Short abstract
Objective
Nitrate has been reported to protect cells via the nitrate-nitrite-nitric oxide (NO) pathway. Most studies tend to use nitrite to investigate the mechanisms of this pathway. However, the latest studies have confirmed that mammals can directly degrade nitrate via xanthine oxidoreductase (XOR). The hypothesis is that nitrate could play a protective role in inflammatory responses independent of bacterial nitrate reductases.
Methods
Mouse RAW264.7 macrophages were pre-incubated with sodium nitrate (10, 100, and 500 µM) for 2 hours, and then treated with lipopolysaccharide (LPS) for 2 hours to induce inflammation. The Quantikine Immunoassay was used to measure interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) concentrations in the supernatant. The fluorescence intensity ratio of red/green from JC-1 was used to assay mitochondrial membrane potential. The fluorescence intensity of MitoSOX Red was used to indicate the generation of mitochondrial reactive oxygen species.
Results
Nitrate partially reduced IL-6 and TNF-α secretion via reducing NF-κB signaling in LPS-induced macrophages. Nitrate also reduced the generation of mitochondrial reactive oxygen species by regulating mitochondrial function. These effects depended on XOR-derived NO but were independent of inducible nitric oxide synthase-derived NO.
Conclusion
Nitrate regulates mitochondrial function via XOR-derived NO to partially inhibit LPS-induced inflammation.
Keywords: Nitrate, nitric oxide, lipopolysaccharide, mitochondrial reactive oxygen species, mitochondrial function, xanthine oxidoreductase
Introduction
Nitrate is a common inorganic molecule that is widely found in green vegetables. Over the past decades, nitrate has been confirmed to have protective effects in many diseases.1–10 The bioavailability of nitrate mainly depends on a unique group of bacterial nitrate reductases in the mouth and gut of mammals that reduce nitrate to nitrite.1–4 The effects of nitrite have been extensively studied. For instance, nitrite can be converted into many bioactive nitrides.5–10 In general, researchers have tended to elucidate the mechanisms of dietary nitrate therapy by focusing on nitrite. However, recent studies have confirmed that mammals can directly degrade nitrate via xanthine oxidoreductase (XOR).11–13 When volunteers were given dietary nitrate therapy, the in vivo concentrations of both nitrate and nitrite increased significantly.3 Because nitrate can be directly degraded in mammals by XOR, the in vivo protective mechanisms of nitrate cannot be ignored.
Inflammation is a defensive response to stimuli during an immune response. Macrophages are the primary participants in an inflammatory response.14 When macrophages are stimulated, such as by lipopolysaccharide (LPS), they generate superfluous reactive oxygen species (ROS).15 The superfluous ROS leads to decreased ROS clearance and increased ROS production, which disrupts the homeostasis of ROS production and clearance, leading to severe oxidative stress.16 Oxidative stress can further aggravate inflammatory responses, causing more serious consequences.17 Preventing superfluous ROS production is key to relieving oxidative stress.
Mitochondria are a primary source of ROS.18 Normally, mitochondrial ROS (mtROS) is in a state of dynamic balance between production and elimination.19 It is widely believed that mitochondria can produce a large number of harmful ROS in many different pathological environments.20,21 A recent hypothesis was that superfluous mtROS could, either directly or indirectly, regulate inflammatory responses.22 High mtROS levels from abnormal stimuli could directly activate NF-κB signaling.23,24 Therefore, reducing mtROS production is a potential treatment that could alleviate inflammatory responses. Many studies have found that dietary nitrate therapy can decrease the cellular generation of mtROS, relieving oxidative stress and secondary injury via the nitrate-nitrite-nitric oxide (NO) pathway.5–7
The nitrate-nitrite-NO pathway is considered supplemental to the classic nitric oxide synthase (NOS) pathway for NO homeostasis.11–13 Originally, nitrate was considered the endpoint product of endogenous NO metabolism; however, several studies have found that nitrate can be used as a potential donor of NO under specific circumstances.25–27 The beneficial effects of nitrate have been demonstrated in a variety of animal and clinical trials. The nitrate-nitrite-NO pathway could modulate mitochondrial function and nicotinamide adenine dinucleotide phosphate (NADPH) oxidase to alleviate cardiorenal disease and ischemia-reperfusion injury.4–7 However, the mechanism of nitrate, rather than being converted to nitrite, has not been adequately studied. Thus, the hypothesis of this study was that nitrate could relieve inflammatory responses via XOR in vitro.
Materials and methods
Ethics approval was not required because no humans or animals were used in this study.
Cell culture and treatment
Mouse RAW264.7 macrophages were purchased from the National Infrastructure of Cell Line Resource (Beijing, China). Cells were incubated in Dulbecco’s Modified Eagle’s Medium (DMEM; SH30022.01, Hyclone, Logan, UT, USA) with 10% Fetal Bovine Serum (FBS; No.10099, GIBCO, Carlsbad, CA, USA), 100 IU/mL penicillin, and 100 mg/mL streptomycin. The culture was maintained in a humidified atmosphere containing 5% CO2 at 37°C. Cells were passaged using a sterile cell scraper. After reaching 90% confluence, the cells were gently scraped off, counted, spread onto 6- or 96-well plates, and then incubated overnight. The next day, the medium was pre-incubated with phenol-free red DMEM and various concentrations of sodium nitrate (10, 100, and 500 µM) or sodium chloride (10, 100, and 500 µM) for 2 hours. The NO scavenger carboxy-PTIO (1 mM; S1546, Beyotime, Shanghai, China), the iNOS inhibitor L-canavanine (1 mM; S0007, Beyotime), or the XOR inhibitor allopurinol (100 µM; A8003, Sigma-Aldrich, Saint Louis, MO, USA) were added 30 minutes before the LPS incubation. Finally, LPS (10 ng/mL, from E. coli O111:B4, Invivogen, San Diego, CA, USA) was added to the medium at various times to collect different samples or to measure different indicators.
Measuring interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) levels
IL-6 and TNF-α were measured using Quantikine Immunoassay Kits according to the manufacturer’s instructions (431304 and 430904, BioLegend, San Diego, CA, USA). Cells were counted and spread onto a 6-well plate at a concentration of 5 × 105 cells/mL. After pre-incubation, LPS was added to stimulate the samples for 2 hours. The medium was collected and centrifuged for 10 minutes at 1000 ×g, and then the supernatant was removed to measure the concentrations of IL-6 and TNF-α.
Western blot analysis
The western blotting protocol was adapted from Zollbrecht et al.29 Proteins extracted from cells were lysed in Radio Immunoprecipitation Assay (RIPA) buffer with 1% protease inhibitor mixture (PIC) and 1% phenylmethylsulphonyl fluoride (PMSF). The samples were separated on a 10% SDS polyacrylamide gel and transferred to polyvinylidene difluoride membranes in a semi-dry transfer apparatus (4561033, Bio-Rad, Hercules, CA, USA). The membranes were pre-incubated at room temperature in 5% dehydrated milk for 2 hours. Then each membrane was incubated with specific primary antibodies against IKKα (2684, Cell Signaling Technology, Danvers, MA, USA), p-IKKα (2694, Cell Signaling Technology), NF-κB (p65) (3034, Cell Signaling Technology), p-NF-κB (p65) (3033, Cell Signaling Technology), iNOS (13120, Cell Signaling Technology), XOR (ab109235, Abcam, Cambridge, UK), and β-actin (AC026, Abclone, Wuhan, China) overnight. Next, the membranes were incubated with secondary antibodies conjugated to horseradish peroxidase (AS014, Abclone) and visualized using enhanced chemiluminescence reagents (1705062, Bio-Rad).
Determining cytoplasmic ROS and mtROS
Cytoplasmic ROS and mtROS were determined according the protocol used by Li et al.24 Briefly, cells were counted and spread onto a 96-well black opaque plate at a concentration of 1 × 105 cells/mL. After a 2-hour LPS challenge, the cells were incubated for 10 minutes with 5 mM MitoSOX Red (M36008, Invitrogen, Carlsbad, CA, USA) to determine mtROS. The cytoplasmic ROS content of the cells was determined by incubating them for 20 minutes with 10 mM DCFH-DA (D6470, Solarbio, Beijing, China). Next, the cells were washed twice with PBS, and fluorescence intensities were measured at 510/590 nm or 488/528 nm excitation/emission wavelengths on a multi-well spectrophotometer.
Determining mitochondrial membrane potential (ΔΨm)
The pre-treatment step was the same as that described for determining mtROS. The ΔΨm were determined according to the protocol developed by Prathapan et al.28 Briefly, the same volume of JC-1 (C2006, Beyotime) was added and incubated with the cells for 20 minutes. Cells were then washed twice with PBS. To measure the fluorescence intensity with a multi-well spectrophotometer, the excitation/emission wavelengths were set at 490/530 nm for the JC-1 monomer and 525/590 nm for the JC-1 aggregate.
Measuring the activity of mitochondrial respiratory chain complexes I and III
These methods were adapted from Shiva et al. and Dezfulian et al.7,9 Briefly, cells were collected and disrupted in a Tris-HCl (pH 7.4) buffer. The buffer was centrifuged at 800 ×g for 10 minutes at 4°C. Then, the supernatant was centrifuged at 20,000 ×g for 10 minutes at 4°C. The supernatant was discarded, then the pellet was re-suspended in 0.2 mM ice-cold sodium citrate. Mitochondria were lysed by ultrasound. Protein concentrations were measured by the BCA Kit (P0011, Beyotime). The activity of complex I was determined by spectrophotometry through monitoring the oxidation of NADH at 340 nm (BC0515, Solarbio). The activity of complex III was measured by monitoring the increased rate of light absorption at 550 nm (BC3245, Solarbio).
Reverse transcription-PCR
The reverse transcription-PCR determination protocol is referenced from Li et al.24 Briefly, total RNA was extracted from cells using TRIzol reagent according to the manufacturer’s instructions (CW0581, CWbiotech, Jiangsu, China). The RNA purity (the 260/280 nm ratio) was between 1.8 and 2.0 for all samples. The RNA concentrations were approximately 300 nM. Next, cDNA was synthesized using a Prime Script RT Reagent Kit (RR047A, Takara, Otsu, Japan). Gene transcripts were quantified via RT-PCR, which was performed with a SYBR Green PCR Kit (204054, Qiagen, Limburg, Germany). The sequences of related genes were searched from the primer bank. The product sizes were approximately 150 bp. The sequences for the primers used were as follows: iNOS, 5′-GGAGTGACG G CAAACATGACT-3′ (forward) and 5′-TCGATGCACAACTGGGTGAAC-3′ (reverse); XOR, 5′-ATGACGAGGACAACGGTAGAT-3′ (forward) and 5′-TCATACTTGGAGATCATCACGGT-3′ (reverse); β-actin, 5′-GGCTGTATTCCCCTCCATCG-3′ (forward) and 5′-CCAGTTGGTAACAATGCCATGT-3′ (reverse). The PCR conditions were 15 minutes at 95°C, followed by 40 cycles of 94°C for 15 seconds, 60°C for 30 seconds and 72°C for 30 seconds. Gene expression was determined by the 2−ΔΔCT method.
Determining NO production
The pre-treatment was the same as that described for determining mtROS. NO production was determined according to the protocol published by Zollbrecht et al.29 Briefly, the cells were incubated for 20 minutes with 2.5 µM diaminofluorescein-FM diacetate (DAF-FM DA; S0019, Beyotime). Next, the cells were washed twice with PBS and the fluorescence intensity was measured at 495/525 nm excitation/emission wavelengths using a multi-well spectrophotometer.
Statistical analysis
Data are expressed as mean ± standard error of the mean (SEM). Statistical significance was determined using one-way ANOVA (followed by the Student’s t test) for multiple comparisons. Analyses and graphs were performed using GraphPad Prism 5 software (GraphPad Software Inc., La Jolla, CA, USA). Differences were considered statistically significant if p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***).
Results
Nitrate relieved LPS-induced inflammation
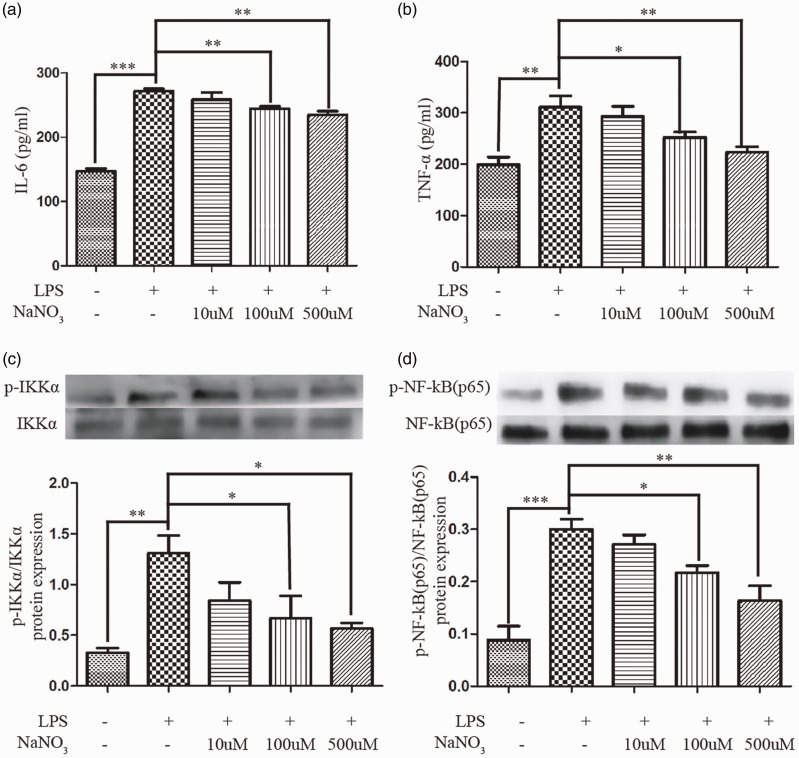
We found that nitrate could gradually reduce the levels of IL-6 (Figure 1a) and TNF-α (Figure 1b) secretion in a concentration gradient of sodium nitrate from 100 to 500 µM after a 2-hour LPS-induction. Then the levels of total and phosphorylated IKKα and NF-κB (p65) were measured at 1 hour (Figure 1c-d). The ratio of phosphorylated protein to total protein was used to indicate the degree of NF-κB pathway activation. LPS significantly activated the NF-κB pathway, while nitrate relieved this form of activation.
Figure 1.
Nitrate relieved LPS-induced inflammation. The concentration of IL-6 (a) and TNF-α (b) in the supernatant after 2-hour LPS treatment (n = 4). Western blot of total and phosphorylated levels of IKKα (c) and NF-κB (p65) (d) after 1-hour LPS treatment (n = 3). Data are expressed as mean ± SEM; p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
Nitrate reduced mtROS generation and enhanced mitochondrial respiration under inflammatory conditions
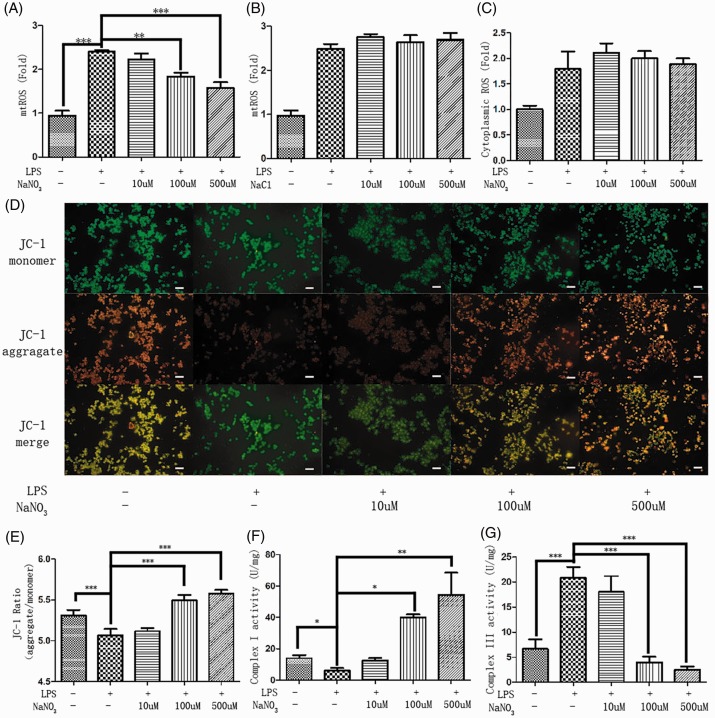
The MitoSOX Red probe was used to monitor mtROS generation. After a 2-hour LPS challenge, mitochondria in the LPS-only group produced about 2.4-fold higher ROS levels than the control group (p < 0.001). The pattern of reduced mtROS generation after LPS challenge was dependent on the sodium nitrate concentration from 100 to 500 µM (Figure 2a). In contrast, changing the sodium chloride concentration did not affect mtROS levels following LPS challenge (Figure 2b). These data suggest that increasing nitrate, rather than sodium, concentrations led to reduced levels of mtROS. Cytoplasmic ROS content was determined by DCFHDA (Figure 2c). Sodium nitrate did not change cytoplasmic ROS production. These results suggest that sodium nitrate may regulate mtROS generation in response to oxidative stress.
Figure 2.
Nitrate reduced mtROS generation and enhanced mitochondrial respiration under an inflammatory environment. Mitochondrial ROS levels (a) in medium loaded with LPS and various concentrations of sodium nitrate (10, 100, and 500 µM). Mitochondrial ROS levels (b) in medium loaded with LPS and various concentrations of sodium chloride (10, 100, and 500 µM). Cytoplasmic ROS levels (c) in medium loaded with LPS and various concentrations of sodium nitrate (10, 100, and 500 µM). Fluorescence micrographs (d) of JC-1 aggregate, monomer, and the merge (scale bar: 20 µm). The fluorescence intensity ratio (e) of JC-1 aggregate to monomer (red to green). Mitochondria were collected 2 hours after LPS stimulation, and the activity of complexes I (f) and III (g) were detected after ultrasonic fragmentation (n = 3). Data are expressed as mean ± SEM (n = 4); p < 0.01 (**) and p < 0.001 (***).
The ΔΨm was determined by the fluorescence intensity of JC-1 (Figure 2d). Pictures of the red fluorescent JC-1 aggregate and the green fluorescent JC-1 monomer were taken by fluorescence microscopy. ImageJ software was used to mix the channels. ΔΨm was evaluated by the fluorescence intensity ratio of red to green (Figure 2e). Nitrate restored the decreased ΔΨm, or in some cases even exceeded the normal levels in the control group (p < 0.001).
To determine how nitrate reduces mtROS generation and enhances ΔΨm, mitochondrial respiratory function was determined by measuring the activity of mitochondrial respiratory chain complex I (Figure 2f) and III (Figure 2g). Nitrate significantly enhanced the activity of complex I (p < 0.01). Additionally, nitrate further inhibited the activity of complex III more than in the LPS-challenged condition. It appeared that nitrate improved mitochondrial respiratory capacity by activating complex I activity and inhibiting complex III activity.
Nitrate modulated mitochondrial function via NO but did not change iNOS or XOR expression
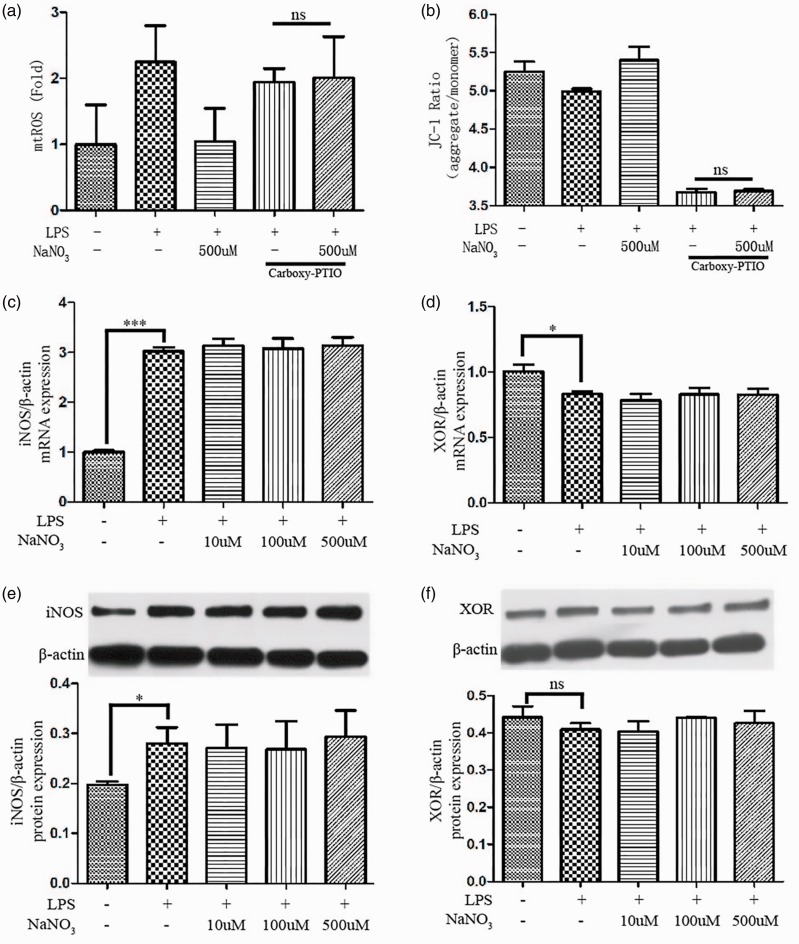
To determine the mechanism of how nitrate regulates mitochondrial function, the NO scavenger Carboxy-PITO was used to detect mtROS generation and ΔΨm (Figure 3a-b). When the scavenger was applied, nitrate could not reduce mtROS production or enhance ΔΨm. These data may explain the mechanism of how these effects of nitrate depend on NO.
Figure 3.
Nitrate modulated mitochondrial function via NO and did not alter the expression of iNOS or XOR. The mtROS (a) and ΔΨm (b) after addition of the NO scavenger Carboxy-PITO (1 mM) (n = 4). The mRNA expression of iNOS (c) and XOR (d) in medium loaded with LPS and various concentrations of sodium nitrate (10, 100, and 500 µM). The protein expression of iNOS (e) and XOR (f) in medium loaded with LPS and various concentrations of sodium nitrate (10, 100, and 500 µM). Data are expressed as mean ± SEM (n = 3); p < 0.05 (*) and p < 0.001 (***).
Additionally, we measured the expression of iNOS and XOR at the mRNA and protein levels (Figure 3c-f). The results indicated that nitrate levels did not alter the high expression of iNOS following LPS stimulation. After adding LPS, a slight reduction in XOR mRNA expression was observed; however, this was not observed for XOR protein. Nitrate levels did not change XOR expression.
The effects of nitrate were dependent on XOR but independent of iNOS
To further explore the protective mechanism of nitrate, the NO content was monitored by loading the iNOS inhibitor L-canavanine or the XOR inhibitor allopurinol. Under the iNOS inhibitor, there was increased NO generation following the concentration gradient of sodium nitrate from 100 to 500 µM (Figure 4a). In contrast, sodium nitrate did not change NO generation in the presence of the XOR inhibitor (Figure 4b). These data suggest that sodium nitrate might act through XOR-derived NO.
Figure 4.
The effects of nitrate were dependent on XOR but independent of iNOS. NO levels in medium loaded with LPS, various concentrations of sodium nitrate (10, 100, and 500 µM), and the iNOS inhibitor L-canavanine (1 mM) (a) or the XOR inhibitor allopurinol (100 µM) (b). The mtROS (c) and ΔΨm (d) after L-canavanine (1 mM) and allopurinol (100 µM) treatment. Data are expressed as mean ± SEM (n=3); p < 0.01 (**) and p < 0.001 (***).
Next, mtROS generation and ΔΨm were determined in the presence of the iNOS inhibitor or XOR inhibitor (Figure 4c-d). There were a statistical differences between the nitrate with L-canavanine group and the L-canavanine only group regarding mtROS generation and ΔΨm. However, allopurinol simultaneously and significantly blocked the effects of nitrate on both mtROS generation and ΔΨm. These results further support that the effects of nitrate depend on XOR-derived NO but are independent of iNOS-derived NO.
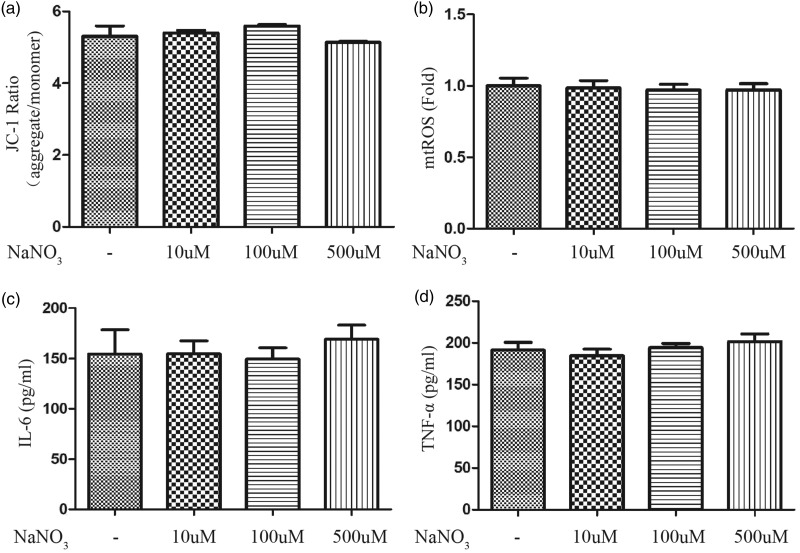
Nitrate does not modulate mitochondrial function or the inflammatory response of macrophages without LPS
The ΔΨm (Figure 5a) and production of mtROS (Figure 5b), IL-6 (Figure 5c), and TNF-α (Figure 5d) were determined in the supernatant to assay the influence of nitrate in the absence of LPS. After a 2-hour treatment with various nitrate concentrations (10, 100, and 500 µM), none of these indicators changed.
Figure 5.
Nitrate did not modulate mitochondrial function or the inflammatory response of macrophages without LPS. After a 2-hour treatment with various concentrations of nitrate (10, 100, and 500 µM), the ΔΨm (a), mtROS (b), and concentrations of IL-6 (c) and TNF-α (d) in the supernatant were detected (n = 3). Data are expressed as mean ± SEM.
Discussion
Emerging evidence has shown that nitrate has many beneficial effects on a variety of diseases.1–10 Although the bioactivity of inorganic nitrate primarily depends on oral commensal bacteria,1–4 some nitrate is also reduced by mammalian enzymes.11–13 However, the effects of nitrate on mammalian enzymes is unclear. This may be an important part of the mechanism of nitrate therapy. Based on these findings and the paucity of studies on the effects of nitrate in vitro, nitrate could partially reduce secretion of the inflammatory factors IL-6 and TNF-α by reducing NF-κB signaling in activated macrophages. The results presented herein support this hypothesis.
NADPH oxidase and other cytoplasmic and mitochondrial proteins can generate ROS.16,17 High levels of ROS from abnormal stimuli can directly activate the NF-κB pathway.23,24 Based on the above theories, nitrate may reduce the production of superfluous ROS. The content of mtROS and cytoplasmic ROS were determined to explore the mechanism of nitrate activity. Nitrate reduced mtROS generation, but did not reduce cytoplasmic ROS generation following LPS stimulation. These data suggest that nitrate may not effect NADPH oxidase and other cytoplasmic proteins. Thus, the effect of nitrate on mitochondrial function was the focus of the rest of this study.
Normally, mitochondria play an important role in cellular energy production.18 And mtROS generation is primarily through the mitochondrial respiratory chain.20 In the respiratory chain, electrons are transferred to molecular oxygen by four complexes. At the same time, complexes I (NADH dehydrogenase), III (cytochrome c reductase) and IV (cytochrome c oxidase) pump protons (H+) into the intermembrane space, maintaining the ΔΨm.20,21 During an inflammatory response, complexes I and III are the main sources of harmful mtROS.21,22 Meanwhile, the generation of harmful mtROS is accompanied by a large amount of electron leakage, which results in loss of the normal ΔΨm.30,31
In this study, LPS stimulation led to a decrease in ΔΨm and the activity of complex I. However, LPS increased the activity of complex III. Decreased complex I activity leads to decreased NADH generation, increased mitochondrial electron leakage, and further increased mtROS generation. Decreased NADH production, which increases the FADH2/NADH ratio, will promote the Q-cycle of complex III, increasing its activity as well as the formation of mtROS.32
Moreover, this research suggested that in the pre-inflammatory state, nitrate greatly improves the activity of complex I. At the same time, the output of proton pumping was also increased, which increased ΔΨm. These two results are consistent. Higher complex I activity led to more NADH generation and a lower FADH2/NADH ratio. The reduced FADH2/NADH ratio restored complex III activity to normal levels. These results showed that nitrate attenuated the increased mtROS following LPS challenge. Nitrate appeared to primarily regulate mitochondrial function by increasing the activity of complex I under inflammatory circumstances.
The above experiments showed that nitrate alleviated LPS-induced inflammation by regulating mitochondrial function; however, the mechanism was still unclear. Considering that nitrate may act through the nitrate-nitrite-NO pathway,1–4 the NO scavenger Carboxy-PITO was used to block the effects of nitrate. These results confirmed that NO played a key role. When NO was removed, nitrate could not reduce mtROS generation nor enhance ΔΨm. These results suggest that nitrate may act through the nitrate-nitrite-NO pathway via XOR.11–13,25–27
XOR catalyzes the metabolism of xanthine into uric acid and has been found to degrade nitrate to NO in biochemical experiments.26 XOR is believed to be the primary source of nitrite-derived NO in living organisms.33 Recently, mammals have been found to have the ability to degrade nitrate through XOR.11–13 It appears that XOR may be an important participant in the effects of nitrate on mitochondrial function. However, considering how NO homeostasis modulates both the nitrate-nitrite-NO and the nitric oxide synthase (NOS) pathways, we cannot ignore the possibility that it can also be regulated by the NOS pathway.12,13
There are three kinds of NOS: neuronal (n)NOS, endothelial (e)NOS and iNOS. In macrophages, iNOS plays a crucial role in inflammatory responses.25,34 Thus, the mRNA and protein expression of iNOS and XOR were measured. Nitrate did not alter the expression of iNOS or XOR following LPS induction.
Next, iNOS or XOR inhibitors were combined with LPS to eliminate interference by iNOS or XOR on NO generation while determining the NO content of the cells. These data suggested that sodium nitrate increased NO generation via XOR. Mitochondrial function was tested by determining mtROS generation and ΔΨm to confirm the previous findings. The XOR inhibitor interfered with the ability of nitrate to regulate mitochondrial function under LPS challenge. This indicated that the mitochondrial regulatory capacity of nitrate was dependent on XOR.
XOR is thought to have two subtypes, xanthine oxidase (XO) and xanthine dehydrogenase (XDH).35 XOR primarily exists in the XDH form, which results from the specific transcription and translation of XDH gene. XDH cannot participate in the nitrate-nitrite-NO pathway. Under pathological conditions such as inflammation and hypoxia, XDH can be reversibly transformed into XO, which has the capability to convert nitrate and nitrite.36,37 Based on the above studies, it can be inferred that although nitrate can change mitochondrial function under LPS induction, it cannot change mitochondrial function under normal conditions. The ΔΨm, mtROS, and secretion of IL-6 and TNF-α were measured after nitrate treatment without LPS-induction. Nitrate had no influence on the levels of these indicators. These results suggest that nitrate might only work in pathological conditions.
In conclusion, many studies on inorganic nitrate and nitrite have confirmed their beneficial roles in vivo.1–10 Most studies have tended to use nitrite to study the mechanism of this pathway. The mechanism of nitrate through XOR in vitro has not been studied. This study was the first to show that nitrate could alleviate inflammation by regulating mitochondrial function in a manner dependent on XOR in vitro. Nitrate affects mitochondrial function and the mechanism might be similar to boron and other compounds.38 The most important finding was that a nitrate concentration of 500 µM, which is slightly above the normal physiological concentration range,39 could significantly alleviate inflammation. This means that the protective mechanism of nitrate is not only through what is converted to nitrite by bacterial nitrate reductases, but also through what is converted to NO by XOR. This study suggests that nitrate in cells may bypass nitrite through the nitrate-NO pathway. These results provide a new research direction through which to evaluate nitrate therapy. It also suggests a new pathway for understanding the mechanism of nitrate.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This study was supported by the National Natural Science Foundation of China (81570991 and 81570957).
ORCID iD
Lizheng Qin https://orcid.org/0000-0003-2384-7514
References
- 1.Carlstrom M, Montenegro MF. Therapeutic value of stimulating the nitrate-nitrite-nitric oxide pathway to attenuate oxidative stress and restore nitric oxide bioavailability in cardiorenal disease. J Intern Med 2019; 285: 2–18. [DOI] [PubMed] [Google Scholar]
- 2.Koch CD, Gladwin MT, Freeman BAet al. Enterosalivary nitrate metabolism and the microbiome: intersection of microbial metabolism, nitric oxide and diet in cardiac and pulmonary vascular health. Free Radic Biol Med 2017; 105: 48–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlstrom M, Lundberg JO, Weitzberg E. Mechanisms underlying blood pressure reduction by dietary inorganic nitrate. Acta Physiol (Oxf) 2018; 224: e13080. [DOI] [PubMed] [Google Scholar]
- 4.Waltz P, Escobar D, Botero AMet al. Nitrate/nitrite as critical mediators to limit oxidative injury and inflammation. Antioxid Redox Signal 2015; 23: 328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingram TE, Fraser AG, Bleasdale RAet al. Low-dose sodium nitrite attenuates myocardial ischemia and vascular ischemia-reperfusion injury in human models. J Am Coll Cardiol 2013; 61: 2534–2541. [DOI] [PubMed] [Google Scholar]
- 6.Liu M, Zollbrecht C, Peleli Met al. Nitrite-mediated renal vasodilatation is increased during ischemic conditions via cGMP-independent signaling. Free Radic Biol Med 2015; 84: 154–160. [DOI] [PubMed] [Google Scholar]
- 7.Shiva S, Sack MN, Greer JJet al. Nitrite augments tolerance to ischemia/reperfusion injury via the modulation of mitochondrial electron transfer. J Exp Med 2007; 204: 2089–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vitturi DA, Patel RP. Current perspectives and challenges in understanding the role of nitrite as an integral player in nitric oxide biology and therapy. Free Radic Biol Med 2011; 51: 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dezfulian C, Shiva S, Alekseyenko Aet al. Nitrite therapy after cardiac arrest reduces reactive oxygen species generation, improves cardiac and neurological function, and enhances survival via reversible inhibition of mitochondrial complex I. Circulation 2009; 120: 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin L, Qin L, Xia Det al. Active secretion and protective effect of salivary nitrate against stress in human volunteers and rats. Free Radic Biol Med 2013; 57: 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L, Borniquel S, Lundberg JO. Enhanced xanthine oxidoreductase expression and tissue nitrate reduction in germ free mice. Nitric Oxide 2010; 22: 191–195. [DOI] [PubMed] [Google Scholar]
- 12.Carlstrom M, Liu M, Yang Tet al. Cross-talk between nitrate-nitrite-NO and NO synthase pathways in control of vascular NO homeostasis. Antioxid Redox Signal 2015; 23: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peleli M, Zollbrecht C, Montenegro MFet al. Enhanced XOR activity in eNOS-deficient mice: effects on the nitrate-nitrite-NO pathway and ROS homeostasis. Free Radic Biol Med 2016; 99: 472–484. [DOI] [PubMed] [Google Scholar]
- 14.Kabat AM, Pearce EJ. Inflammation by way of macrophage metabolism. Science 2017; 356: 488–489. [DOI] [PubMed] [Google Scholar]
- 15.Hamidzadeh K, Christensen SM, Dalby Eet al. Macrophages and the recovery from acute and chronic inflammation. Annu Rev Physiol 2017; 79: 567–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Wang X, Cueto Ret al. Biochemical basis and metabolic interplay of redox regulation. Redox Biol 2019; 26: 101284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umeno A, Biju V, Yoshida Y. In vivo ROS production and use of oxidative stress-derived biomarkers to detect the onset of diseases such as Alzheimer’s disease, Parkinson’s disease, and diabetes. Free Radic Res 2017; 51: 413–427. [DOI] [PubMed] [Google Scholar]
- 18.Henze K, Martin W. Evolutionary biology: essence of mitochondria. Nature 2003; 426: 127–128. [DOI] [PubMed] [Google Scholar]
- 19.Handy DE, Loscalzo J. Redox regulation of mitochondrial function. Antioxid Redox Signal 2012; 16: 1323–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 2009; 417: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brand MD. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic Biol Med 2016; 100: 14–31. [DOI] [PubMed] [Google Scholar]
- 22.Banoth B, Cassel SL. Mitochondria in innate immune signaling. Transl Res 2018; 202: 52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West AP, Brodsky IE, Rahner Cet al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 2011; 472: 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Wang X, Zheng Met al. Mitochondrial reactive oxygen species mediate the lipopolysaccharide-induced pro-inflammatory response in human gingival fibroblasts. Exp Cell Res 2016; 347: 212–221. [DOI] [PubMed] [Google Scholar]
- 25.Lundberg JO, Weitzberg E, Lundberg JMet al. Intragastric nitric oxide production in humans: measurements in expelled air. Gut 1994; 35: 1543–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millar TM, Stevens CR, Benjamin Net al. Xanthine oxidoreductase catalyses the reduction of nitrates and nitrite to nitric oxide under hypoxic conditions. FEBS Lett 1998; 427: 225–228. [DOI] [PubMed] [Google Scholar]
- 27.Carlsson S, Wiklund NP, Engstrand Let al. Effects of pH, nitrite, and ascorbic acid on nonenzymatic nitric oxide generation and bacterial growth in urine. Nitric Oxide 2001; 5: 580–586. [DOI] [PubMed] [Google Scholar]
- 28.Prathapan A, Vineetha VP, Raghu KG. Protective effect of Boerhaavia diffusa L. against mitochondrial dysfunction in angiotensin II induced hypertrophy in H9c2 cardiomyoblast cells. PLoS One 2014; 9: e96220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zollbrecht C, Persson AE, Lundberg JOet al. Nitrite-mediated reduction of macrophage NADPH oxidase activity is dependent on xanthine oxidoreductase-derived nitric oxide but independent of S-nitrosation. Redox Biol 2016; 10: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rigoulet M, Yoboue ED, Devin A. Mitochondrial ROS generation and its regulation: mechanisms involved in H(2)O(2) signaling. Antioxid Redox Signal 2011; 14: 459–468. [DOI] [PubMed] [Google Scholar]
- 31.Szczepanek K, Chen Q, Larner ACet al. Cytoprotection by the modulation of mitochondrial electron transport chain: the emerging role of mitochondrial STAT3. Mitochondrion 2012; 12: 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speijer D. Can All Major ROS Forming Sites of the Respiratory Chain Be Activated By High FADH2/NADH Ratios?: ancient evolutionary constraints determine mitochondrial ROS formation. Bioessays 2019; 41: e1800180. [DOI] [PubMed] [Google Scholar]
- 33.Cantu-Medellin N, Kelley EE. Xanthine oxidoreductase-catalyzed reduction of nitrite to nitric oxide: insights regarding where, when and how. Nitric Oxide 2013; 34: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowenstein CJ, Padalko E. iNOS (NOS2) at a glance. J Cell Sci 2004; 117: 2865–2867. [DOI] [PubMed] [Google Scholar]
- 35.Asai R, Nishino T, Matsumura Tet al. Two mutations convert mammalian xanthine oxidoreductase to highly superoxide-productive xanthine oxidase. J Biochem 2007; 141: 525–534. [DOI] [PubMed] [Google Scholar]
- 36.Nishino T, Okamoto K, Eger BTet al. Mammalian xanthine oxidoreductase - mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J 2008; 275: 3278–3289. [DOI] [PubMed] [Google Scholar]
- 37.Kelley EE. A new paradigm for XOR-catalyzed reactive species generation in the endothelium. Pharmacol Rep 2015; 67: 669–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Routray I, Ali S. Boron induces lymphocyte proliferation and modulates the priming effects of lipopolysaccharide on macrophages. PLoS One 2016; 11: e0150607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montenegro MF, Sundqvist ML, Nihlen Cet al. Profound differences between humans and rodents in the ability to concentrate salivary nitrate: implications for translational research. Redox Biol 2016; 10: 206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]