pathogenesis.
Acknowledgements
We apologize to colleagues whose work we could not cite owing to length restrictions. We thank Jacky Vonderscher and Karin Jorga for review of the manuscript and Mark Misenheimer for help with the graphic design.
References
- 1.Ratajczak J., Miekus K., Kucia M., Zhang J., Reca R., Dvorak P. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 2.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 3.Skog J., Wurdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathivanan S., Ji H., Simpson R.J. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Trams E.G., Lauter C.J., Salem N., Heine U., Jr. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta. 1981;645:63–70. doi: 10.1016/0005-2736(81)90512-5. [DOI] [PubMed] [Google Scholar]
- 6.Johnstone R.M., Bianchini A., Teng K. Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions. Blood. 1989;74:1844–1851. [PubMed] [Google Scholar]
- 7.Fevrier B., Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Lakkaraju A., Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008;18:199–209. doi: 10.1016/j.tcb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocucci E., Racchetti G., Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Balaj L., Lessard R., Dai L., Cho Y.J., Pomeroy S.L., Breakefield X.O. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heijnen H.F., Schiel A.E., Fijnheer R., Geuze H.J., Sixma J.J. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–3799. [PubMed] [Google Scholar]
- 12.Simpson R.J., Lim J.W., Moritz R.L., Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 13.Caby M.P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 14.Mathivanan S., Simpson R.J. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 15.Iguchi H., Kosaka N., Ochiya T. Secretory microRNAs as a versatile communication tool. Commun Integr Biol. 2010;3:478–481. doi: 10.4161/cib.3.5.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raimondo F., Morosi L., Chinello C., Magni F., Pitto M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. 2011;11:709–720. doi: 10.1002/pmic.201000422. [DOI] [PubMed] [Google Scholar]
- 17.Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 18.Rana S., Zoller M. Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochem Soc Trans. 2011;39:559–562. doi: 10.1042/BST0390559. [DOI] [PubMed] [Google Scholar]
- 19.Irion U., St Johnston D. bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature. 2007;445:554–558. doi: 10.1038/nature05503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siomi H., Siomi M.C. RISC hitches onto endosome trafficking. Nat Cell Biol. 2009;11:1049–1051. doi: 10.1038/ncb0909-1049. [DOI] [PubMed] [Google Scholar]
- 21.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011; 13:423–433. [DOI] [PMC free article] [PubMed]
- 23.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 2011; 29: 341-345. [DOI] [PubMed]
- 24.Feng D., Zhao W.L., Ye Y.Y., Bai X.C., Liu R.Q., Chang L.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 25.Raposo G., Nijman H.W., Stoorvogel W., Liejendekker R., Harding C.V., Melief C.J. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tulp A., Verwoerd D., Dobberstein B., Ploegh H.L., Pieters J. Isolation and characterization of the intracellular MHC class II compartment. Nature. 1994;369:120–126. doi: 10.1038/369120a0. [DOI] [PubMed] [Google Scholar]
- 27.Sprent J. Direct stimulation of naive T cells by antigen-presenting cell vesicles. Blood Cells Mol Dis. 2005;35:17–20. doi: 10.1016/j.bcmd.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Van Niel G., Mallegol J., Bevilacqua C., Candalh C., Brugiere S., Tomaskovic-Crook E. Intestinal epithelial exosomes carry MHC class II/peptides able to inform the immune system in mice. Gut. 2003;52:1690–1697. doi: 10.1136/gut.52.12.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thery C., Duban L., Segura E., Veron P., Lantz O., Amigorena S. Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes. Nat Immunol. 2002;3:1156–1162. doi: 10.1038/ni854. [DOI] [PubMed] [Google Scholar]
- 30.Andre F., Chaput N., Schartz N.E., Flament C., Aubert N., Bernard J. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J Immunol. 2004;172:2126–2136. doi: 10.4049/jimmunol.172.4.2126. [DOI] [PubMed] [Google Scholar]
- 31.Segura E., Guerin C., Hogg N., Amigorena S., Thery C. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J Immunol. 2007;179:1489–1496. doi: 10.4049/jimmunol.179.3.1489. [DOI] [PubMed] [Google Scholar]
- 32.Menager M.M., Menasche G., Romao M., Knapnougel P., Ho C.H., Garfa M. Secretory cytotoxic granule maturation and exocytosis require the effector protein hMunc13-4. Nat Immunol. 2007;8:257–267. doi: 10.1038/ni1431. [DOI] [PubMed] [Google Scholar]
- 33.Skokos D., Botros H.G., Demeure C., Morin J., Peronet R., Birkenmeier G. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J Immunol. 2003;170:3037–3045. doi: 10.4049/jimmunol.170.6.3037. [DOI] [PubMed] [Google Scholar]
- 34.Qu Y., Franchi L., Nunez G., Dubyak G.R. Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J Immunol. 2007;179:1913–1925. doi: 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]
- 35.Obregon C., Rothen-Rutishauser B., Gerber P., Gehr P., Nicod L.P. Active uptake of dendritic cell-derived exovesicles by epithelial cells induces the release of inflammatory mediators through a TNF-alpha-mediated pathway. Am J Pathol. 2009;175:696–705. doi: 10.2353/ajpath.2009.080716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viaud S., Thery C., Ploix S., Tursz T., Lapierre V., Lantz O. Dendritic cell-derived exosomes for cancer immunotherapy: what's next? Cancer Res. 2010;70:1281–1285. doi: 10.1158/0008-5472.CAN-09-3276. [DOI] [PubMed] [Google Scholar]
- 37.Wiley R.D., Gummuluru S. Immature dendritic cell-derived exosomes can mediate HIV-1 trans infection. Proc Natl Acad Sci U S A. 2006;103:738–743. doi: 10.1073/pnas.0507995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izquierdo-Useros N., Naranjo-Gomez M., Archer J., Hatch S.C., Erkizia I., Blanco J. Capture and transfer of HIV-1 particles by mature dendritic cells converges with the exosome-dissemination pathway. Blood. 2009;113:2732–2741. doi: 10.1182/blood-2008-05-158642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gould S.J., Booth A.M., Hildreth J.E. The Trojan exosome hypothesis. Proc Natl Acad Sci U S A. 2003;100:10592–10597. doi: 10.1073/pnas.1831413100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnstone R.M. Exosomes biological significance: a concise review. Blood Cells Mol Dis. 2006;36:315–321. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Riteau B., Faure F., Menier C., Viel S., Carosella E.D., Amigorena S. Exosomes bearing HLA-G are released by melanoma cells. Hum Immunol. 2003;64:1064–1072. doi: 10.1016/j.humimm.2003.08.344. [DOI] [PubMed] [Google Scholar]
- 42.Kim J.W., Wieckowski E., Taylor D.D., Reichert T.E., Watkins S., Whiteside T.L. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11:1010–1020. [PubMed] [Google Scholar]
- 43.Valenti R., Huber V., Iero M., Filipazzi P., Parmiani G., Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67:2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- 44.Filipazzi P., Valenti R., Huber V., Pilla L., Canese P., Iero M. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol. 2007;25:2546–2553. doi: 10.1200/JCO.2006.08.5829. [DOI] [PubMed] [Google Scholar]
- 45.Gesierich S., Berezovskiy I., Ryschich E., Zoller M. Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res. 2006;66:7083–7094. doi: 10.1158/0008-5472.CAN-06-0391. [DOI] [PubMed] [Google Scholar]
- 46.Hood J.L., Pan H., Lanza G.M., Wickline S.A. Paracrine induction of endothelium by tumor exosomes. Lab Invest. 2009;89:1317–1328. doi: 10.1038/labinvest.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nazarenko I., Rana S., Baumann A., McAlear J., Hellwig A., Trendelenburg M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010;70:1668–1678. doi: 10.1158/0008-5472.CAN-09-2470. [DOI] [PubMed] [Google Scholar]
- 48.Higginbotham J.N., Demory Beckler M., Gephart J.D., Franklin J.L., Bogatcheva G., Kremers G.J. Amphiregulin exosomes increase cancer cell invasion. Curr Biol. 2011;21:779–786. doi: 10.1016/j.cub.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meckes D.G., Jr., Shair K.H., Marquitz A.R., Kung C.P., Edwards R.H., Raab-Traub N. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A. 2010;107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bulloj A., Leal M.C., Xu H., Castano E.M., Morelli L. Insulin-degrading enzyme sorting in exosomes: a secretory pathway for a key brain amyloid-beta degrading protease. J Alzheimers Dis. 2010;19:79–95. doi: 10.3233/JAD-2010-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghidoni R., Benussi L., Binetti G. Exosomes: the Trojan horses of neurodegeneration. Med Hypotheses. 2008;70:1226–1227. doi: 10.1016/j.mehy.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Vella L.J., Sharples R.A., Nisbet R.M., Cappai R., Hill A.F. The role of exosomes in the processing of proteins associated with neurodegenerative diseases. Eur Biophys J. 2008;37:323–332. doi: 10.1007/s00249-007-0246-z. [DOI] [PubMed] [Google Scholar]
- 53.Rajendran L., Honsho M., Zahn T.R., Keller P., Geiger K.D., Verkade P. Alzheimer's disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci U S A. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Looze C., Yui D., Leung L., Ingham M., Kaler M., Yao X. Proteomic profiling of human plasma exosomes identifies PPARgamma as an exosome-associated protein. Biochem Biophys Res Commun. 2009;378:433–438. doi: 10.1016/j.bbrc.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mathivanan S., Lim J.W., Tauro B.J., Ji H., Moritz R.L., Simpson R.J. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coren L.V., Shatzer T., Ott D.E. CD45 immunoaffinity depletion of vesicles from Jurkat T cells demonstrates that exosomes contain CD45: no evidence for a distinct exosome/HIV-1 budding pathway. Retrovirology. 2008;5:64. doi: 10.1186/1742-4690-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keller S., Konig A.K., Marme F., Runz S., Wolterink S., Koensgen D. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 2009;278:73–81. doi: 10.1016/j.canlet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 58.Pisitkun T., Johnstone R., Knepper M.A. Discovery of urinary biomarkers. Mol Cell Proteomics. 2006;5:1760–1771. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Cheruvanky A., Zhou H., Pisitkun T., Kopp J.B., Knepper M.A., Yuen P.S. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol. 2007;292:F1657–F1661. doi: 10.1152/ajprenal.00434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzales P.A., Pisitkun T., Hoffert J.D., Tchapyjnikov D., Star R.A., Kleta R. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welton J.L., Khanna S., Giles P.J., Brennan P., Brewis I.A., Staffurth J. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smalley D.M., Sheman N.E., Nelson K., Theodorescu D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J Proteome Res. 2008;7:2088–2096. doi: 10.1021/pr700775x. [DOI] [PubMed] [Google Scholar]
- 63.Nilsson J., Skog J., Nordstrand A., Baranov V., Mincheva-Nilsson L., Breakefield X.O. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitchell P.J., Welton J., Staffurth J., Court J., Mason M.D., Tabi Z. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med. 2009;7:4. doi: 10.1186/1479-5876-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu Q., Zhang J., Allison R., Gay H., Yang W.X., Bhowmick N.A. Identification of extracellular delta-catenin accumulation for prostate cancer detection. Prostate. 2009;69:411–418. doi: 10.1002/pros.20902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barreto A., Rodriguez L.S., Rojas O.L., Wolf M., Greenberg H.B., Franco M.A. Membrane vesicles released by intestinal epithelial cells infected with rotavirus inhibit T-cell function. Viral Immunol. 2010;23:595–608. doi: 10.1089/vim.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou H., Pisitkun T., Aponte A., Yuen P.S., Hoffert J.D., Yasuda H. Exosomal Fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int. 2006;70:1847–1857. doi: 10.1038/sj.ki.5001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou H., Cheruvanky A., Hu X., Matsumoto T., Hiramatsu N., Cho M.E. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008;74:613–621. doi: 10.1038/ki.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miranda K.C., Bond D.T., McKee M., Skog J., Paunescu T.G., Da Silva N. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010;78:191–199. doi: 10.1038/ki.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michael A., Bajracharya S.D., Yuen P.S., Zhou H., Star R.A., Illei G.G. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heneghan H.M., Miller N., Lowery A.J., Sweeney K.J., Kerin M.J. MicroRNAs as novel biomarkers for breast cancer. J Oncol. 2009;2009:950201. doi: 10.1155/2010/950201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 73.Rabinowits G., Gercel-Taylor C., Day J.M., Taylor D.D., Kloecker G.H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 74.Li J., Sherman-Baust C.A., Tsai-Turton M., Bristow R.E., Roden R.B., Morin P.J. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer. 2009;9:244. doi: 10.1186/1471-2407-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Corcoran C., Friel A.M., Duffy M.J., Crown J., O’Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57:18–32. doi: 10.1373/clinchem.2010.150730. [DOI] [PubMed] [Google Scholar]
- 76.Gourzones C., Gelin A., Bombik I., Klibi J., Verillaud B., Guigay J. Extra-cellular release and blood diffusion of BART viral micro-RNAs produced by EBV-infected nasopharyngeal carcinoma cells. Virol J. 2010;7:271. doi: 10.1186/1743-422X-7-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silva J., Garcia V., Zaballos A., Provencio M., Lombardia L., Almonacid L. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J. 2011;37:617–623. doi: 10.1183/09031936.00029610. [DOI] [PubMed] [Google Scholar]
- 78.Reich C.F., 3rd, Pisetsky D.S. The content of DNA and RNA in microparticles released by Jurkat and HL-60 cells undergoing in vitro apoptosis. Exp Cell Res. 2009;315:760–768. doi: 10.1016/j.yexcr.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 79.Sonoda H., Yokota-Ikeda N., Oshikawa S., Kanno Y., Yoshinaga K., Uchida K. Decreased abundance of urinary exosomal aquaporin-1 in renal ischemia-reperfusion injury. Am J Physiol Renal Physiol. 2009;297:F1006–F1016. doi: 10.1152/ajprenal.00200.2009. [DOI] [PubMed] [Google Scholar]
- 80.Rood I.M., Deegens J.K., Merchant M.L., Tamboer W.P., Wilkey D.W., Wetzels J.F. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010;78:810–816. doi: 10.1038/ki.2010.262. [DOI] [PubMed] [Google Scholar]
- 81.Lawrie C.H., Gal S., Dunlop H.M., Pushkaran B., Liggins A.P., Pulford K. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 82.Wittmann J., Jack H.M. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta. 2010;1806:200–207. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 83.Kosaka N., Iguchi H., Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Swarup V., Rajeswari M.R. Circulating (cell-free) nucleic acids—a promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007;581:795–799. doi: 10.1016/j.febslet.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 85.Hu Z., Chen X., Zhao Y., Tian T., Jin G., Shu Y. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 86.Lebanony D., Benjamin H., Gilad S., Ezagouri M., Dov A., Ashkenazi K. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 87.De Smaele E., Ferretti E., Gulino A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010;1338:100–111. doi: 10.1016/j.brainres.2010.03.103. [DOI] [PubMed] [Google Scholar]
- 88.Resnick K.E., Alder H., Hagan J.P., Richardson D.L., Croce C.M., Cohn D.E. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 89.Wang K., Zhang S., Marzolf B., Troisch P., Brightman A., Hu Z. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cogswell J.P, Ward J., Taylor I.A., Waters M., Shi Y., Cannon B. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 91.Corsten M.F., Dennert R., Jochems S., Kuznetsova T., Devaux Y., Hofstra L. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 92.Tijsen A.J., Creemers E.E., Moerland P.D., de Windt L.J., van der Wal A.C., Kok W.E. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]


