Abstract
Heptad repeat regions (HR1 and HR2) are highly conserved sequences located in the glycoproteins of enveloped viruses. They form a six-helix bundle structure and are important in the process of virus fusion. Peptides derived from the HR regions of some viruses have been shown to inhibit the entry of these viruses. SARS-CoV was also predicted to have HR1 and HR2 regions in the S2 protein. Based on this prediction, we designed 25 peptides and screened them using a HIV-luc/SARS pseudotyped virus assay. Two peptides, HR1-1 and HR2-18, were identified as potential inhibitors, with EC50 values of 0.14 and 1.19 μM, respectively. The inhibitory effects of these peptides were validated by the wild-type SARS-CoV assay. HR1-1 and HR2-18 can serve as functional probes for dissecting the fusion mechanism of SARS-CoV and also provide the potential of further identifying potent inhibitors for SARS-CoV entry.
Keywords: SARS-CoV, Entry inhibitor, Peptides, Spike protein, Pseudotyped virus
Severe acute respiratory syndrome (SARS) is a life-threatening form of atypical pneumonia [1], [2] caused by a coronavirus—SARS coronavirus (SARS-CoV) [3], [4]. Although the first outbreak is over, concerns remain over the possibility of future recurrences. Despite the early success in SARS-CoV genome sequencing [5] and later the identification of receptor ACE2 [6], [7], neither efficacious therapies nor preventive treatments have yet been available. Therefore, effective drugs are needed for the treatment and prevention of this disease.
Viral entry process is an attractive drug target for blocking infection [8]. During this process, some enveloped viruses, such as retrovirus HIV-1, filovirus Ebola, paramyxovirus MV (measles virus), and coronavirus MHV (murine hepatitis virus), adopt a similar molecular fusion model [9], [10], [11], [12], [13], [14], [15], [16]. Under this model, it is believed that envelope glycoprotein initially binds to cell surface receptor, which leads to its conformational changes, thus the fusion peptide is exposed to insert into the target cell membrane. Subsequently, the highly conserved heptad repeat (HR) regions (HR1 and HR2) in the glycoprotein interact with each other to form a six-helix bundle structure, which facilitates the juxtaposition of the virus and cell membranes, leading to membrane fusion [9]. At the intermediate state, soluble synthesized HR1, HR2 or homologues can bind the exposed HR1 or HR2 region and block the formation of the six-helix bundle, thus inhibiting virus fusion with the target cell [11], [17]. Based on this model, entry inhibitors have been successfully identified for some viruses, such as T20 for HIV-1 [18], [19], GP610 for Ebola virus [15], and HR2 for MHV [16].
As a member of typical class I enveloped virus, SARS-CoV was presumed to use a similar membrane fusion mechanism for virus entry. Spike (S) protein, the viral membrane protein responsible for cell entry, is believed to consist of two subunits—the surface subunit S1 and the transmembrane subunit S2 [3], [5]. The S1 subunit has been identified to bind to ACE2, the cell receptor for SARS-CoV [6], [7], [20], [21]. S2 is also suggested to be involved in the virus-cell fusion process [22], [23]. S2 modeling shows that S2 contains two heptad repeat regions (HRs) similar to other enveloped viruses [23], [24]. Based on the fusion mechanism described above, we used an approach that was successfully used for studying other enveloped viruses, such as HIV-1 and MHV, to identify the potent inhibitor for virus entry [16], [18], [19], [25]. A series of peptides overlapping the predicted HR regions in the S2 subunit were designed. Screened through the pseudotyped virus infection assay [7], [26], [27], [28] and validated with wild-typed virus infection, two peptides were identified, HR1-1 and HR2-18, which were able to inhibit the SARS-CoV entry process.
Materials and methods
Cell lines. 293T cells used in pseudotyped virus generation and VERO-E6 cells used in virus infection were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum.
Plasmids. DNA fragments corresponding to the designed peptides were amplified through polymerase chain reaction (PCR) from the full length S constructs (strain BJ01, GenBank Accession No. AY278488, a gift from Beijing Municipal Center for Disease Control and Prevention). Primers were designed to incorporate a BamHI restriction site upstream, a stop codon, and an EcoRI site downstream of each target fragment. These fragments were then subcloned into the BamHI/EcoRI site of pGEX-4T-1 (Amersham–Pharmacia Biotech) and confirmed by DNA sequencing.
To produce the SARS-CoV pseudovirus, pNL-4-3E-R-Luc (HIV-luc) and the codon optimized SARS-CoV S protein expression plasmid pcTSh were utilized [7], [27], [28].
Expression and purification of peptides. The N-terminal GST fused peptides were expressed in Escherichia coli JM109 (DE3) strain and purified with glutathione–Sepharose 4B (Pharmacia Biotech) according to the manufacturer’s instructions with some minor modifications. Briefly, cells were grown in 500 ml Luria Broth (LB) containing ampicillin agitating at 37 °C until the OD600 (optical density) reached 0.4. Isopropyl-β-d-thiogalactoside (IPTG) was then added to a final concentration of 0.2 mM. Three hours later the induced cells were pelleted, resuspended in 30 ml Lysis Buffer (1% Triton X-100, PBS, pH 7.3), and shaken at 4 °C for 30 min. After sonication, the suspension was centrifuged at 12,000 rpm for 15 min. The supernatant was collected and bonded with glutathione–Sepharose 4B at 4 °C for 30 min with shaking. The mixture was then loaded into a column, washed by 10 column volumes of PBS (pH 7.3), and cleaved by 80 U thrombin at 22 °C for 3 h. The peptides were eluted with 4 ml PBS, which were collected and further purified by RP FPLC. Fractions containing the peptides were vacuum dried overnight and dissolved in PBS. At last, these products were detected by SDS–PAGE in a 15% Tris/Glycine gel and the concentrations were determined through bicinchoninic acid protein analysis (Micro BCA assay kit; Pierce).
HIV-luc/SARS pseudotyped virus entry inhibition assay. To produce HIV-luc/SARS pseudotyped virus, 10 μg of pNL-4-3E-R-Luc (HIV-luc) and 10 μg codon optimized SARS-CoV S protein expression plasmids pcTSh were co-transfected into 2 × 106 293T cells with calcium phosphate. The medium was replaced 16 h posttransfection. After further 48 h incubation, the supernatant containing the pseudotyped virus was collected and filtered through a 0.45 μm Millipore-sized membrane. For the inhibition assay, 0.5 ng of HIV-luc/SARS pseudotyped virus was incubated with serially diluted peptides at 37 °C for 30 min. The virus/peptide mixture was then transferred to 96-well plates seeded with Vero E6 cells (3 × 103 cells/well). Each concentration was tested in octuple. After overnight incubation, the medium was replaced and the sample was incubated for an additional 36 h. The cells were then lysed and luciferase activities were measured by a Wallac Mutilabel 1420 Counter (Perkin–Elmer) using the Luciferase Assay System (Promega).
Wild-type SARS-CoV infection inhibition assay. BJ01 strain (SARS-CoV wild-type virus) was a gift of Beijing Genomics Institute. Two hundred TCID50 of wild-type SARS-CoV were co-incubated with 50 μl peptides of different concentrations at 37 °C for 30 min. The mixture was then transferred to 96-well plates, eight wells for each dilution. After incubation for 60 h, a MTT assay was then performed as described previously [29]. Briefly, 10 μl dimethylthiazol diphenyltetrazolium (MTT) (5 mg/ml) (Sigma–Aldrich) was added to each well and incubated for 4 h. The medium in each well was then replaced with 100 μl DMSO and the plates stayed at room temperature for 10 min for color development before being read by a BioRad Model 550 ELISA reader (using a test wavelength of 570 nm and a reference wavelength of 630 nm).
CD spectra analysis. CD spectra scans of peptides (25 μM in PBS) were recorded on a Jobin Yvon CD 6 spectrometer at 20 °C, using a 0.1-mm path length. Each spectrum was an average of four scans corrected by subtracting a spectrum of the buffer solution in the absence of proteinase/peptide and recorded under identical conditions. Each scan, in the range of 200–260 nm, was obtained by taking data points every 0.5 nm with an integration time of 1 s and a bandwidth of 2 nm. Secondary structure content was calculated using the program K2d (http://www.embl-heidelberg.de/~andrade/k2d.html).
Results and discussion
Production of recombinant peptides
To identify the heptad repeat (HR) region within the S protein of the SARS-CoV, we utilized the LearnCoil-VMF program to predict the coiled-coil region [25]. The graphical output result is shown in Fig. 1A . As the prediction shows, the S2 subunit ectodomain of SARS-CoV, like MHV, contains two HR domains, the HR1 region (residues 900–1005) and the HR2 region (residues 1151–1185). HR2 is located near the transmembrane domain and HR1 is about 145 amino acid residues upstream of HR2.
Fig. 1.
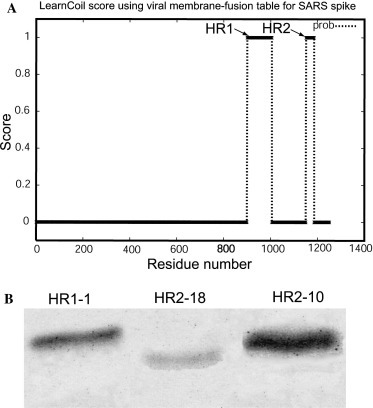
Peptide design and purification. (A) Prediction output of Learncoil-VMF program for SARS-CoV spike protein. The HR1 and HR2 are indicated by arrowhead. (B) Fifteen percent SDS–PAGE analysis of purified peptides. Samples (7.5 μl) were loaded for each lane. The gel was stained by Coomassie blue.
We then designed 25 peptides corresponding to the HR1 and HR2 regions in order to quickly and effectively identify potent inhibitors. For other class I enveloped viruses, effective peptide entry inhibitors were mostly derived from the C terminal of the helix region, thus we have designed 20 peptides (Table 1 ; HR2-1 to HR2-20) covering the HR2 region and five peptides overlapping the HR1 region (Table 1; HR1-1 to HR1-5).
Table 1.
Sequences of peptides derived from HR1 and HR2 regions of SARS-CoV spike proteina
| Peptideb | Residues |
|---|---|
| HR1-1(889–926) | NGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTA |
| HR1-2(909–948) | FNKAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQL |
| HR1-3(940–978) | ALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRL |
| HR1-4(975–1010) | IDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATK |
| HR1-5(989–1028) | YVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKG |
| HR2-1(1119–1159) | VYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVV |
| HR2-2(1124–1162) | QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQ |
| HR2-3(1129–1170) | SFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNE |
| HR2-4(1134–1173) | LDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAK |
| HR2-5(1138–1178) | FKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNES |
| HR2-6(1144–1183) | PDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQ |
| HR2-7(1149–1188) | GDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKY |
| HR2-8(1151–1196) | ISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPW |
| HR2-9(1151–1187) | ISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGK |
| HR2-10(1151–1185) | ISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL |
| HR2-11(1153–1187) | GINASVVNIQKEIDRLNEVAKNLNESLIDLQELGK |
| HR2-12(1154–1196) | INASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPW |
| HR2-13(1154–1192) | INASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYI |
| HR2-14(1154–1187) | INASVVNIQKEIDRLNEVAKNLNESLIDLQELGK |
| HR2-15(1154–1182) | INASVVNIQKEIDRLNEVAKNLNESLIDL |
| HR2-16(1158–1196) | VVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPW |
| HR2-17(1158–1187) | VVNIQKEIDRLNEVAKNLNESLIDLQELGK |
| HR2-18(1161–1187) | IQKEIDRLNEVAKNLNESLIDLQELGK |
| HR2-19(1161–1196) | IQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPW |
| HR2-20(1165–1196) | IDRLNEVAKNLNESLIDLQELGKYEQYIKWPW |
The residues in bold are the peptides that show inhibition activity.
Numbers in the brackets are the beginning and end residues for each peptide corresponding to the spike protein.
To produce these long peptides, we used the recombinant E. coli expression system, which was able to provide peptide products in large quantities at low cost. Each recombinant peptide was expressed in E. coli and then purified through chromatography. The products were analyzed by 15% SDS–PAGE. Fig. 1B shows the purified HR1-1, HR2-10, and HR2-18, as examples of these 25 peptides.
Inhibition activity of the peptides
We then detected the entry inhibition activity of this panel of peptides by HIV-luc/SARS pseudotyped virus [7], [27], [28]. Two peptides, HR1-1 and HR2-18, were found to have anti-viral activities. As Fig. 2 shows, HR1-1 locates in the N-terminal of HR1 (residues 889–926; Fig. 2A) and its EC50 value was 0.14 μM, while HR2-18 corresponds to the HR2 region 1161–1187 (Fig. 2A) with an EC50 value of 1.19 μM. All other peptides such as HR2-10 showed little anti-virus activities (Fig. 2B).
Fig. 2.
The inhibitory activities of the peptides for HIV-luc/SARS pseudotyped virus infection. (A) Sequence location of the peptides HR1-1 and HR2-18 (italics and underlined). (B) Peptides HR1-1 and HR2-18 can inhibit the HIV-luc/SARS pseudotyped virus infection with EC50 value of 0.14 and 1.19 μM while HR2-10 showed little effect. The experiments were repeated three times and the data here show the average values.
We further tested the inhibitory effect by wild-type SARS-CoV using a MTT assay as previously described [29]. HR1-1 and HR2-18 also showed inhibition activities against wild-type virus infection, with EC50 values 3.68 and 5.22 μM, respectively (Fig. 3 ). The result was similar to the pseudotyped virus assay. None of the other peptides showed anti-virus activity (data not shown).
Fig. 3.
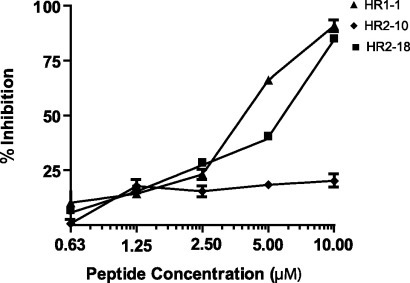
The inhibition activities of HR1-1 and HR2-18 for wild-typed SARS-CoV infection. Inhibitory effects of HR1-1, HR2-10, and HR2-18 were tested by MTT assay. The EC50 of HR1-1 (triangle) was 3.68 μM and the EC50 of HR2-18 (square) was 5.22 μM; HR2-10 (diamond) showed no effect. Similar results were obtained in three independent experiments.
Through the screening of the pseudotyped virus assay, we found HR1-1 was more effective than HR2-18 in inhibiting viral entry, whereas in the wild-type assay, their difference was shown to be less distinct. This discrepancy may be attributed to the more sensitive and quantitative nature of the pseudotyped virus assay for analyzing the inhibition effect of virus entry with its single-round infection and the use of a quantitative luciferase assay system, compared with the wild-type virus assay.
CD spectra analysis
As the peptide inhibitors, such as T-20 for HIV-1, bind to the fusogenic intermediate through interaction of α-helixes, we investigated whether HR1-1 and HR2-18 also have α-helical structure and whether these two peptides were able to form an oligomeric complex. Circular dichroism (CD) was used to study the second structure of the HR1-1, HR2-18, and HR1-1/HR2-18 complexes. The CD spectra of HR1-1, HR2-18, and the equimolar mixture of the two peptides are shown in Fig. 4 . The spectra of HR1-1, HR2-18, and HR1-1/HR2-18 mixture all showed clear negative peaks at 208 nm and small negative peaks at 220 nm, thus suggesting the atypical-helix characteristics.
Fig. 4.
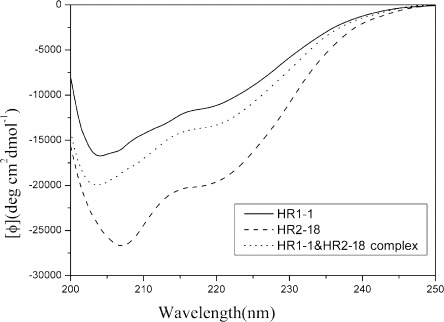
Secondary structure of HR1-1, HR2-18, and HR1-1 and HR2-18 complex by circular dichroism (CD) analysis. CD spectra of the peptides were analyzed at 25 °C in PBS. Spectra of HR1-1 (solid line) and HR2-18 (dashed line) showed a clear negative peak at 208 nm; the spectra of HR1-1 and HR2-18 complex (dotted line) showed a clear negative peak at 208 nm and small negative peaks at 220 nm, which is indicative of α-helix characteristics.
In this study, we found that none of the 20 different peptides corresponding to HR2 region had a higher inhibition activity than HR1-1. This result is in contrast to most other enveloped viruses, such as HIV-1 and MHV, whose peptides in the HR2 region outperform those in HR1 in inhibiting virus fusion [30], [31]. In the enveloped virus fusion model, the HR1 region forms an inner, trimeric, coiled-coil core surrounded by three helical HR2 regions outside [9], [10], [11], [12], [17]. In this way, HR1 interacts not only with HR2, but also with other HR1 molecules. It has been proposed that HR1 are poor inhibitors for some viruses because they can form self-associated oligomers and aggregate in isolation, therefore interfering with their ability to efficiently block the formation of a six-helix bundle [17], [31]. In the case of SARS-CoV, the more potent inhibition activity of HR1-1 may correlate with its atypical-helix structure (as the CD spectra show in Fig. 4). This conformation, with its low coiled-coil tendency, might inhibit the self-associated oligomerization, and thus increase the inhibition potency of HR1-1. Our analysis of the HR1-1 structure was consistent with the recent report by Liu et al. [32]. It has been reported that the major localization of helical content in HR1 is downstream of HR1-1 [33], such as HR-N13 (residues 927–973) with typical α-helix structure. However, we found HR1-3 similar to HR-N13 did not show any viral entry inhibition activity. Similarly, in the case of MHV, it has been demonstrated that the HR1 region with a typical α-helix structure showed low inhibitory activity [16]. These evidences support our proposition above. It has been reported that HR2 peptide of HIV could induce the formation of the coiled-coil HR1 core [34]. Therefore, this suggests, in the presence of the endogenous HR2 region, that the HR1-1 may undergo conformational changes from an atypical-helix structure to a typical-helix structure and bind to the HR2 region to block fusion in the intermediate state.
Although we designed a series of peptides overlapping the HR2 region, only one peptide HR2-18 was identified to inhibit the entry of SARS-CoV with low inhibitor activity. In the recent report, a peptide corresponding to HR2 had a stronger α-helical spectrum than HR2-18, but this also showed low inhibitory activity [32]. Therefore, potent inhibitors for the entry of SARS-CoV are still under identification. In addition, during screening the peptides we found slight differences between each peptide could affect their inhibitory activities. For example the peptide HR2-17, with just three amino acids more than HR2-18, showed no inhibition activity. Moreover, our peptide HR2-11 did not show any activity to inhibit virus entry, but with the addition of only two amino acids in the C-terminal, peptide CP-1 was identified to have inhibition activity by Liu et al. [32]. It is possible that the addition or deletion of several amino acids may change the conformational structure of the peptides and consequently influence their inhibitory effects. Based on this, we will further refine the mapping of the peptides to detect the changes in their conformation and inhibitory activity in order to identify possible correlations.
In summary, our data suggest that the HR1 and HR2 regions are crucial in the entry of SARS-CoV, and support the assumption that SARS-CoV may employ the same fusion mechanism as HIV-1 and other viruses with class I envelope proteins. HR1-1 and HR2-18 can be used as functional probes to further study the fusion process of SARS-CoV, which may finally lead to the finding of potent inhibitor peptides for SARS-CoV.
Acknowledgements
We are grateful to Dr. Taisheng Li and Dr. Yang Han for their assistance with the P3 laboratory and we thank Wang Zai for assistance in full-length S gene construction. We also thank Xiaojun zhao, Xiangpeng Kong, Guangwen Wang, and Hong Zhang for critical reading of the manuscript. This research was supported by the National Nature Science Foundation of China Grant (30340027), the Ministry of Science and Technology Grant (2003CB514116), the National Nature Science Foundation of China for Outstanding Young Scientist Award (30125022) to H. Deng, and the Ministry of Science and Technology Grant (G1999011904) to M. Ding.
References
- 1.Tsang K.W, Ho P.L, Ooi G.C, Yee W.K, Wang T, Chan-Yeung M, Lam W.K, Seto W.H, Yam L.Y, Cheung T.M, Wong P.C, Lam B, Ip M.S, Chan J, Yuen K.Y, Lai K.N. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 2.Poutanen S.M, Low D.E, Henry B, Finkelstein S, Rose D, Green K, Tellier R, Draker R, Adachi D, Ayers M, Chan A.K, Skowronski D.M, Salit I, Simor A.E, Slutsky A.S, Doyle P.W, Krajden M, Petric M, Brunham R.C, McGeer A.J. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 3.Rota P.A, Oberste M.S, Monroe S.S, Nix W.A, Campagnoli R, Icenogle J.P, Penaranda S, Bankamp B, Maher K, Chen M.H, Tong S, Tamin A, Lowe L, Frace M, DeRisi J.L, Chen Q, Wang D, Erdman D.D, Peret T.C, Burns C, Ksiazek T.G, Rollin P.E, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus A.D, Drosten C, Pallansch M.A, Anderson L.J, Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt H.R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier R.A, Berger A, Burguiere A.M, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra J.C, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk H.D, Osterhaus A.D, Schmitz H, Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Marra M.A, Jones S.J, Astell C.R, Holt R.A, Brooks-Wilson A, Butterfield Y.S, Khattra J, Asano J.K, Barber S.A, Chan S.Y, Cloutier A, Coughlin S.M, Freeman D, Girn N, Griffith O.L, Leach S.R, Mayo M, McDonald H, Montgomery S.B, Pandoh P.K, Petrescu A.S, Robertson A.G, Schein J.E, Siddiqui A, Smailus D.E, Stott J.M, Yang G.S, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth T.F, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples G.A, Tyler S, Vogrig R, Ward D, Watson B, Brunham R.C, Krajden M, Petric M, Skowronski D.M, Upton C, Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 6.Li W, Moore M.J, Vasilieva N, Sui J, Wong S.K, Berne M.A, Somasundaran M, Sullivan J.L, Luzuriaga K, Greenough T.C, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang P, Chen J, Zheng A, Nie Y, Shi X, Wang W, Wang G, Luo M, Liu H, Tan L, Song X, Wang Z, Yin X, Qu X, Wang X, Qing T, Ding M, Deng H. Expression cloning of functional receptor used by SARS coronavirus. Biochem. Biophys. Res. Commun. 2004;315:439–444. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore J.P, Doms R.W. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. USA. 2003;100:10598–10602. doi: 10.1073/pnas.1932511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckert D.M, Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 10.Weissenhorn W, Dessen A, Calder L.J, Harrison S.C, Skehel J.J, Wiley D.C. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 11.Skehel J.J, Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 12.Bentz J. Membrane fusion mediated by coiled coils: a hypothesis. Biophys. J. 2000;78:886–900. doi: 10.1016/S0006-3495(00)76646-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb R.A, Joshi S.B, Dutch R.E. The paramyxovirus fusion protein forms an extremely stable core trimer: structural parallels to influenza virus haemagglutinin and HIV-1 gp41. Mol. Membr. Biol. 1999;16:11–19. doi: 10.1080/096876899294715. [DOI] [PubMed] [Google Scholar]
- 14.Peisajovich S.G, Shai Y. New insights into the mechanism of virus-induced membrane fusion. Trends Biochem. Sci. 2002;27:183–190. doi: 10.1016/s0968-0004(01)02050-3. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe S, Takada A, Watanabe T, Ito H, Kida H, Kawaoka Y. Functional importance of the coiled-coil of the Ebola virus glycoprotein. J. Virol. 2000;74:10194–10201. doi: 10.1128/jvi.74.21.10194-10201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch B.J, van der Zee R, de Haan C.A, Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Root M.J, Kay M.S, Kim P.S. Protein design of an HIV-1 entry inhibitor. Science. 2001;291:884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- 18.Kilby J.M, Hopkins S, Venetta T.M, DiMassimo B, Cloud G.A, Lee J.Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M.R, Nowak M.A, Shaw G.M, Saag M.S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 19.Imai M, Okada N, Okada H. Inhibition of HIV-1 infection by an intramolecular antisense peptide to T20 in gp160. Microbiol. Immunol. 2000;44:205–212. doi: 10.1111/j.1348-0421.2000.tb02485.x. [DOI] [PubMed] [Google Scholar]
- 20.Wong S.K, Li W, Moore M.J, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao X, Chakraborti S, Dimitrov A.S, Gramatikoff K, Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang E, Sun X, Qian Y, Zhao L, Tien P, Gao G.F. Both heptad repeats of human respiratory syncytial virus fusion protein are potent inhibitors of viral fusion. Biochem. Biophys. Res. Commun. 2003;302:469–475. doi: 10.1016/s0006-291x(03)00197-9. [DOI] [PubMed] [Google Scholar]
- 23.Kliger Y, Levanon E.Y. Cloaked similarity between HIV-1 and SARS-CoV suggests an anti-SARS strategy. BMC Microbiol. 2003;3:20. doi: 10.1186/1471-2180-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spiga O, Bernini A, Ciutti A, Chiellini S, Menciassi N, Finetti F, Causarono V, Anselmi F, Prischi F, Niccolai N. Molecular modelling of S1 and S2 subunits of SARS coronavirus spike glycoprotein. Biochem. Biophys. Res. Commun. 2003;310:78–83. doi: 10.1016/j.bbrc.2003.08.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh M, Berger B, Kim P.S. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J. Mol. Biol. 1999;290:1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons G, Reeves J.D, Rennekamp A.J, Amberg S.M, Piefer A.J, Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA. 2004 doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuchun Nie, Guangwen Wang, Xuanling Shi, Hong Zhang, Yan Qiu, Zhongping He, Wei Wang, Gewei Lian, L.D. Xiaolei Yin, Lili Ren, Jianwei Wang, Xiong He, Taisheng Li, Hongkui Deng, M. Ding, Neutralizing antibodies in the SARS-CoV-infected patients, J. Infect. Dis. (2004) in press [DOI] [PMC free article] [PubMed]
- 28.Hong Zhang, Guangwen Wang, Jian Li, Yuchun Nie, Xuanling Shi, Gewei Lian, Wei Wang, Xiaolei Yin, Yang Zhao, Xiuxia Qu, M. Ding, H. Deng, Identification of an antigenic determinant on the S2 domain of the SARS-CoV spike glycoprotein capable of inducing neutralizing antibodies, J. Virol. (2004) in press [DOI] [PMC free article] [PubMed]
- 29.Hussain R.F, Nouri A.M, Oliver R.T. A new approach for measurement of cytotoxicity using colorimetric assay. J. Immunol. Methods. 1993;160:89–96. doi: 10.1016/0022-1759(93)90012-v. [DOI] [PubMed] [Google Scholar]
- 30.Wild C.T, Shugars D.C, Greenwell T.K, McDanal C.B, Matthews T.J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu M, Blacklow S.C, Kim P.S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 32.Liu S, Xiao G, Chen Y, He Y, Niu J, Escalante C.R, Xiong H, Farmar J, Debnath A.K, Tien P, Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripet B, Howard M.W, Jobling M, Holmes R.K, Holmes K.V, Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004 doi: 10.1074/jbc.M400759200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dwyer J.J, Hasan A, Wilson K.L, White J.M, Matthews T.J, Delmedico M.K. The hydrophobic pocket contributes to the structural stability of the N-terminal coiled coil of HIV gp41 but is not required for six-helix bundle formation. Biochemistry. 2003;42:4945–4953. doi: 10.1021/bi027283n. [DOI] [PubMed] [Google Scholar]


